

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0162b.5
(220 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
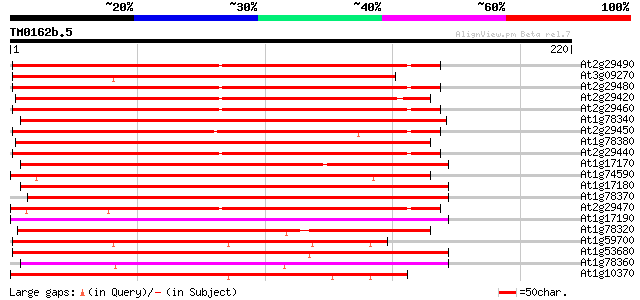
Sequences producing significant alignments: (bits) Value
At2g29490 putative glutathione S-transferase 171 4e-43
At3g09270 putative glutathione transferase 165 2e-41
At2g29480 putative glutathione S-transferase 163 6e-41
At2g29420 putative glutathione S-transferase 161 2e-40
At2g29460 putative glutathione S-transferase 159 8e-40
At1g78340 GST7 like protein 157 3e-39
At2g29450 glutathione S-transferase 157 5e-39
At1g78380 glutathione S-transferase like protein 157 5e-39
At2g29440 putative glutathione S-transferase 156 9e-39
At1g17170 putative glutathione transferase 155 1e-38
At1g74590 glutathione S-transferase like protein 151 3e-37
At1g17180 glutathione transferase like protein 151 3e-37
At1g78370 unknown protein 150 7e-37
At2g29470 glutathione S-transferase like protein 149 9e-37
At1g17190 putative glutathione transferase 149 9e-37
At1g78320 2,4-D inducible glutathione S-transferase like protein 148 2e-36
At1g59700 glutathione S-transferase, putative 145 1e-35
At1g53680 glutathione transferase, putative 145 2e-35
At1g78360 hypothetical protein 135 1e-32
At1g10370 glutathione S-transferase (GST30b) 135 1e-32
>At2g29490 putative glutathione S-transferase
Length = 224
Score = 171 bits (432), Expect = 4e-43
Identities = 86/168 (51%), Positives = 120/168 (71%), Gaps = 2/168 (1%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHG 61
+ + VKL+ FWAS + ++VE ALKLKGV YEY+EED+ NK+ LLLELNP+HKKVPVLVH
Sbjct: 4 KEESVKLLGFWASPFSRRVEMALKLKGVPYEYLEEDLPNKTPLLLELNPLHKKVPVLVHN 63
Query: 62 NKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDE 121
+K + ES +ILEYID+TWK+ P+ PQDPYE+A+ARFWA I+ ++L + L +
Sbjct: 64 DKILLESHLILEYIDQTWKNS-PILPQDPYEKAMARFWAKFIDDQILTLGFRSLVKAEKG 122
Query: 122 QENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGR 169
+E A++E E L +E+++ GK FFGG T+ L + + GS+ F R
Sbjct: 123 REVAIEETRELLMFLEKEVTGKDFFGGKTIGFLDM-IAGSMIPFCLAR 169
>At3g09270 putative glutathione transferase
Length = 224
Score = 165 bits (417), Expect = 2e-41
Identities = 75/151 (49%), Positives = 110/151 (72%), Gaps = 1/151 (0%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIF-NKSNLLLELNPVHKKVPVLVH 60
+ + VKL+ W S + K+VE LKLKG+ YEY+EED++ N+S +LL+ NP+HKKVPVL+H
Sbjct: 3 QEEHVKLLGLWGSPFSKRVEMVLKLKGIPYEYIEEDVYGNRSPMLLKYNPIHKKVPVLIH 62
Query: 61 GNKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGD 120
+ IAESL+I+EYI++TWK + + PQDPYERA+ARFWA ++ K++ A +
Sbjct: 63 NGRSIAESLVIVEYIEDTWKTTHTILPQDPYERAMARFWAKYVDEKVMLAVKKACWGPES 122
Query: 121 EQENAMKEATEGLEMVEEQIKGKKFFGGITL 151
E+E +KEA EGL+ +E+++ K FFGG T+
Sbjct: 123 EREKEVKEAYEGLKCLEKELGDKLFFGGETI 153
>At2g29480 putative glutathione S-transferase
Length = 225
Score = 163 bits (413), Expect = 6e-41
Identities = 81/168 (48%), Positives = 116/168 (68%), Gaps = 2/168 (1%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHG 61
+ + VKL+ FW S + ++VE ALKLKGV YEY+EED+ KS LLLELNPVHKKVPVLVH
Sbjct: 4 KEESVKLLGFWISPFSRRVEMALKLKGVPYEYLEEDLPKKSTLLLELNPVHKKVPVLVHN 63
Query: 62 NKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDE 121
+K ++ES +ILEYID+TW + P+ P DPYE+A+ RFWA ++ ++L ++ L +
Sbjct: 64 DKLLSESHVILEYIDQTWNNN-PILPHDPYEKAMVRFWAKFVDEQILPVGFMPLVKAEKG 122
Query: 122 QENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGR 169
+ A++E E L +E+++ GK FFGG T+ L + + GS+ F R
Sbjct: 123 IDVAIEEIREMLMFLEKEVTGKDFFGGKTIGFLDM-VAGSMIPFCLAR 169
>At2g29420 putative glutathione S-transferase
Length = 227
Score = 161 bits (408), Expect = 2e-40
Identities = 78/163 (47%), Positives = 115/163 (69%), Gaps = 3/163 (1%)
Query: 3 SKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHGN 62
S+EVKL+ WAS + +++E AL LKGV YE++E+DI NKS+LLL+LNPVHK +PVLVH
Sbjct: 7 SEEVKLLGMWASPFSRRIEIALTLKGVSYEFLEQDITNKSSLLLQLNPVHKMIPVLVHNG 66
Query: 63 KPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDEQ 122
KPI+ESL+ILEYIDETW+ P+ PQDPYER +ARFW+ ++ ++ A + +G E+
Sbjct: 67 KPISESLVILEYIDETWRDN-PILPQDPYERTMARFWSKFVDEQIYVTAMKVVGKTGKER 125
Query: 123 ENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGF 165
+ ++ + L +E+++ GK F GG +L + +V +L F
Sbjct: 126 DAVVEATRDLLMFLEKELVGKDFLGGKSLG--FVDIVATLVAF 166
>At2g29460 putative glutathione S-transferase
Length = 224
Score = 159 bits (403), Expect = 8e-40
Identities = 81/168 (48%), Positives = 118/168 (70%), Gaps = 2/168 (1%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHG 61
+ ++VKL+ FWAS + ++VE A KLKGV YEY+E+DI NKS LLL++NPV+KKVPVLV+
Sbjct: 4 KEEDVKLLGFWASPFTRRVEMAFKLKGVPYEYLEQDIVNKSPLLLQINPVYKKVPVLVYK 63
Query: 62 NKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDE 121
K ++ES +ILEYID+ WK+ P+ PQDPYE+A+A FWA ++ ++ A++ + +
Sbjct: 64 GKILSESHVILEYIDQIWKNN-PILPQDPYEKAMALFWAKFVDEQVGPVAFMSVAKAEKG 122
Query: 122 QENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGR 169
E A+KEA E +E+++ GK FFGG T+ L L + GS+ F R
Sbjct: 123 VEVAIKEAQELFMFLEKEVTGKDFFGGKTIGFLDL-VAGSMIPFCLAR 169
>At1g78340 GST7 like protein
Length = 218
Score = 157 bits (398), Expect = 3e-39
Identities = 75/167 (44%), Positives = 109/167 (64%)
Query: 5 EVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHGNKP 64
EV L+ FW S +G + AL+ KGVE+EY EE++ +KS LLL++NPVHKK+PVL+H KP
Sbjct: 4 EVILLDFWPSPFGVRARIALREKGVEFEYREENLRDKSPLLLQMNPVHKKIPVLIHNGKP 63
Query: 65 IAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDEQEN 124
+ ES+ +++YIDE W + P+ P DPY+RA ARFW ++ KL + A T G+EQE
Sbjct: 64 VCESMNVVQYIDEVWSDKNPILPSDPYQRAQARFWVDFVDTKLFEPADKIWQTKGEEQET 123
Query: 125 AMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGRKL 171
A KE E L+++E ++ K +FGG T + + + G + F KL
Sbjct: 124 AKKEYIEALKILETELGDKPYFGGDTFGFVDIAMTGYYSWFEASEKL 170
>At2g29450 glutathione S-transferase
Length = 224
Score = 157 bits (396), Expect = 5e-39
Identities = 82/169 (48%), Positives = 113/169 (66%), Gaps = 3/169 (1%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHG 61
E +EVKL+ WAS + ++VE ALKLKG+ YEYVEE + NKS LLL LNP+HKKVPVLVH
Sbjct: 3 EKEEVKLLGIWASPFSRRVEMALKLKGIPYEYVEEILENKSPLLLALNPIHKKVPVLVHN 62
Query: 62 NKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDE 121
K I ES +ILEYIDETW Q P+ PQDPYER+ ARF+A ++ +++ ++ + + ++
Sbjct: 63 GKTILESHVILEYIDETWP-QNPILPQDPYERSKARFFAKLVDEQIMNVGFISMARADEK 121
Query: 122 QENAMKEATEGLEM-VEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGR 169
+ E L M +E+++ GK +FGG T+ L + GSL F R
Sbjct: 122 GREVLAEQVRELIMYLEKELVGKDYFGGKTVGFLDF-VAGSLIPFCLER 169
>At1g78380 glutathione S-transferase like protein
Length = 219
Score = 157 bits (396), Expect = 5e-39
Identities = 76/163 (46%), Positives = 105/163 (63%)
Query: 3 SKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHGN 62
+ EV L+ FW S +G + AL+ KGVE+EY EED+ NKS LLL++NP+HKK+PVL+H
Sbjct: 2 ANEVILLDFWPSMFGMRTRIALREKGVEFEYREEDLRNKSPLLLQMNPIHKKIPVLIHNG 61
Query: 63 KPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDEQ 122
KP+ ES+I ++YIDE W H+ P+ P DPY RA ARFWA I+ KL A T G+EQ
Sbjct: 62 KPVNESIIQVQYIDEVWSHKNPILPSDPYLRAQARFWADFIDKKLYDAQRKVWATKGEEQ 121
Query: 123 ENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGF 165
E K+ E L+ +E ++ K +F G + + L+G T F
Sbjct: 122 EAGKKDFIEILKTLESELGDKPYFSGDDFGYVDIALIGFYTWF 164
>At2g29440 putative glutathione S-transferase
Length = 223
Score = 156 bits (394), Expect = 9e-39
Identities = 77/168 (45%), Positives = 115/168 (67%), Gaps = 2/168 (1%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHG 61
+++EVKL+ WAS + +++E ALKLKGV YEY+EED+ NKS+LLL L+P+HKK+PVLVH
Sbjct: 3 KNEEVKLLGIWASPFSRRIEMALKLKGVPYEYLEEDLENKSSLLLALSPIHKKIPVLVHN 62
Query: 62 NKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDE 121
K I ES +ILEYIDETWKH P+ PQDP++R+ AR A ++ K++ + L +
Sbjct: 63 GKTIIESHVILEYIDETWKHN-PILPQDPFQRSKARVLAKLVDEKIVNVGFASLAKTEKG 121
Query: 122 QENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGR 169
+E +++ E + +E+++ GK +FGG T+ L + GS+ F R
Sbjct: 122 REVLIEQTRELIMCLEKELAGKDYFGGKTVGFLDF-VAGSMIPFCLER 168
>At1g17170 putative glutathione transferase
Length = 218
Score = 155 bits (393), Expect = 1e-38
Identities = 81/168 (48%), Positives = 109/168 (64%), Gaps = 1/168 (0%)
Query: 5 EVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHGNKP 64
EV L+ FWAS +G + AL K V+Y++ EED++NKS+LLLE+NPVHKK+PVL+H KP
Sbjct: 4 EVILLDFWASMFGMRTRIALAEKRVKYDHREEDLWNKSSLLLEMNPVHKKIPVLIHNGKP 63
Query: 65 IAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDEQEN 124
+ ESLI +EYIDETW PL P DPY+RA A+FWA I+ K+ A G+EQE
Sbjct: 64 VCESLIQIEYIDETWPDNNPLLPSDPYKRAHAKFWADFIDKKVNVTARRIWAVKGEEQE- 122
Query: 125 AMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGRKLG 172
A KE E L+ +E ++ KK+FG T + + L+G + F K G
Sbjct: 123 AAKELIEILKTLESELGDKKYFGDETFGYVDIALIGFHSWFAVYEKFG 170
>At1g74590 glutathione S-transferase like protein
Length = 232
Score = 151 bits (381), Expect = 3e-37
Identities = 76/171 (44%), Positives = 111/171 (64%), Gaps = 6/171 (3%)
Query: 1 MESKEVKLV--SFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVL 58
ME K+ K++ W S+Y K+VE ALKLKGV YEY+EED+ NKS L++LNPVHKK+PVL
Sbjct: 1 MEEKKSKVILHGTWISTYSKRVEIALKLKGVLYEYLEEDLQNKSESLIQLNPVHKKIPVL 60
Query: 59 VHGNKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTS 118
VH KP+AESL+ILEYIDETW + FP+DPYERA RFW + I ++ + +
Sbjct: 61 VHDGKPVAESLVILEYIDETWTNSPRFFPEDPYERAQVRFWVSYINQQVFEVMGQVMSQE 120
Query: 119 GDEQENAMKEATEGLEMVEEQIK----GKKFFGGITLATLTLQLVGSLTGF 165
G+ Q +++EA + ++++E +K K + L + ++ +L G+
Sbjct: 121 GEAQAKSVEEARKRFKVLDEGLKKHFPNKNIRRNDDVGLLEITIIATLGGY 171
>At1g17180 glutathione transferase like protein
Length = 221
Score = 151 bits (381), Expect = 3e-37
Identities = 73/168 (43%), Positives = 106/168 (62%)
Query: 5 EVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHGNKP 64
EV L+ FW S +G + AL+ K V+++Y E+D++NKS +LLE+NPVHKK+PVL+H P
Sbjct: 4 EVILLDFWPSMFGMRTRIALEEKNVKFDYREQDLWNKSPILLEMNPVHKKIPVLIHNGNP 63
Query: 65 IAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDEQEN 124
+ ESLI +EYIDE W + PL P DPY+RA A+FW I+ K+ +A + G+E E
Sbjct: 64 VCESLIQIEYIDEVWPSKTPLLPSDPYQRAQAKFWGDFIDKKVYASARLIWGAKGEEHEA 123
Query: 125 AMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGRKLG 172
KE E L+ +E ++ K +FGG T + + L+G + F K G
Sbjct: 124 GKKEFIEILKTLESELGDKTYFGGETFGYVDIALIGFYSWFEAYEKFG 171
>At1g78370 unknown protein
Length = 217
Score = 150 bits (378), Expect = 7e-37
Identities = 71/165 (43%), Positives = 102/165 (61%)
Query: 8 LVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHGNKPIAE 67
L+ +W S +G + AL+ KGVE+EY EED NKS LLL+ NP+HKK+PVLVH KP+ E
Sbjct: 7 LLDYWPSMFGMRARVALREKGVEFEYREEDFSNKSPLLLQSNPIHKKIPVLVHNGKPVCE 66
Query: 68 SLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGDEQENAMK 127
SL +++Y+DE W + P FP DPY RA ARFWA ++ K A + G+EQE K
Sbjct: 67 SLNVVQYVDEAWPEKNPFFPSDPYGRAQARFWADFVDKKFTDAQFKVWGKKGEEQEAGKK 126
Query: 128 EATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGRKLG 172
E E ++++E ++ K +FGG + + + L+ + F K G
Sbjct: 127 EFIEAVKILESELGDKPYFGGDSFGYVDISLITFSSWFQAYEKFG 171
>At2g29470 glutathione S-transferase like protein
Length = 225
Score = 149 bits (377), Expect = 9e-37
Identities = 82/172 (47%), Positives = 114/172 (65%), Gaps = 5/172 (2%)
Query: 1 MESKE--VKLVSFWASSYGKKVEWALKLKGVEYEYVEED-IFNKSNLLLELNPVHKKVPV 57
M KE VKL+ WAS + ++VE ALKLKGV Y+Y++ED + KS LLL+LNPV+KKVPV
Sbjct: 1 MAEKEEGVKLIGSWASPFSRRVEMALKLKGVPYDYLDEDYLVVKSPLLLQLNPVYKKVPV 60
Query: 58 LVHGNKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFT 117
LVH K + ES +ILEYID+TW + P+ PQ PY++A+ARFWA ++ ++ L
Sbjct: 61 LVHNGKILPESQLILEYIDQTWTNN-PILPQSPYDKAMARFWAKFVDEQVTMIGLRSLVK 119
Query: 118 SGDEQENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGR 169
S + A++E E + ++E QI GKK FGG T+ L + +VGS+ F R
Sbjct: 120 SEKRIDVAIEEVQELIMLLENQITGKKLFGGETIGFLDM-VVGSMIPFCLAR 170
>At1g17190 putative glutathione transferase
Length = 220
Score = 149 bits (377), Expect = 9e-37
Identities = 72/172 (41%), Positives = 103/172 (59%)
Query: 1 MESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVH 60
M + +V L+ +W S +G + + AL KGV+YEY E D + K+ LL+E+NP+HKK+PVL+H
Sbjct: 1 MANDQVILLDYWPSMFGMRTKMALAEKGVKYEYKETDPWVKTPLLIEMNPIHKKIPVLIH 60
Query: 61 GNKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELFTSGD 120
KPI ESLI LEYIDE W P+ P DPY+++ ARFWA I+ K +W T G+
Sbjct: 61 NGKPICESLIQLEYIDEVWSDASPILPSDPYQKSRARFWAEFIDKKFYDPSWKVWATMGE 120
Query: 121 EQENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGRKLG 172
E KE E + +E ++ K ++GG L + L+G + F K G
Sbjct: 121 EHAAVKKELLEHFKTLETELGDKPYYGGEVFGYLDIALMGYYSWFKAMEKFG 172
>At1g78320 2,4-D inducible glutathione S-transferase like protein
Length = 220
Score = 148 bits (374), Expect = 2e-36
Identities = 75/165 (45%), Positives = 106/165 (63%), Gaps = 6/165 (3%)
Query: 4 KEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHGNK 63
+E+ L+ +WAS YG + AL+ K V+YEY EED+ NKS LLL++NP+HKK+PVL+H K
Sbjct: 3 EEIILLDYWASMYGMRTRIALEEKKVKYEYREEDLSNKSPLLLQMNPIHKKIPVLIHEGK 62
Query: 64 PIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKL---LKAAWVELFTSGD 120
PI ES+I ++YIDE W P+ P DPY+RA ARFWA I+ K KA W E SG+
Sbjct: 63 PICESIIQVQYIDELWPDTNPILPSDPYQRAQARFWADYIDKKTYVPCKALWSE---SGE 119
Query: 121 EQENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGF 165
+QE A E E L+ ++ ++ K +FGG + + +G + F
Sbjct: 120 KQEAAKIEFIEVLKTLDSELGDKYYFGGNEFGLVDIAFIGFYSWF 164
>At1g59700 glutathione S-transferase, putative
Length = 234
Score = 145 bits (367), Expect = 1e-35
Identities = 80/154 (51%), Positives = 103/154 (65%), Gaps = 7/154 (4%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIF-NKSNLLLELNPVHKKVPVLVH 60
E +EVKL+ W S Y + + AL+LK V+Y+YVEE++F +KS LLL+ NPVHKKVPVL+H
Sbjct: 3 EKEEVKLLGVWYSPYAIRPKIALRLKSVDYDYVEENLFGSKSELLLKSNPVHKKVPVLLH 62
Query: 61 GNKPIAESLIILEYIDETWKHQYP-LFPQDPYERALARFWAASIEPKLLKAAWVELFT-S 118
NKPI ESL I+EYIDETW P + P PY+RALARFW+ ++ K A + T S
Sbjct: 63 NNKPIVESLNIVEYIDETWNSSAPSILPSHPYDRALARFWSDFVDNKWFPALRMAAITKS 122
Query: 119 GDEQENAMKEATEGLEMVEEQI----KGKKFFGG 148
D + AM+E EGL +E+ KGK FFGG
Sbjct: 123 EDAKAKAMEEVEEGLLQLEDAFVSISKGKPFFGG 156
>At1g53680 glutathione transferase, putative
Length = 224
Score = 145 bits (366), Expect = 2e-35
Identities = 76/173 (43%), Positives = 106/173 (60%), Gaps = 2/173 (1%)
Query: 2 ESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVHG 61
E+ +V ++ FWAS Y + + AL+ KGVE+E EED++NKS LLL+ NPVHKKVPVL+H
Sbjct: 4 ENSKVVVLDFWASPYAMRTKVALREKGVEFEVQEEDLWNKSELLLKSNPVHKKVPVLIHN 63
Query: 62 NKPIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPKLLKAAWVELF--TSG 119
N PI+ESLI ++YIDETW P DP RA ARFWA + + +++ G
Sbjct: 64 NTPISESLIQVQYIDETWTDAASFLPSDPQSRATARFWADYADKTISFEGGRKIWGNKKG 123
Query: 120 DEQENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGRKLG 172
+EQE KE E L+++E ++ K +FGG T + + LV + F K G
Sbjct: 124 EEQEKGKKEFLESLKVLEAELGDKSYFGGETFGYVDITLVPFYSWFYALEKCG 176
>At1g78360 hypothetical protein
Length = 222
Score = 135 bits (341), Expect = 1e-32
Identities = 75/171 (43%), Positives = 101/171 (58%), Gaps = 3/171 (1%)
Query: 5 EVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFN-KSNLLLELNPVHKKVPVLVHGNK 63
EV L+ FW S +G + AL+ KGV+YEY EED+ N KS LLLE+NP+HK +PVL+H K
Sbjct: 4 EVILLGFWPSMFGMRTMIALEEKGVKYEYREEDVINNKSPLLLEMNPIHKTIPVLIHNGK 63
Query: 64 PIAESLIILEYIDETWKHQYPLFPQDPYERALARFWAASIEPK--LLKAAWVELFTSGDE 121
P+ ESLI ++YIDE W P DPY RA A FWA I+ K L T G+E
Sbjct: 64 PVLESLIQIQYIDEVWSDNNSFLPSDPYHRAQALFWADFIDKKEQLYVCGRKTWATKGEE 123
Query: 122 QENAMKEATEGLEMVEEQIKGKKFFGGITLATLTLQLVGSLTGFLFGRKLG 172
E A KE E L+ ++ ++ K +FGG + + L+G + F +K G
Sbjct: 124 LEAANKEFIEILKTLQCELGEKPYFGGDKFGFVDIVLIGFYSWFPAYQKFG 174
>At1g10370 glutathione S-transferase (GST30b)
Length = 170
Score = 135 bits (341), Expect = 1e-32
Identities = 74/162 (45%), Positives = 102/162 (62%), Gaps = 6/162 (3%)
Query: 1 MESKEVKLVSFWASSYGKKVEWALKLKGVEYEYVEEDIFNKSNLLLELNPVHKKVPVLVH 60
M S +VKL+ WAS + + AL LK V YE+++E +KS LLL+ NPVHKK+PVL+H
Sbjct: 1 MASSDVKLIGAWASPFVMRPRIALNLKSVPYEFLQETFGSKSELLLKSNPVHKKIPVLLH 60
Query: 61 GNKPIAESLIILEYIDETWKHQYP-LFPQDPYERALARFWAASIEPKLLKAAWVELFTSG 119
+KP++ES II+EYID+TW P + P DPY+RA+ARFWAA I+ K A L G
Sbjct: 61 ADKPVSESNIIVEYIDDTWSSSGPSILPSDPYDRAMARFWAAYIDEKWFVALRGFLKAGG 120
Query: 120 DEQENA-MKEATEGLEMVEEQI----KGKKFFGGITLATLTL 156
+E++ A + + EG +E+ KGK FF G + TL
Sbjct: 121 EEEKKAVIAQLEEGNAFLEKAFIDCSKGKPFFNGDNIGYSTL 162
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.144 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,785,768
Number of Sequences: 26719
Number of extensions: 195470
Number of successful extensions: 737
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 668
Number of HSP's gapped (non-prelim): 53
length of query: 220
length of database: 11,318,596
effective HSP length: 95
effective length of query: 125
effective length of database: 8,780,291
effective search space: 1097536375
effective search space used: 1097536375
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0162b.5


