

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0162a.11
(401 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
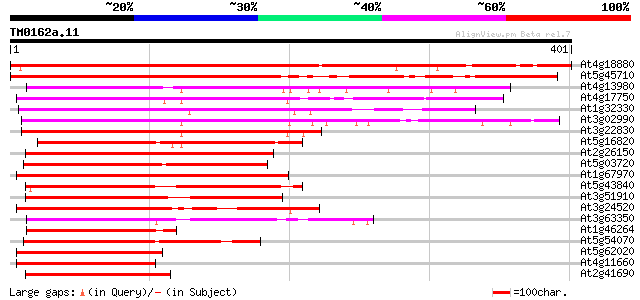
Score E
Sequences producing significant alignments: (bits) Value
At4g18880 heat shock transcription factor 21 (AtHSF21) 391 e-109
At5g45710 heat shock transcription factor 355 2e-98
At4g13980 unknown protein 208 4e-54
At4g17750 heat shock transcription factor HSF1 201 8e-52
At1g32330 heat shock transcription factor HSF8 like protein 201 8e-52
At3g02990 putative heat shock transcription factor 200 1e-51
At3g22830 putative heat shock protein 186 2e-47
At5g16820 Heat Shock Factor 3 178 5e-45
At2g26150 heat shock transcription factor like protein 169 2e-42
At5g03720 heat shock transcription factor -like protein 169 3e-42
At1g67970 putative heat shock transcription factor 167 9e-42
At5g43840 heat shock transcription factor-like protein 158 4e-39
At3g51910 putative heat shock transcription factor 155 4e-38
At3g24520 heat shock transcription factor HSF1, putative 154 6e-38
At3g63350 heat shock transcription factor-like protein 149 3e-36
At1g46264 heat shock transcription factor, putative 148 6e-36
At5g54070 putative protein 146 2e-35
At5g62020 heat shock factor 6 138 6e-33
At4g11660 heat shock transcription factor - like protein 138 6e-33
At2g41690 putative heat shock transcription factor 133 2e-31
>At4g18880 heat shock transcription factor 21 (AtHSF21)
Length = 401
Score = 391 bits (1005), Expect = e-109
Identities = 213/411 (51%), Positives = 282/411 (67%), Gaps = 20/411 (4%)
Query: 1 MDEAQN--SSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFK 58
MDE + SSSSLPPFL KTYEMVDD SSD IVSWS +N+SF+VWNPP+F+R+LLPRFFK
Sbjct: 1 MDENNHGVSSSSLPPFLTKTYEMVDDSSSDSIVSWSQSNKSFIVWNPPEFSRDLLPRFFK 60
Query: 59 HNNFSSFIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHA 118
HNNFSSFIRQLNTYGFRK DPEQWEFAND FVRG+PHLMKNIHRRKPVHSHS+ NLQA
Sbjct: 61 HNNFSSFIRQLNTYGFRKADPEQWEFANDDFVRGQPHLMKNIHRRKPVHSHSLPNLQAQL 120
Query: 119 -PLGESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKM 177
PL +SER +N++IE+L KE LL EL + E + +E+Q+ LKE+L+ +E++Q+ M
Sbjct: 121 NPLTDSERVRMNNQIERLTKEKEGLLEELHKQDEEREVFEMQVKELKERLQHMEKRQKTM 180
Query: 178 VSSVSQVLHKPVIALNLLP-PTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEG 236
VS VSQVL KP +ALNL P ET +RKRR P +F DE +E+ + + ++ RE
Sbjct: 181 VSFVSQVLEKPGLALNLSPCVPETNERKRRFPRIEFFPDEPMLEE--NKTCVVVREEGST 238
Query: 237 ASVLTLNMERLDQLESSMLFWETIAHESGETFIHIHTNM--DVDESTSCADSLSISCPQL 294
+ +++QLESS+ WE + +S E+ + + M DVDES++ +S +SC QL
Sbjct: 239 SPSSHTREHQVEQLESSIAIWENLVSDSCESMLQSRSMMTLDVDESSTFPESPPLSCIQL 298
Query: 295 EVEVRSKSSG----IDMNSEPTAAAVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGA 350
V+ R KS IDMN EP + VA+ P G ND FW+QF +E+PG+
Sbjct: 299 SVDSRLKSPPSPRIIDMNCEPDGSKEQNTVAAPPPP----PVAGANDGFWQQFFSENPGS 354
Query: 351 SEAQEVQSERKEYNGRKSEGKPTDHGRFWWNARNANNLPEQVGHVSQAEKT 401
+E +EVQ ERK+ + G T+ + WWN+RN N + EQ+GH++ +E++
Sbjct: 355 TEQREVQLERKD--DKDKAGVRTE--KCWWNSRNVNAITEQLGHLTSSERS 401
>At5g45710 heat shock transcription factor
Length = 345
Score = 355 bits (911), Expect = 2e-98
Identities = 198/391 (50%), Positives = 257/391 (65%), Gaps = 48/391 (12%)
Query: 1 MDEAQNSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHN 60
MDE SSSLPPFL KTYEMVDD SSD +V+WS N+SF+V NP +F+R+LLPRFFKH
Sbjct: 1 MDENNGGSSSLPPFLTKTYEMVDDSSSDSVVAWSENNKSFIVKNPAEFSRDLLPRFFKHK 60
Query: 61 NFSSFIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPL 120
NFSSFIRQLNTYGFRKVDPE+WEF ND FVRG P+LMKNIHRRKPVHSHS+ NLQA PL
Sbjct: 61 NFSSFIRQLNTYGFRKVDPEKWEFLNDDFVRGRPYLMKNIHRRKPVHSHSLVNLQAQNPL 120
Query: 121 GESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSS 180
ESER+S+ D+IE+LK+ KE LL ELQ + E + +E+Q+ LK++L+ +EQ Q+ +V+
Sbjct: 121 TESERRSMEDQIERLKNEKEGLLAELQNQEQERKEFELQVTTLKDRLQHMEQHQKSIVAY 180
Query: 181 VSQVLHKPVIALNLLPPTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEGASVL 240
VSQVL KP ++LNL E +R++R F + + LP ++
Sbjct: 181 VSQVLGKPGLSLNL----ENHERRKRR-----FQENS-----------LPPSSS------ 214
Query: 241 TLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCADSLSISCPQLEVEVRS 300
++E++++LESS+ FWE + ES E ++MD D + S SLSI + R
Sbjct: 215 --HIEQVEKLESSLTFWENLVSESCEKSGLQSSSMDHDAAES---SLSIG------DTRP 263
Query: 301 KSSGIDMNSEPTAAAVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGASEAQEVQSER 360
KSS IDMNSEP PA K TGVND FWEQ LTE+PG++E QEVQSER
Sbjct: 264 KSSKIDMNSEPPVTVTA--------PAPK---TGVNDDFWEQCLTENPGSTEQQEVQSER 312
Query: 361 KEYNGRKSEGKPTDHGRFWWNARNANNLPEQ 391
++ + K + +WWN+ N NN+ E+
Sbjct: 313 RDVGNDNNGNKIGNQRTYWWNSGNVNNITEK 343
>At4g13980 unknown protein
Length = 466
Score = 208 bits (530), Expect = 4e-54
Identities = 140/419 (33%), Positives = 208/419 (49%), Gaps = 79/419 (18%)
Query: 13 PFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNTY 72
PFL KTYEMVDD S+D+IVSWSA N SF+VWN +F+R LLP +FKHNNFSSFIRQLNTY
Sbjct: 23 PFLVKTYEMVDDSSTDQIVSWSANNNSFIVWNHAEFSRLLLPTYFKHNNFSSFIRQLNTY 82
Query: 73 GFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLG--ESERKSLND 130
GFRK+DPE+WEF ND F++ + HL+KNIHRRKP+HSHS H P + ER L +
Sbjct: 83 GFRKIDPERWEFLNDDFIKDQKHLLKNIHRRKPIHSHS------HPPASSTDQERAVLQE 136
Query: 131 EIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVI 190
+++KL K + +L + + + + Q + E ++ +E +Q+K+++ + + P
Sbjct: 137 QMDKLSREKAAIEAKLLKFKQQKVVAKHQFEEMTEHVDDMENRQKKLLNFLETAIRNPTF 196
Query: 191 ALNL------------------------LPPTE------TMDRKRRLPGGGY---FNDEA 217
N PP+E + RR G + F+++
Sbjct: 197 VKNFGKKVEQLDISAYNKKRRLPEVEQSKPPSEDSHLDNSSGSSRRESGNIFHQNFSNKL 256
Query: 218 SIE------DAVDTSQMLPRENAEGASV------------------LTLNMERLDQLESS 253
+E D S + N EGAS L E L+ ++
Sbjct: 257 RLELSPADSDMNMVSHSIQSSNEEGASPKGILSGGDPNTTLTKREGLPFAPEALELADTG 316
Query: 254 MLFWETIAHESGETFI---HIHTNMDVDESTSCADSLSISCPQLEVEVRS-------KSS 303
+ +++ + ++ + D S SC +L+++ L + S KS
Sbjct: 317 TCPRRLLLNDNTRVETLQQRLTSSEETDGSFSCHLNLTLASAPLPDKTASQIAKTTLKSQ 376
Query: 304 GIDMNSEPTAAAVL----EPVASKEQPAGKAAATGVNDVFWEQFLTEDPGASEAQEVQS 358
++ NS T+A+ + +A A A VNDVFWEQFLTE PG+S+ +E S
Sbjct: 377 ELNFNSIETSASEKNRGRQEIAVGGSQANAAPPARVNDVFWEQFLTERPGSSDNEEASS 435
>At4g17750 heat shock transcription factor HSF1
Length = 495
Score = 201 bits (510), Expect = 8e-52
Identities = 122/370 (32%), Positives = 199/370 (52%), Gaps = 36/370 (9%)
Query: 6 NSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSF 65
N++S PPFL+KTY+MV+DP++D IVSWS TN SF+VW+PP+F+R+LLP++FKHNNFSSF
Sbjct: 45 NANSLPPPFLSKTYDMVEDPATDAIVSWSPTNNSFIVWDPPEFSRDLLPKYFKHNNFSSF 104
Query: 66 IRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSH--------SMQNLQAH 117
+RQLNTYGFRKVDP++WEFAN+GF+RG+ HL+K I RRK V H S Q Q
Sbjct: 105 VRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKKISRRKSVQGHGSSSSNPQSQQLSQGQ 164
Query: 118 APLG------ESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLE 171
+ E + L +E+E+LK +K L+ EL + + + Q + +L L + L+ +E
Sbjct: 165 GSMAALSSCVEVGKFGLEEEVEQLKRDKNVLMQELVKLRQQQQTTDNKLQVLVKHLQVME 224
Query: 172 QKQQKMVSSVSQVLHKPVIALNLLPP-------TETMDRKRRLPGGGYFNDEASIEDAVD 224
Q+QQ+++S +++ + P + ++KRRL D + ++
Sbjct: 225 QRQQQIMSFLAKAVQNPTFLSQFIQKQTDSNMHVTEANKKRRLR-----EDSTAATESNS 279
Query: 225 TSQMLPRENAEGASVLTLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCA 284
S L E ++G V + L + + W + + F+ ++ + +
Sbjct: 280 HSHSL--EASDGQIV------KYQPLRNDSMMWNMMKTDDKYPFLDGFSSPNQVSGVTLQ 331
Query: 285 DSLSISCPQLEVEVRSKSSGIDMNSEP-TAAAVLEPVASKEQPAGKAAATGVNDVFWEQF 343
+ L I+ Q + S SG ++ P T+ ++ + + + + +ND E F
Sbjct: 332 EVLPITSGQSQA-YASVPSGQPLSYLPSTSTSLPDTIMPETSQIPQLTRESINDFPTENF 390
Query: 344 LTEDPGASEA 353
+ + EA
Sbjct: 391 MDTEKNVPEA 400
>At1g32330 heat shock transcription factor HSF8 like protein
Length = 482
Score = 201 bits (510), Expect = 8e-52
Identities = 121/349 (34%), Positives = 185/349 (52%), Gaps = 50/349 (14%)
Query: 7 SSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFI 66
SS++ PPFL+KTY+MVDD ++D IVSWSA N SF+VW PP+FAR+LLP+ FKHNNFSSF+
Sbjct: 31 SSNAPPPFLSKTYDMVDDHNTDSIVSWSANNNSFIVWKPPEFARDLLPKNFKHNNFSSFV 90
Query: 67 RQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERK 126
RQLNTYGFRKVDP++WEFAN+GF+RG+ HL+++I RRKP H + ++ G++
Sbjct: 91 RQLNTYGFRKVDPDRWEFANEGFLRGQKHLLQSITRRKPAHGQGQGHQRSQHSNGQNSSV 150
Query: 127 S---------LNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKM 177
S L +E+E+LK +K L+ EL R + + Q + QL + ++L+ +E +QQ++
Sbjct: 151 SACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQSTDNQLQTMVQRLQGMENRQQQL 210
Query: 178 VSSVSQVLHKPVIALNLLPPTETMD----------RKRRLPGGGYF--NDEASIEDAVDT 225
+S +++ + P L + +KRR G ND A+ + +
Sbjct: 211 MSFLAKAVQSPHFLSQFLQQQNQQNESNRRISDTSKKRRFKRDGIVRNNDSATPDGQIVK 270
Query: 226 SQMLPRENAEGASVLTLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCAD 285
Q E A+ + ME +++G+ + E T
Sbjct: 271 YQPPMHEQAKAMFKQLMKME---------------PYKTGDDGFLLGNGTSTTEGT---- 311
Query: 286 SLSISCPQLEVEVRSKS-SGIDMNSEPTAAAVLEPVASKEQPAGKAAAT 333
E+E S SGI + PTA+ + + P +AA+
Sbjct: 312 ---------EMETSSNQVSGITLKEMPTASEIQSSSPIETTPENVSAAS 351
>At3g02990 putative heat shock transcription factor
Length = 468
Score = 200 bits (508), Expect = 1e-51
Identities = 149/450 (33%), Positives = 223/450 (49%), Gaps = 74/450 (16%)
Query: 9 SSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQ 68
SS+PPFL+KTY+MVDDP +D +VSWS+ N SFVVWN P+FA++ LP++FKHNNFSSF+RQ
Sbjct: 19 SSIPPFLSKTYDMVDDPLTDDVVSWSSGNNSFVVWNVPEFAKQFLPKYFKHNNFSSFVRQ 78
Query: 69 LNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQA-HAPLG---ESE 124
LNTYGFRKVDP++WEFAN+GF+RG+ ++K+I RRKP Q Q H+ +G E
Sbjct: 79 LNTYGFRKVDPDRWEFANEGFLRGQKQILKSIVRRKPAQVQPPQQPQVQHSSVGACVEVG 138
Query: 125 RKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQV 184
+ L +E+E+L+ +K L+ EL R + + Q E L + +++ +EQ+QQ+M+S +++
Sbjct: 139 KFGLEEEVERLQRDKNVLMQELVRLRQQQQVTEHHLQNVGQKVHVMEQRQQQMMSFLAKA 198
Query: 185 LHKPVIALNLLPPT-------ETMDRKRRLPGGGYFND---------------EASIEDA 222
+ P + ++KRRLP N ++S+ DA
Sbjct: 199 VQSPGFLNQFSQQSNEANQHISESNKKRRLPVEDQMNSGSHGVNGLSRQIVRYQSSMNDA 258
Query: 223 VDT--SQMLPRENAEGASVLTLNMER--LDQLESSML----------------------- 255
+T Q+ NA L+ N L + +S +
Sbjct: 259 TNTMLQQIQQMSNAPSHESLSSNNGSFLLGDVPNSNISDNGSSSNGSPEVTLADVSSIPA 318
Query: 256 -FWETIA-HESGETFIHIHTNMDVDESTSCADSLSISCPQLEVEVRSKSSGIDM-NSEPT 312
F+ + HE ET + TN+ + D L P + S SS D+ E
Sbjct: 319 GFYPAMKYHEPCETNQVMETNLPFSQ----GDLL----PPTQGAAASGSSSSDLVGCETD 370
Query: 313 AAAVLEPVASKEQPAGKAAATGVN-------DVFWEQFLTEDPGASEAQEV--QSERKEY 363
L+P+ + A + A +N D FWEQF+ E P E E+ S E
Sbjct: 371 NGECLDPIMAVLDGALELEADTLNELLPEVQDSFWEQFIGESPVIGETDELISGSVENEL 430
Query: 364 NGRKSEGKPTDHGRFWWNARNANNLPEQVG 393
+ E + T W + N+L EQ+G
Sbjct: 431 ILEQLELQST-LSNVWSKNQQMNHLTEQMG 459
>At3g22830 putative heat shock protein
Length = 406
Score = 186 bits (473), Expect = 2e-47
Identities = 101/242 (41%), Positives = 150/242 (61%), Gaps = 27/242 (11%)
Query: 9 SSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQ 68
S PPFL KTY++V+D ++ +VSWS +N SF+VW+P F+ LLPRFFKHNNFSSF+RQ
Sbjct: 57 SGPPPFLTKTYDLVEDSRTNHVVSWSKSNNSFIVWDPQAFSVTLLPRFFKHNNFSSFVRQ 116
Query: 69 LNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPV-HSHSMQNLQAHAPLG------ 121
LNTYGFRKV+P++WEFAN+GF+RG+ HL+KNI RRK +S+ MQ Q+
Sbjct: 117 LNTYGFRKVNPDRWEFANEGFLRGQKHLLKNIRRRKTSNNSNQMQQPQSSEQQSLDNFCI 176
Query: 122 ESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSV 181
E R L+ E++ L+ +K+ L+MEL R + + Q ++ L ++E+L+K E KQ++M+S +
Sbjct: 177 EVGRYGLDGEMDSLRRDKQVLMMELVRLRQQQQSTKMYLTLIEEKLKKTESKQKQMMSFL 236
Query: 182 SQVLHKPVIALNLLPP-------TETMDRKRRLP-------------GGGYFNDEASIED 221
++ + P L+ E + +KR+ P GY ND A+
Sbjct: 237 ARAMQNPDFIQQLVEQKEKRKEIEEAISKKRQRPIDQGKRNVEDYGDESGYGNDVAASSS 296
Query: 222 AV 223
A+
Sbjct: 297 AL 298
>At5g16820 Heat Shock Factor 3
Length = 447
Score = 178 bits (451), Expect = 5e-45
Identities = 92/196 (46%), Positives = 135/196 (67%), Gaps = 10/196 (5%)
Query: 21 MVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNTYGFRKVDPE 80
MVDDP ++ +VSWS+ N SFVVW+ P+F++ LLP++FKHNNFSSF+RQLNTYGFRKVDP+
Sbjct: 1 MVDDPLTNEVVSWSSGNNSFVVWSAPEFSKVLLPKYFKHNNFSSFVRQLNTYGFRKVDPD 60
Query: 81 QWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQ----AHAPLG---ESERKSLNDEIE 133
+WEFAN+GF+RG L+K+I RRKP SH QN Q + +G E + + +E+E
Sbjct: 61 RWEFANEGFLRGRKQLLKSIVRRKP--SHVQQNQQQTQVQSSSVGACVEVGKFGIEEEVE 118
Query: 134 KLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVIALN 193
+LK +K L+ EL R + + Q E QL + ++++ +EQ+QQ+M+S +++ + P LN
Sbjct: 119 RLKRDKNVLMQELVRLRQQQQATENQLQNVGQKVQVMEQRQQQMMSFLAKAVQSPGF-LN 177
Query: 194 LLPPTETMDRKRRLPG 209
L D R++PG
Sbjct: 178 QLVQQNNNDGNRQIPG 193
>At2g26150 heat shock transcription factor like protein
Length = 345
Score = 169 bits (428), Expect = 2e-42
Identities = 79/177 (44%), Positives = 123/177 (68%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTYEMV+DP++D +VSWS SFVVW+ F+ LLPR+FKH+NFSSFIRQLNT
Sbjct: 43 PPFLTKTYEMVEDPATDTVVSWSNGRNSFVVWDSHKFSTTLLPRYFKHSNFSSFIRQLNT 102
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLNDE 131
YGFRK+DP++WEFAN+GF+ G+ HL+KNI RR+ + ++ + E + + E
Sbjct: 103 YGFRKIDPDRWEFANEGFLAGQKHLLKNIKRRRNMGLQNVNQQGSGMSCVEVGQYGFDGE 162
Query: 132 IEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKP 188
+E+LK + L+ E+ R + + + Q+ ++++L E++QQ+M++ +++ L+ P
Sbjct: 163 VERLKRDHGVLVAEVVRLRQQQHSSKSQVAAMEQRLLVTEKRQQQMMTFLAKALNNP 219
>At5g03720 heat shock transcription factor -like protein
Length = 476
Score = 169 bits (427), Expect = 3e-42
Identities = 77/174 (44%), Positives = 122/174 (69%), Gaps = 2/174 (1%)
Query: 11 LPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLN 70
+PPFL+KT+++VDDP+ D ++SW T SFVVW+P +FAR +LPR FKHNNFSSF+RQLN
Sbjct: 117 IPPFLSKTFDLVDDPTLDPVISWGLTGASFVVWDPLEFARIILPRNFKHNNFSSFVRQLN 176
Query: 71 TYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLND 130
TYGFRK+D ++WEFAN+ F+RG+ HL+KNIHRR+ S+ Q + + +
Sbjct: 177 TYGFRKIDTDKWEFANEAFLRGKKHLLKNIHRRRSPQSN--QTCCSSTSQSQGSPTEVGG 234
Query: 131 EIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQV 184
EIEKL+ + L+ E+ Q + + ++ + ++L+ EQ+Q++++S ++++
Sbjct: 235 EIEKLRKERRALMEEMVELQQQSRGTARHVDTVNQRLKAAEQRQKQLLSFLAKL 288
>At1g67970 putative heat shock transcription factor
Length = 374
Score = 167 bits (423), Expect = 9e-42
Identities = 85/195 (43%), Positives = 132/195 (67%), Gaps = 1/195 (0%)
Query: 6 NSSSSLPPFLAKTYEMVDDPSSDRIVSWS-ATNRSFVVWNPPDFARELLPRFFKHNNFSS 64
+SSSS+ PFL K Y+MVDD ++D I+SWS + + SFV+ + F+ +LLP++FKH+NFSS
Sbjct: 12 SSSSSVAPFLRKCYDMVDDSTTDSIISWSPSADNSFVILDTTVFSVQLLPKYFKHSNFSS 71
Query: 65 FIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESE 124
FIRQLN YGFRKVD ++WEFANDGFVRG+ L+KN+ RRK V S ++ + E
Sbjct: 72 FIRQLNIYGFRKVDADRWEFANDGFVRGQKDLLKNVIRRKNVQSSEQSKHESTSTTYAQE 131
Query: 125 RKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQV 184
+ L E++ LK +K+ L EL + + + + ++ L+++++ +E+ QQ+M+S + V
Sbjct: 132 KSGLWKEVDILKGDKQVLAQELIKVRQYQEVTDTKMLHLEDRVQGMEESQQEMLSFLVMV 191
Query: 185 LHKPVIALNLLPPTE 199
+ P + + LL P E
Sbjct: 192 MKNPSLLVQLLQPKE 206
>At5g43840 heat shock transcription factor-like protein
Length = 251
Score = 158 bits (400), Expect = 4e-39
Identities = 86/200 (43%), Positives = 124/200 (62%), Gaps = 24/200 (12%)
Query: 12 PP--FLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQL 69
PP FL KTY +V+D S++ IVSWS N SF+VW P FA LPR FKHNNFSSF+RQL
Sbjct: 16 PPTAFLTKTYNIVEDSSTNNIVSWSRDNNSFIVWEPETFALICLPRCFKHNNFSSFVRQL 75
Query: 70 NTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLN 129
NTYGF+K+D E+WEFAN+ F++GE HL+KNI RRK +++ +SL
Sbjct: 76 NTYGFKKIDTERWEFANEHFLKGERHLLKNIKRRK--------------TSSQTQTQSLE 121
Query: 130 DEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPV 189
EI +L+ ++ L +EL R + + + + L+ ++E+L+ E KQ+ M++ + + + KP
Sbjct: 122 GEIHELRRDRMALEVELVRLRRKQESVKTYLHLMEEKLKVTEVKQEMMMNFLLKKIKKPS 181
Query: 190 IALNLLPPTETMDRKRRLPG 209
+L RKR L G
Sbjct: 182 FLQSL--------RKRNLQG 193
>At3g51910 putative heat shock transcription factor
Length = 272
Score = 155 bits (392), Expect = 4e-38
Identities = 78/184 (42%), Positives = 116/184 (62%), Gaps = 15/184 (8%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KT+EMVDDP++D IVSW+ SFVVW+ F+ LLPR FKH+NFSSFIRQLNT
Sbjct: 28 PPFLTKTFEMVDDPNTDHIVSWNRGGTSFVVWDLHSFSTILLPRHFKHSNFSSFIRQLNT 87
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLNDE 131
YGFRK++ E+WEFAN+ F+ G+ L+KNI RR P S + +D
Sbjct: 88 YGFRKIEAERWEFANEEFLLGQRQLLKNIKRRNPFTPSSSPS---------------HDA 132
Query: 132 IEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVIA 191
+L+ K+ L+ME+ + + Q + + +++++E E+KQ++M+S +++ + P
Sbjct: 133 CNELRREKQVLMMEIVSLRQQQQTTKSYIKAMEQRIEGTERKQRQMMSFLARAMQSPSFL 192
Query: 192 LNLL 195
LL
Sbjct: 193 HQLL 196
>At3g24520 heat shock transcription factor HSF1, putative
Length = 330
Score = 154 bits (390), Expect = 6e-38
Identities = 83/224 (37%), Positives = 135/224 (60%), Gaps = 31/224 (13%)
Query: 6 NSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSF 65
N+++ + PF+ KTY+MV+DPS+D +++W + SF+V +P DF++ +LP +FKHNNFSSF
Sbjct: 10 NNNNVIAPFIVKTYQMVNDPSTDWLITWGPAHNSFIVVDPLDFSQRILPAYFKHNNFSSF 69
Query: 66 IRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESER 125
+RQLNTYGFRKVDP++WEFAN+ F+RG+ HL+ NI RRK Q+L+ GE R
Sbjct: 70 VRQLNTYGFRKVDPDRWEFANEHFLRGQKHLLNNIARRKHARGMYGQDLED----GEIVR 125
Query: 126 KSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVL 185
EIE+LK + L E+QR + ++E E++ ++M++ + +V+
Sbjct: 126 -----EIERLKEEQRELEAEIQR--------------MNRRIEATEKRPEQMMAFLYKVV 166
Query: 186 HKPVIALNLLPPTE--------TMDRKRRLPGGGYFNDEASIED 221
P + ++ E + +KRR+ ++E +E+
Sbjct: 167 EDPDLLPRMMLEKERTKQQQQVSDKKKRRVTMSTVKSEEEEVEE 210
>At3g63350 heat shock transcription factor-like protein
Length = 282
Score = 149 bits (376), Expect = 3e-36
Identities = 89/260 (34%), Positives = 142/260 (54%), Gaps = 34/260 (13%)
Query: 13 PFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNTY 72
PFL KT+EMV DP+++ IVSW+ SFVVW+P F+ +LP +FKHNNFSSF+RQLNTY
Sbjct: 28 PFLTKTFEMVGDPNTNHIVSWNRGGISFVVWDPHSFSATILPLYFKHNNFSSFVRQLNTY 87
Query: 73 GFRKVDPEQWEFANDGFVRGEPHLMKNIHRR-----KPVHSHSMQNLQAHAPLGESERKS 127
GFRK++ E+WEF N+GF+ G+ L+K+I RR P ++S +AH P
Sbjct: 88 GFRKIEAERWEFMNEGFLMGQRDLLKSIKRRTSSSSPPSLNYSQSQPEAHDP-------- 139
Query: 128 LNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHK 187
E+ +L+ + L+ME+ + E Q + +++++ E+KQ+ M+S + + +
Sbjct: 140 -GVELPQLREERHVLMMEISTLRQEEQRARGYVQAMEQRINGAEKKQRHMMSFLRRAVEN 198
Query: 188 PVIALNLLPPTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEGASVLTLNM--- 244
P + + ++KR +EA++ D +M E+ L L M
Sbjct: 199 PSLL------QQIFEQKRD-------REEAAMIDQAGLIKMEEVEHLSELEALALEMQGY 245
Query: 245 --ERLDQLESSM--LFWETI 260
+R D +E + FWE +
Sbjct: 246 GRQRTDGVERELDDGFWEEL 265
>At1g46264 heat shock transcription factor, putative
Length = 348
Score = 148 bits (373), Expect = 6e-36
Identities = 66/107 (61%), Positives = 83/107 (76%), Gaps = 4/107 (3%)
Query: 13 PFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNTY 72
PFL KTY++VDDP++D +VSW + +FVVW PP+FAR+LLP +FKHNNFSSF+RQLNTY
Sbjct: 34 PFLTKTYQLVDDPATDHVVSWGDDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVRQLNTY 93
Query: 73 GFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAP 119
GFRK+ P++WEFAN+ F RGE HL+ IHRRK S Q H+P
Sbjct: 94 GFRKIVPDRWEFANEFFKRGEKHLLCEIHRRKT----SQMIPQQHSP 136
>At5g54070 putative protein
Length = 331
Score = 146 bits (369), Expect = 2e-35
Identities = 72/169 (42%), Positives = 111/169 (65%), Gaps = 9/169 (5%)
Query: 11 LPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLN 70
+ PFL KT+E+VDD +D +VSWS T +SF++W+ +F+ LLP++FKH NFSSFIRQLN
Sbjct: 69 ITPFLRKTFEIVDDKVTDPVVSWSPTRKSFIIWDSYEFSENLLPKYFKHKNFSSFIRQLN 128
Query: 71 TYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLND 130
+YGF+KVD ++WEFAN+GF G+ HL+KNI RR + N +A E+E +SL +
Sbjct: 129 SYGFKKVDSDRWEFANEGFQGGKKHLLKNIKRRS--KNTKCCNKEASTTTTETEVESLKE 186
Query: 131 EIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVS 179
E ++ +L + + QH Q+ ++E++ ++ +QQ M+S
Sbjct: 187 EQSPMRLEMLKLKQQQEESQH-------QMVTVQEKIHGVDTEQQHMLS 228
>At5g62020 heat shock factor 6
Length = 299
Score = 138 bits (347), Expect = 6e-33
Identities = 61/105 (58%), Positives = 81/105 (77%), Gaps = 1/105 (0%)
Query: 6 NSSSSLP-PFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSS 64
+S S+P PFL KT+ +V+D S D ++SW+ SF+VWNP DFA++LLP+ FKHNNFSS
Sbjct: 15 SSQRSIPTPFLTKTFNLVEDSSIDDVISWNEDGSSFIVWNPTDFAKDLLPKHFKHNNFSS 74
Query: 65 FIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSH 109
F+RQLNTYGF+KV P++WEF+ND F RGE L++ I RRK +H
Sbjct: 75 FVRQLNTYGFKKVVPDRWEFSNDFFKRGEKRLLREIQRRKITTTH 119
>At4g11660 heat shock transcription factor - like protein
Length = 377
Score = 138 bits (347), Expect = 6e-33
Identities = 60/100 (60%), Positives = 81/100 (81%), Gaps = 1/100 (1%)
Query: 6 NSSSSLP-PFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSS 64
+S S+P PFL KTY++V+DP D ++SW+ +F+VW P +FAR+LLP++FKHNNFSS
Sbjct: 51 DSQRSIPTPFLTKTYQLVEDPVYDELISWNEDGTTFIVWRPAEFARDLLPKYFKHNNFSS 110
Query: 65 FIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRK 104
F+RQLNTYGFRKV P++WEF+ND F RGE L+++I RRK
Sbjct: 111 FVRQLNTYGFRKVVPDRWEFSNDCFKRGEKILLRDIQRRK 150
>At2g41690 putative heat shock transcription factor
Length = 244
Score = 133 bits (335), Expect = 2e-31
Identities = 61/105 (58%), Positives = 78/105 (74%), Gaps = 1/105 (0%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTY++V+DP++D ++SW+ FVVW P +FAR+LLP FKH NFSSF+RQLNT
Sbjct: 39 PPFLVKTYKVVEDPTTDGVISWNEYGTGFVVWQPAEFARDLLPTLFKHCNFSSFVRQLNT 98
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVH-SHSMQNLQ 115
YGFRKV +WEF+N+ F +G+ LM NI RRK H SH+ N Q
Sbjct: 99 YGFRKVTTIRWEFSNEMFRKGQRELMSNIRRRKSQHWSHNKSNHQ 143
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,412,286
Number of Sequences: 26719
Number of extensions: 412608
Number of successful extensions: 1466
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 1323
Number of HSP's gapped (non-prelim): 172
length of query: 401
length of database: 11,318,596
effective HSP length: 101
effective length of query: 300
effective length of database: 8,619,977
effective search space: 2585993100
effective search space used: 2585993100
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0162a.11


