

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0162b.4
(228 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
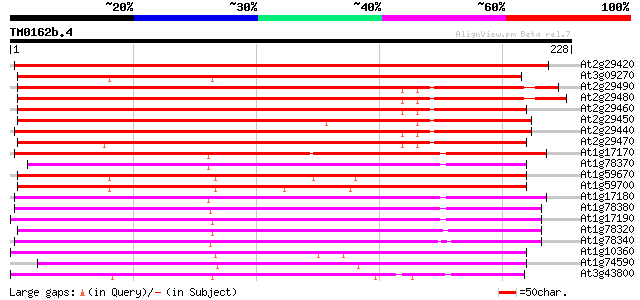
Score E
Sequences producing significant alignments: (bits) Value
At2g29420 putative glutathione S-transferase 219 9e-58
At3g09270 putative glutathione transferase 214 3e-56
At2g29490 putative glutathione S-transferase 209 1e-54
At2g29480 putative glutathione S-transferase 204 4e-53
At2g29460 putative glutathione S-transferase 199 1e-51
At2g29450 glutathione S-transferase 199 1e-51
At2g29440 putative glutathione S-transferase 194 4e-50
At2g29470 glutathione S-transferase like protein 181 2e-46
At1g17170 putative glutathione transferase 174 3e-44
At1g78370 unknown protein 170 5e-43
At1g59670 glutathione S-transferase, putative 170 7e-43
At1g59700 glutathione S-transferase, putative 169 1e-42
At1g17180 glutathione transferase like protein 168 2e-42
At1g78380 glutathione S-transferase like protein 167 3e-42
At1g17190 putative glutathione transferase 165 2e-41
At1g78320 2,4-D inducible glutathione S-transferase like protein 164 5e-41
At1g78340 GST7 like protein 163 8e-41
At1g10360 putative glutathione S-transferase TSI-1 161 2e-40
At1g74590 glutathione S-transferase like protein 159 9e-40
At3g43800 glutathione transferase-like protein 159 2e-39
>At2g29420 putative glutathione S-transferase
Length = 227
Score = 219 bits (558), Expect = 9e-58
Identities = 99/217 (45%), Positives = 147/217 (67%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
S++VKLL W SPFS+R+E AL LKGV YE++E+DI NKSSLLL+LNPVHK IPVLVH
Sbjct: 7 SEEVKLLGMWASPFSRRIEIALTLKGVSYEFLEQDITNKSSLLLQLNPVHKMIPVLVHNG 66
Query: 63 KTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQE 122
K I+ES +I+EYIDETW+ P+LP DPYER +ARFW+ F ++++ T + TG E++
Sbjct: 67 KPISESLVILEYIDETWRDNPILPQDPYERTMARFWSKFVDEQIYVTAMKVVGKTGKERD 126
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFP 182
+ R+ + +E+ + GK F GG+++G++DI +++W+ EE+ + ++ KFP
Sbjct: 127 AVVEATRDLLMFLEKELVGKDFLGGKSLGFVDIVATLVAFWLMRTEEIVGVKVVPVEKFP 186
Query: 183 ATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLS 219
W+ N L + VIK +PP D+ + Y R + L+
Sbjct: 187 EIHRWVKNLLGNDVIKKCIPPEDEHLKYIRARMEKLN 223
>At3g09270 putative glutathione transferase
Length = 224
Score = 214 bits (545), Expect = 3e-56
Identities = 98/207 (47%), Positives = 147/207 (70%), Gaps = 2/207 (0%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIF-NKSSLLLELNPVHKKIPVLVHGH 62
+ VKLL W SPFSKRVE LKLKG+ YEY+EED++ N+S +LL+ NP+HKK+PVL+H
Sbjct: 5 EHVKLLGLWGSPFSKRVEMVLKLKGIPYEYIEEDVYGNRSPMLLKYNPIHKKVPVLIHNG 64
Query: 63 KTIAESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
++IAES +I+EYI++TWK + +LP DPYERA+ARFWA + ++K++ V A E+
Sbjct: 65 RSIAESLVIVEYIEDTWKTTHTILPQDPYERAMARFWAKYVDEKVMLAVKKACWGPESER 124
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
EK + EA E ++ +E+ + K FFGGE IG++DIA +I YW+ I++E + I+ +F
Sbjct: 125 EKEVKEAYEGLKCLEKELGDKLFFGGETIGFVDIAADFIGYWLGIFQEASGVTIMTAEEF 184
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMI 208
P W +F+ + IK+ LPP++K++
Sbjct: 185 PKLQRWSEDFVGNNFIKEVLPPKEKLV 211
>At2g29490 putative glutathione S-transferase
Length = 224
Score = 209 bits (531), Expect = 1e-54
Identities = 105/222 (47%), Positives = 148/222 (66%), Gaps = 7/222 (3%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+ VKLL FW SPFS+RVE ALKLKGV YEY+EED+ NK+ LLLELNP+HKK+PVLVH K
Sbjct: 6 ESVKLLGFWASPFSRRVEMALKLKGVPYEYLEEDLPNKTPLLLELNPLHKKVPVLVHNDK 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
+ ES +I+EYID+TWK P+LP DPYE+A+ARFWA F + ++L F ++ +E
Sbjct: 66 ILLESHLILEYIDQTWKNSPILPQDPYEKAMARFWAKFIDDQILTLGFRSLVKAEKGREV 125
Query: 124 ALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVG-WISYWI-HIWEEVGSIHIIDPLKF 181
A+ E RE + +E+ + GK FFGG+ IG+LD+ G I + + +W+ +G I +I KF
Sbjct: 126 AIEETRELLMFLEKEVTGKDFFGGKTIGFLDMIAGSMIPFCLARLWKGIG-IDMIPEEKF 184
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSETFR 223
P W+ N ++ +PPR+K I+ R ++ET +
Sbjct: 185 PELNRWIKNLEEVEAVRGCIPPREKQIE----RMTKIAETIK 222
>At2g29480 putative glutathione S-transferase
Length = 225
Score = 204 bits (518), Expect = 4e-53
Identities = 103/225 (45%), Positives = 147/225 (64%), Gaps = 7/225 (3%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+ VKLL FW SPFS+RVE ALKLKGV YEY+EED+ KS+LLLELNPVHKK+PVLVH K
Sbjct: 6 ESVKLLGFWISPFSRRVEMALKLKGVPYEYLEEDLPKKSTLLLELNPVHKKVPVLVHNDK 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
++ES +I+EYID+TW P+LPHDPYE+A+ RFWA F ++++L F+ + +
Sbjct: 66 LLSESHVILEYIDQTWNNNPILPHDPYEKAMVRFWAKFVDEQILPVGFMPLVKAEKGIDV 125
Query: 124 ALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVG-WISYWI-HIWEEVGSIHIIDPLKF 181
A+ E RE + +E+ + GK FFGG+ IG+LD+ G I + + WE +G I + F
Sbjct: 126 AIEEIREMLMFLEKEVTGKDFFGGKTIGFLDMVAGSMIPFCLARAWECLG-IDMTPEDTF 184
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSETFRGRF 226
P W+ N ++++ +PP++K I+ R K + E + F
Sbjct: 185 PELNRWIKNLNEVEIVRECIPPKEKHIE----RMKKIIERAKSTF 225
>At2g29460 putative glutathione S-transferase
Length = 224
Score = 199 bits (506), Expect = 1e-51
Identities = 96/209 (45%), Positives = 143/209 (67%), Gaps = 3/209 (1%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+DVKLL FW SPF++RVE A KLKGV YEY+E+DI NKS LLL++NPV+KK+PVLV+ K
Sbjct: 6 EDVKLLGFWASPFTRRVEMAFKLKGVPYEYLEQDIVNKSPLLLQINPVYKKVPVLVYKGK 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
++ES +I+EYID+ WK P+LP DPYE+A+A FWA F ++++ F+++ E
Sbjct: 66 ILSESHVILEYIDQIWKNNPILPQDPYEKAMALFWAKFVDEQVGPVAFMSVAKAEKGVEV 125
Query: 124 ALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVG-WISYWI-HIWEEVGSIHIIDPLKF 181
A+ EA+E +E+ + GK FFGG+ IG+LD+ G I + + WE +G I +I KF
Sbjct: 126 AIKEAQELFMFLEKEVTGKDFFGGKTIGFLDLVAGSMIPFCLARGWEGMG-IDMIPEEKF 184
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
P W+ N ++++ +PPR++ I++
Sbjct: 185 PELNRWIKNLKEIEIVRECIPPREEQIEH 213
>At2g29450 glutathione S-transferase
Length = 224
Score = 199 bits (506), Expect = 1e-51
Identities = 97/212 (45%), Positives = 142/212 (66%), Gaps = 4/212 (1%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
++VKLL W SPFS+RVE ALKLKG+ YEYVEE + NKS LLL LNP+HKK+PVLVH K
Sbjct: 5 EEVKLLGIWASPFSRRVEMALKLKGIPYEYVEEILENKSPLLLALNPIHKKVPVLVHNGK 64
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
TI ES +I+EYIDETW Q P+LP DPYER+ ARF+A +++++ F++M ++ +
Sbjct: 65 TILESHVILEYIDETWPQNPILPQDPYERSKARFFAKLVDEQIMNVGFISMARADEKGRE 124
Query: 124 ALNE-AREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWI--HIWEEVGSIHIIDPLK 180
L E RE + +E+ + GK +FGG+ +G+LD G + + WE +G + +I K
Sbjct: 125 VLAEQVRELIMYLEKELVGKDYFGGKTVGFLDFVAGSLIPFCLERGWEGIG-LEVITEEK 183
Query: 181 FPATTAWMTNFLSHPVIKDTLPPRDKMIDYFH 212
FP W+ N ++KD +PPR++ +++ +
Sbjct: 184 FPEFKRWVRNLEKVEIVKDCVPPREEHVEHMN 215
>At2g29440 putative glutathione S-transferase
Length = 223
Score = 194 bits (492), Expect = 4e-50
Identities = 95/212 (44%), Positives = 140/212 (65%), Gaps = 3/212 (1%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
+++VKLL W SPFS+R+E ALKLKGV YEY+EED+ NKSSLLL L+P+HKKIPVLVH
Sbjct: 4 NEEVKLLGIWASPFSRRIEMALKLKGVPYEYLEEDLENKSSLLLALSPIHKKIPVLVHNG 63
Query: 63 KTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQE 122
KTI ES +I+EYIDETWK P+LP DP++R+ AR A ++K++ F ++ T +E
Sbjct: 64 KTIIESHVILEYIDETWKHNPILPQDPFQRSKARVLAKLVDEKIVNVGFASLAKTEKGRE 123
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVG-WISYWI-HIWEEVGSIHIIDPLK 180
+ + RE + +E+ + GK +FGG+ +G+LD G I + + WE +G + +I K
Sbjct: 124 VLIEQTRELIMCLEKELAGKDYFGGKTVGFLDFVAGSMIPFCLERAWEGMG-VEMITEKK 182
Query: 181 FPATTAWMTNFLSHPVIKDTLPPRDKMIDYFH 212
FP W+ ++ D +P R+K I++ +
Sbjct: 183 FPEYNKWVKKLKEVEIVVDCIPLREKHIEHMN 214
>At2g29470 glutathione S-transferase like protein
Length = 225
Score = 181 bits (460), Expect = 2e-46
Identities = 92/210 (43%), Positives = 138/210 (64%), Gaps = 4/210 (1%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEED-IFNKSSLLLELNPVHKKIPVLVHGH 62
+ VKL+ W SPFS+RVE ALKLKGV Y+Y++ED + KS LLL+LNPV+KK+PVLVH
Sbjct: 6 EGVKLIGSWASPFSRRVEMALKLKGVPYDYLDEDYLVVKSPLLLQLNPVYKKVPVLVHNG 65
Query: 63 KTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQE 122
K + ES +I+EYID+TW P+LP PY++A+ARFWA F ++++ ++ + +
Sbjct: 66 KILPESQLILEYIDQTWTNNPILPQSPYDKAMARFWAKFVDEQVTMIGLRSLVKSEKRID 125
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVG-WISYWI-HIWEEVGSIHIIDPLK 180
A+ E +E + +E I GK+ FGGE IG+LD+ VG I + + WE +G I +I K
Sbjct: 126 VAIEEVQELIMLLENQITGKKLFGGETIGFLDMVVGSMIPFCLARAWEGMG-IDMIPEEK 184
Query: 181 FPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
FP W+ N ++++ +P R+K I++
Sbjct: 185 FPELNRWIKNLKEIEIVRECIPDREKHIEH 214
>At1g17170 putative glutathione transferase
Length = 218
Score = 174 bits (441), Expect = 3e-44
Identities = 94/217 (43%), Positives = 131/217 (60%), Gaps = 4/217 (1%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
+ +V LL FW S F R AL K V+Y++ EED++NKSSLLLE+NPVHKKIPVL+H
Sbjct: 2 ADEVILLDFWASMFGMRTRIALAEKRVKYDHREEDLWNKSSLLLEMNPVHKKIPVLIHNG 61
Query: 63 KTIAESFIIIEYIDETW-KQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
K + ES I IEYIDETW PLLP DPY+RA A+FWA+F ++K+ T + G+EQ
Sbjct: 62 KPVCESLIQIEYIDETWPDNNPLLPSDPYKRAHAKFWADFIDKKVNVTARRIWAVKGEEQ 121
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
E A E E ++ +E + K++FG E GY+DIA+ W ++E+ G++ I +
Sbjct: 122 E-AAKELIEILKTLESELGDKKYFGDETFGYVDIALIGFHSWFAVYEKFGNVSI--ESEC 178
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
AW L + LP +K+I + R+K L
Sbjct: 179 SKLVAWAKRCLERESVAKALPESEKVITFISERRKKL 215
>At1g78370 unknown protein
Length = 217
Score = 170 bits (431), Expect = 5e-43
Identities = 85/204 (41%), Positives = 122/204 (59%), Gaps = 3/204 (1%)
Query: 8 LLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTIAE 67
LL +WPS F R AL+ KGVE+EY EED NKS LLL+ NP+HKKIPVLVH K + E
Sbjct: 7 LLDYWPSMFGMRARVALREKGVEFEYREEDFSNKSPLLLQSNPIHKKIPVLVHNGKPVCE 66
Query: 68 SFIIIEYIDETW-KQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKALN 126
S +++Y+DE W ++ P P DPY RA ARFWA+F ++K + F G+EQE
Sbjct: 67 SLNVVQYVDEAWPEKNPFFPSDPYGRAQARFWADFVDKKFTDAQFKVWGKKGEEQEAGKK 126
Query: 127 EAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPATTA 186
E EA++ +E + K +FGG++ GY+DI++ S W +E+ G+ I + P A
Sbjct: 127 EFIEAVKILESELGDKPYFGGDSFGYVDISLITFSSWFQAYEKFGNFSI--ESESPKLIA 184
Query: 187 WMTNFLSHPVIKDTLPPRDKMIDY 210
W + + +LP +K++ Y
Sbjct: 185 WAKRCMEKESVSKSLPDSEKIVAY 208
>At1g59670 glutathione S-transferase, putative
Length = 233
Score = 170 bits (430), Expect = 7e-43
Identities = 90/215 (41%), Positives = 133/215 (61%), Gaps = 8/215 (3%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIF-NKSSLLLELNPVHKKIPVLVHGH 62
++VKLL W SP R + AL+LK V+Y+YVEED+F +KS LLL+ NP+ KK+PVL+H
Sbjct: 5 EEVKLLGTWYSPVVIRAKIALRLKSVDYDYVEEDLFGSKSELLLKSNPIFKKVPVLIHNT 64
Query: 63 KTIAESFIIIEYIDETWKQY--PLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDE 120
K + S I+EYIDETW +LP PY+RALARFW+ F + K L T+ A+ +E
Sbjct: 65 KPVCVSLNIVEYIDETWNSSGSSILPSHPYDRALARFWSVFVDDKWLPTLMAAVVAKSEE 124
Query: 121 QE-KALNEAREAMEKIEEVI----EGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHI 175
+ K + E E + ++E +GK FFGGE IG++DI +G + E++ + I
Sbjct: 125 AKAKGMEEVEEGLLQLEAAFIALSKGKSFFGGETIGFIDICLGSFLVLLKAREKLKNEKI 184
Query: 176 IDPLKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+D LK P+ W FLS+ ++K+ +P DK+ +
Sbjct: 185 LDELKTPSLYRWANQFLSNEMVKNVVPDIDKVAKF 219
>At1g59700 glutathione S-transferase, putative
Length = 234
Score = 169 bits (428), Expect = 1e-42
Identities = 89/215 (41%), Positives = 135/215 (62%), Gaps = 8/215 (3%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIF-NKSSLLLELNPVHKKIPVLVHGH 62
++VKLL W SP++ R + AL+LK V+Y+YVEE++F +KS LLL+ NPVHKK+PVL+H +
Sbjct: 5 EEVKLLGVWYSPYAIRPKIALRLKSVDYDYVEENLFGSKSELLLKSNPVHKKVPVLLHNN 64
Query: 63 KTIAESFIIIEYIDETWKQY--PLLPHDPYERALARFWANFTEQKLLETV-FVAMCITGD 119
K I ES I+EYIDETW +LP PY+RALARFW++F + K + A+ + D
Sbjct: 65 KPIVESLNIVEYIDETWNSSAPSILPSHPYDRALARFWSDFVDNKWFPALRMAAITKSED 124
Query: 120 EQEKALNEAREAMEKIEE----VIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHI 175
+ KA+ E E + ++E+ + +GK FFGGE IG++DI G + E+ + +
Sbjct: 125 AKAKAMEEVEEGLLQLEDAFVSISKGKPFFGGEAIGFMDICFGSFVVLLKAREKFKAEKL 184
Query: 176 IDPLKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+D K P+ W FLS +K+ P +K+ ++
Sbjct: 185 LDESKTPSLCKWADRFLSDETVKNVAPEIEKVAEF 219
>At1g17180 glutathione transferase like protein
Length = 221
Score = 168 bits (426), Expect = 2e-42
Identities = 87/217 (40%), Positives = 127/217 (58%), Gaps = 3/217 (1%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
+ +V LL FWPS F R AL+ K V+++Y E+D++NKS +LLE+NPVHKKIPVL+H
Sbjct: 2 ADEVILLDFWPSMFGMRTRIALEEKNVKFDYREQDLWNKSPILLEMNPVHKKIPVLIHNG 61
Query: 63 KTIAESFIIIEYIDETW-KQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
+ ES I IEYIDE W + PLLP DPY+RA A+FW +F ++K+ + + G+E
Sbjct: 62 NPVCESLIQIEYIDEVWPSKTPLLPSDPYQRAQAKFWGDFIDKKVYASARLIWGAKGEEH 121
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
E E E ++ +E + K +FGGE GY+DIA+ W +E+ GS I +
Sbjct: 122 EAGKKEFIEILKTLESELGDKTYFGGETFGYVDIALIGFYSWFEAYEKFGSFSI--EAEC 179
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
P AW + + +LP +K+I + +K L
Sbjct: 180 PKLIAWGKRCVERESVAKSLPDSEKIIKFVPELRKKL 216
>At1g78380 glutathione S-transferase like protein
Length = 219
Score = 167 bits (424), Expect = 3e-42
Identities = 86/215 (40%), Positives = 127/215 (59%), Gaps = 3/215 (1%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
+ +V LL FWPS F R AL+ KGVE+EY EED+ NKS LLL++NP+HKKIPVL+H
Sbjct: 2 ANEVILLDFWPSMFGMRTRIALREKGVEFEYREEDLRNKSPLLLQMNPIHKKIPVLIHNG 61
Query: 63 KTIAESFIIIEYIDETWK-QYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
K + ES I ++YIDE W + P+LP DPY RA ARFWA+F ++KL + G+EQ
Sbjct: 62 KPVNESIIQVQYIDEVWSHKNPILPSDPYLRAQARFWADFIDKKLYDAQRKVWATKGEEQ 121
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
E + E ++ +E + K +F G++ GY+DIA+ W +E+ + I +
Sbjct: 122 EAGKKDFIEILKTLESELGDKPYFSGDDFGYVDIALIGFYTWFPAYEKFANFSI--ESEV 179
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKK 216
P AW+ L + +LP +K+ ++ +K
Sbjct: 180 PKLIAWVKKCLQRESVAKSLPDPEKVTEFVSELRK 214
>At1g17190 putative glutathione transferase
Length = 220
Score = 165 bits (417), Expect = 2e-41
Identities = 86/217 (39%), Positives = 120/217 (54%), Gaps = 3/217 (1%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M + V LL +WPS F R + AL KGV+YEY E D + K+ LL+E+NP+HKKIPVL+H
Sbjct: 1 MANDQVILLDYWPSMFGMRTKMALAEKGVKYEYKETDPWVKTPLLIEMNPIHKKIPVLIH 60
Query: 61 GHKTIAESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGD 119
K I ES I +EYIDE W P+LP DPY+++ ARFWA F ++K + + G+
Sbjct: 61 NGKPICESLIQLEYIDEVWSDASPILPSDPYQKSRARFWAEFIDKKFYDPSWKVWATMGE 120
Query: 120 EQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPL 179
E E E + +E + K ++GGE GYLDIA+ W E+ G I
Sbjct: 121 EHAAVKKELLEHFKTLETELGDKPYYGGEVFGYLDIALMGYYSWFKAMEKFGEFSI--ET 178
Query: 180 KFPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKK 216
+FP T W L + L D++I+Y + +K
Sbjct: 179 EFPILTTWTKRCLERESVVKALADSDRIIEYVYVLRK 215
>At1g78320 2,4-D inducible glutathione S-transferase like protein
Length = 220
Score = 164 bits (414), Expect = 5e-41
Identities = 85/214 (39%), Positives = 124/214 (57%), Gaps = 3/214 (1%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+++ LL +W S + R AL+ K V+YEY EED+ NKS LLL++NP+HKKIPVL+H K
Sbjct: 3 EEIILLDYWASMYGMRTRIALEEKKVKYEYREEDLSNKSPLLLQMNPIHKKIPVLIHEGK 62
Query: 64 TIAESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQE 122
I ES I ++YIDE W P+LP DPY+RA ARFWA++ ++K +G++QE
Sbjct: 63 PICESIIQVQYIDELWPDTNPILPSDPYQRAQARFWADYIDKKTYVPCKALWSESGEKQE 122
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFP 182
A E E ++ ++ + K +FGG G +DIA W +EEV ++ I+ L+FP
Sbjct: 123 AAKIEFIEVLKTLDSELGDKYYFGGNEFGLVDIAFIGFYSWFRTYEEVANLSIV--LEFP 180
Query: 183 ATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKK 216
AW L + LP DK++ +K
Sbjct: 181 KLMAWAQRCLKRESVAKALPDSDKVLKSVSDHRK 214
>At1g78340 GST7 like protein
Length = 218
Score = 163 bits (412), Expect = 8e-41
Identities = 87/215 (40%), Positives = 129/215 (59%), Gaps = 3/215 (1%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
+ +V LL FWPSPF R AL+ KGVE+EY EE++ +KS LLL++NPVHKKIPVL+H
Sbjct: 2 ADEVILLDFWPSPFGVRARIALREKGVEFEYREENLRDKSPLLLQMNPVHKKIPVLIHNG 61
Query: 63 KTIAESFIIIEYIDETWK-QYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
K + ES +++YIDE W + P+LP DPY+RA ARFW +F + KL E G+EQ
Sbjct: 62 KPVCESMNVVQYIDEVWSDKNPILPSDPYQRAQARFWVDFVDTKLFEPADKIWQTKGEEQ 121
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
E A E EA++ +E + K +FGG+ G++DIA+ W E++ + I+P +
Sbjct: 122 ETAKKEYIEALKILETELGDKPYFGGDTFGFVDIAMTGYYSWFEASEKLANFS-IEP-EC 179
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKK 216
P A L + +L +K++ + + +K
Sbjct: 180 PTLMASAKRCLQRESVVQSLHDSEKILAFAYKIRK 214
>At1g10360 putative glutathione S-transferase TSI-1
Length = 227
Score = 161 bits (408), Expect = 2e-40
Identities = 84/217 (38%), Positives = 125/217 (56%), Gaps = 7/217 (3%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M ++DVKL+ W S + R AL LK + YE+++E +KS LLL+ NPVHKK+PVL+H
Sbjct: 1 MATEDVKLIGSWASVYVMRARIALHLKSISYEFLQETYGSKSELLLKSNPVHKKMPVLIH 60
Query: 61 GHKTIAESFIIIEYIDETWKQY--PLLPHDPYERALARFWANFTEQKLLETVFVAMCITG 118
K + ES II+ YIDE W +LP PY+RA+ARFWA + + + +V + G
Sbjct: 61 ADKPVCESNIIVHYIDEAWNSSGPSILPSHPYDRAIARFWAAYIDDQWFISVRSILTAQG 120
Query: 119 DEQEKA----LNEAREAMEK-IEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSI 173
DE++KA + E + +EK + +GK FF G++IGYLDIA+G W + E +
Sbjct: 121 DEEKKAAIAQVEERTKLLEKAFNDCSQGKPFFNGDHIGYLDIALGSFLGWWRVVELDANH 180
Query: 174 HIIDPLKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+D K P+ W F P +K +P K+ ++
Sbjct: 181 KFLDETKTPSLVKWAERFCDDPAVKPIMPEITKLAEF 217
>At1g74590 glutathione S-transferase like protein
Length = 232
Score = 159 bits (403), Expect = 9e-40
Identities = 85/204 (41%), Positives = 119/204 (57%), Gaps = 5/204 (2%)
Query: 12 WPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTIAESFII 71
W S +SKRVE ALKLKGV YEY+EED+ NKS L++LNPVHKKIPVLVH K +AES +I
Sbjct: 14 WISTYSKRVEIALKLKGVLYEYLEEDLQNKSESLIQLNPVHKKIPVLVHDGKPVAESLVI 73
Query: 72 IEYIDETWKQYP-LLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKALNEARE 130
+EYIDETW P P DPYERA RFW ++ Q++ E + M G+ Q K++ EAR+
Sbjct: 74 LEYIDETWTNSPRFFPEDPYERAQVRFWVSYINQQVFEVMGQVMSQEGEAQAKSVEEARK 133
Query: 131 AMEKIEEVIE----GKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPATTA 186
+ ++E ++ K +++G L+I + E + II P+ P
Sbjct: 134 RFKVLDEGLKKHFPNKNIRRNDDVGLLEITIIATLGGYKAHREAIGVDIIGPVNTPTLYN 193
Query: 187 WMTNFLSHPVIKDTLPPRDKMIDY 210
W+ VIK+ P D ++ +
Sbjct: 194 WIERLQDLSVIKEVEVPHDTLVTF 217
>At3g43800 glutathione transferase-like protein
Length = 227
Score = 159 bits (401), Expect = 2e-39
Identities = 88/216 (40%), Positives = 128/216 (58%), Gaps = 11/216 (5%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFN-KSSLLLELNPVHKKIPVLV 59
M ++V +L+FWPS F RV AL+ K +++EY EED+F K+ LLL+ NPV+KKIPVL+
Sbjct: 1 MSEEEVVVLNFWPSMFGARVIMALEEKEIKFEYKEEDVFGQKTDLLLQSNPVNKKIPVLI 60
Query: 60 HGHKTIAESFIIIEYIDETWKQ---YPLLPHDPYERALARFWANFTEQKLLETVFVAMCI 116
H K + ES II+EYIDE WK LLP DPY+++ RFWA+ ++K+ +
Sbjct: 61 HNGKPVCESNIIVEYIDEVWKDDKTLRLLPSDPYQKSQCRFWADLIDKKVFDAGRRTWTK 120
Query: 117 TGDEQEKALNEAREAMEKIEEVIEGKRFFGG-ENIGYLDIAVGWISY--WIHIWEEVGSI 173
G EQE+A E E ++ +E + K +FGG +N+ +D+ + ISY W H WE +G
Sbjct: 121 RGKEQEEAKQEFIEILKVLERELGDKVYFGGNDNVSMVDLVL--ISYYPWFHTWETIGGF 178
Query: 174 HIIDPLKFPATTAWMTNFLSHPVIKDTLPPRDKMID 209
+ D P W+ L+ P I +LP K+ D
Sbjct: 179 SVED--HTPKLMDWIRKCLTRPAISKSLPDPLKIFD 212
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,726,017
Number of Sequences: 26719
Number of extensions: 248090
Number of successful extensions: 717
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 618
Number of HSP's gapped (non-prelim): 53
length of query: 228
length of database: 11,318,596
effective HSP length: 96
effective length of query: 132
effective length of database: 8,753,572
effective search space: 1155471504
effective search space used: 1155471504
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0162b.4


