

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.4
(1003 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
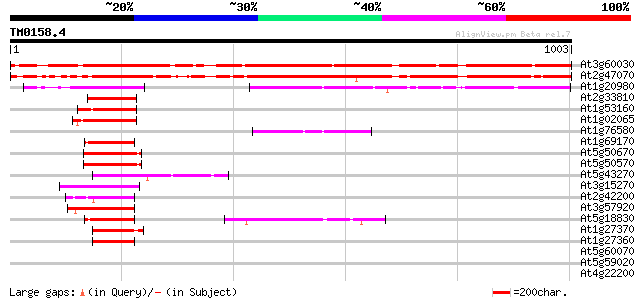
Score E
Sequences producing significant alignments: (bits) Value
At3g60030 squamosa promoter binding protein-like 12 980 0.0
At2g47070 squamosa promoter binding protein-like 1 979 0.0
At1g20980 putative SPL1-related protein 301 1e-81
At2g33810 squamosa-promoter binding protein-like 3 126 5e-29
At1g53160 squamosa promoter binding protein-like 4 (spl4) 125 1e-28
At1g02065 squamosa promoter binding protein-like 8 (SPL8) 123 5e-28
At1g76580 unknown protein 123 6e-28
At1g69170 squamosa promoter binding protein-like 6 (spl6) 123 6e-28
At5g50670 putative protein 121 2e-27
At5g50570 putative protein 121 2e-27
At5g43270 squamosa promoter binding protein-like 2 (emb|CAB56576.1) 120 3e-27
At3g15270 squamosa promoter binding protein-like 5 120 4e-27
At2g42200 squamosa promoter binding protein-like 9 117 2e-26
At3g57920 squamosa promoter-binding protein homolog 112 1e-24
At5g18830 squamosa promoter binding protein-like 7 109 9e-24
At1g27370 squamosa-promoter binding protein-like 10 108 1e-23
At1g27360 squamosa-promoter binding protein-like 11 107 4e-23
At5g60070 ankyrin-like protein 38 0.025
At5g59020 putative protein 35 0.16
At4g22200 potassium channel (AKT2) 34 0.47
>At3g60030 squamosa promoter binding protein-like 12
Length = 927
Score = 980 bits (2534), Expect = 0.0
Identities = 552/1022 (54%), Positives = 703/1022 (68%), Gaps = 114/1022 (11%)
Query: 1 MEARFGAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQQL 60
MEAR E G S + GKRS EWDLN+W+W+GDLF+A+++N
Sbjct: 1 MEARIEGEVE-----GHSLEYGFSGKRSVEWDLNDWKWNGDLFVATQLNH---------- 45
Query: 61 FPLVSGIPASNSSSSCSEEVDLRDPMGSR---EGERKRR---VIVLEDDGLNEEGG-TLS 113
+SNSSS+CS+E ++ R E ++KRR V+ +E+D L ++ L+
Sbjct: 46 -------GSSNSSSTCSDEGNVEIMERRRIEMEKKKKRRAVTVVAMEEDNLKDDDAHRLT 98
Query: 114 LKLGGHAADREVASWDGGNGKKSRVAGGGASNRAVC-QVEDCCADLSRAKDYHRRHKVCE 172
L LGG+ + GNG K GGG +RA+C QV++C ADLS+ KDYHRRHKVCE
Sbjct: 99 LNLGGNNIE--------GNGVKKTKLGGGIPSRAICCQVDNCGADLSKVKDYHRRHKVCE 150
Query: 173 MHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGN 232
+HSKAT ALVG MQRFCQQCSRFH+L+EFDEGKRSCRRRLAGHNKRRRK N + + NG
Sbjct: 151 IHSKATTALVGGIMQRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANPDTIGNGT 210
Query: 233 PLNDDQTSSYLLISLLKILSNMNSDRSDQTTDQDLLTHLLRSLASRNGEQGGNNLSNILR 292
++DDQTS+Y+LI+LLKILSN++S++SDQT DQDLL+HLL+SL S+ GE G NL
Sbjct: 211 SMSDDQTSNYMLITLLKILSNIHSNQSDQTGDQDLLSHLLKSLVSQAGEHIGRNL----- 265
Query: 293 EPENLLREGGSSRESEMVPTL--LSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATD 350
LL+ GG + S+ + L L + Q DI+ H S+++ + V+A+ A+
Sbjct: 266 --VGLLQGGGGLQASQNIGNLSALLSLEQAPREDIKHH---SVSETPWQEVYANSAQE-- 318
Query: 351 QQLMSYIKPSISNTPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHV 410
A D + Q+K+N+FDLNDIYIDSDD + P TN
Sbjct: 319 ------------------RVAPDRSEKQVKVNDFDLNDIYIDSDDTTDIERSSPPPTNPA 360
Query: 411 TSSLDYPWTQQDSHQSSPPQTS-RNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDF 469
TSSLDY QDS QSSPPQTS RNS+S S SPSSS+G+AQ RTDRIV KLFGKEP+DF
Sbjct: 361 TSSLDY---HQDSRQSSPPQTSRRNSDSASDQSPSSSSGDAQSRTDRIVFKLFGKEPNDF 417
Query: 470 PLILRAQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSD 529
P+ LR QIL+WL+H+PTD+ESYIRPGCIVLTIYLRQ EA WEE+C DL+ SL RLLD+SD
Sbjct: 418 PVALRGQILNWLAHTPTDMESYIRPGCIVLTIYLRQDEASWEELCCDLSFSLRRLLDLSD 477
Query: 530 DTFWRTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVK 589
D W GW+++RVQ+Q+AF FNGQVV+DTSLP RS++YS+I+TV P+A +K+ QF+VK
Sbjct: 478 DPLWTDGWLYLRVQNQLAFAFNGQVVLDTSLPLRSHDYSQIITVRPLA--VTKKAQFTVK 535
Query: 590 GVNMIRPATRLMCALEGKYLVCEDAHESMDQYSE--ELDGLHCIQFSCAVPVTNGRGFIE 647
G+N+ RP TRL+C +EG +LV E M++ + E + + + FSC +P+ +GRGF+E
Sbjct: 536 GINLRRPGTRLLCTVEGTHLVQEATQGGMEERDDLKENNEIDFVNFSCEMPIASGRGFME 595
Query: 648 IEDQ-GLSSSFFPFIVAE-EDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHE 705
IEDQ GLSSSFFPFIV+E ED+CS+I LE LE + TD + QAMDFIHE
Sbjct: 596 IEDQGGLSSSFFPFIVSEDEDICSEIRRLESTLEFTGTD----------SAMQAMDFIHE 645
Query: 706 IGWLLHGSQLKSRMVHLS-SSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLL-EGTVS 763
IGWLLH S+LKSR+ + DLF L RFK+L+EFSMD +WC V+KKLLN+L EGTV
Sbjct: 646 IGWLLHRSELKSRLAASDHNPEDLFSLIRFKFLIEFSMDREWCCVMKKLLNILFEEGTVD 705
Query: 764 TGDHPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFL 823
PS D ALSE+ LLHRAVR+NSK +VE+LL + P+ + + + L
Sbjct: 706 ----PSPDAALSELCLLHRAVRKNSKPMVEMLLRFSPKK-------------KNQTLAGL 748
Query: 824 FRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLR 883
FRPDAAGP GLTPLHIAAGKDGSEDVLDALT DP M GI+AWKN+RD+TG TPEDYARLR
Sbjct: 749 FRPDAAGPGGLTPLHIAAGKDGSEDVLDALTEDPGMTGIQAWKNSRDNTGFTPEDYARLR 808
Query: 884 GHYTYIHLVQKKINKRQGA-PHVVVEIPTNLT-ENDTNQKQNESSTTFEIAKTRDQGHCK 941
GH++YIHLVQ+K++++ A HVVV IP + E+ ++ S++ EI + CK
Sbjct: 809 GHFSYIHLVQRKLSRKPIAKEHVVVNIPESFNIEHKQEKRSPMDSSSLEITQI---NQCK 865
Query: 942 LCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFG 1001
LCD K T +S+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PF WE L++G
Sbjct: 866 LCDHKRVFVTTHHKSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLEYG 925
Query: 1002 TS 1003
TS
Sbjct: 926 TS 927
>At2g47070 squamosa promoter binding protein-like 1
Length = 881
Score = 979 bits (2530), Expect = 0.0
Identities = 547/1013 (53%), Positives = 682/1013 (66%), Gaps = 142/1013 (14%)
Query: 1 MEARF--GAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQ 58
MEAR G EA +YG +GKRS EWDLN+W+WDGDLF+A++ G+
Sbjct: 1 MEARIDEGGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR------GR 46
Query: 59 QLFPLVSGIPASNSSSSCSEEVDLRDPMGSREGERKRRVIVLEDDGLNEEGGTLSLKLGG 118
Q FPL + +SNSSSSCS+E + ++KRR + ++ D G L+L L G
Sbjct: 47 QFFPLGN---SSNSSSSCSDEGN----------DKKRRAVAIQGD----TNGALTLNLNG 89
Query: 119 HAADREVASWDGGNGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKAT 178
+ DG K +G AVCQVE+C ADLS+ KDYHRRHKVCEMHSKAT
Sbjct: 90 ES--------DGLFPAKKTKSG------AVCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 179 RALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLNDDQ 238
A VG +QRFCQQCSRFH+LQEFDEGKRSCRRRLAGHNKRRRKTN E NGNP +DD
Sbjct: 136 SATVGGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNP-SDDH 194
Query: 239 TSSYLLISLLKILSNMNSDRSDQTTDQDLLTHLLRSLASRNGEQGGNNLSNILREPENLL 298
+S+YLLI+LLKILSNM++ T DQDL++HLL+SL S GEQ G NL LL
Sbjct: 195 SSNYLLITLLKILSNMHN----HTGDQDLMSHLLKSLVSHAGEQLGKNLVE-------LL 243
Query: 299 REGGSSRESEMVPTLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIK 358
+GG GSQGS +I + + + QE + AR
Sbjct: 244 LQGG--------------GSQGS-LNIGNSALLGIEQAPQEELKQFSARQDG-------- 280
Query: 359 PSISNTPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHVTSSLDYP- 417
+ + Q+KMN+FDLNDIYIDSDD D+ER P TN TSSLDYP
Sbjct: 281 ----------TATENRSEKQVKMNDFDLNDIYIDSDD--TDVERSPPPTNPATSSLDYPS 328
Query: 418 WTQQDSHQSSPPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQI 477
W HQSSPPQTSRNS+S S SPSSS+ +AQ RT RIV KLFGKEP++FP++LR QI
Sbjct: 329 WI----HQSSPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQI 384
Query: 478 LDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGW 537
LDWLSHSPTD+ESYIRPGCIVLTIYLRQAE WEE+ DL SLG+LLD+SDD W TGW
Sbjct: 385 LDWLSHSPTDMESYIRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGW 444
Query: 538 VHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPA 597
+++RVQ+Q+AF++NGQVV+DTSL +S +YS I++V P+A A+++ QF+VKG+N+ +
Sbjct: 445 IYVRVQNQLAFVYNGQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRQRG 504
Query: 598 TRLMCALEGKYLVCEDAHESM----DQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGL 653
TRL+C++EGKYL+ E H+S D + + + + C+ FSC +P+ +GRGF+EIEDQGL
Sbjct: 505 TRLLCSVEGKYLIQETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGL 564
Query: 654 SSSFFPFIVAE-EDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHG 712
SSSFFPF+V E +DVCS+I LE LE + TD + QAMDFIHEIGWLLH
Sbjct: 565 SSSFFPFLVVEDDDVCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLH- 613
Query: 713 SQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDL 772
+S++ + +FPL RF+WL+EFSMD +WCAV++KLLN+ +G V S +
Sbjct: 614 ---RSKLGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NA 669
Query: 773 ALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPA 832
LSE+ LLHRAVR+NSK +VE+LL Y+P+ ++++ LFRPDAAGPA
Sbjct: 670 TLSELCLLHRAVRKNSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPA 713
Query: 833 GLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLV 892
GLTPLHIAAGKDGSEDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+
Sbjct: 714 GLTPLHIAAGKDGSEDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLI 773
Query: 893 QKKINKRQGA-PHVVVEIPTNLTENDTNQ-KQNESSTTFEIAKTRDQGHCKLCDIKLSCR 950
Q+KINK+ HVVV IP + ++ + + K ++ EI Q CKLCD KL
Sbjct: 774 QRKINKKSTTEDHVVVNIPVSFSDREQKEPKSGPMASALEIT----QIPCKLCDHKLVYG 829
Query: 951 TAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
T RS+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PF WE LD+GTS
Sbjct: 830 T-TRRSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 881
>At1g20980 putative SPL1-related protein
Length = 1035
Score = 301 bits (771), Expect = 1e-81
Identities = 189/581 (32%), Positives = 307/581 (52%), Gaps = 35/581 (6%)
Query: 430 QTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIE 489
Q S + SGS +SP S +AQ RT +IV KL K+PS P LR++I +WLS+ P+++E
Sbjct: 481 QQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQLPGTLRSEIYNWLSNIPSEME 540
Query: 490 SYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFI 549
SYIRPGC+VL++Y+ + A WE++ L LG LL S FWR + Q+A
Sbjct: 541 SYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSPSDFWRNARFIVNTGRQLASH 600
Query: 550 FNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYL 609
NG+V S +R+ N ++++VSP+A A + V+G ++ + C G Y+
Sbjct: 601 KNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVVRGRSLTNDGISIRCTHMGSYM 658
Query: 610 VCEDAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCS 669
E Q + ++ + P GR FIE+E+ G FP I+A +C
Sbjct: 659 AMEVTRAVCRQTIFDELNVNSFKVQNVHPGFLGRCFIEVEN-GFRGDSFPLIIANASICK 717
Query: 670 DICGL-----EPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSS 724
++ L +++E G ++ + + F++E+GWL K++ L
Sbjct: 718 ELNRLGEEFHPKSQDMTEEQAQSSNRGP-TSREEVLCFLNELGWLFQ----KNQTSELRE 772
Query: 725 STDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGD--HPSLDLALSEMGLLHR 782
+D F L RFK+L+ S++ D+CA+++ LL++L+E + + +LD+ L+E+ LL+R
Sbjct: 773 QSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREALDM-LAEIQLLNR 830
Query: 783 AVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAG 842
AV+R S ++VELL+ Y+ V P L + F+F P+ GP G+TPLH+AA
Sbjct: 831 AVKRKSTKMVELLIHYL-------VNP----LTLSSSRKFVFLPNITGPGGITPLHLAAC 879
Query: 843 KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGA 902
GS+D++D LTNDP +G+ +W RD+TG TP YA +R ++ Y LV +K+ ++
Sbjct: 880 TSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRNK 939
Query: 903 PHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQ-GHCKLCDIKLSCRTAVGRSLVYKP 961
++ N+ +Q + E+ K+ C +K R + + L P
Sbjct: 940 -----QVSLNIEHEVVDQTGLSKRLSLEMNKSSSSCASCATVALKYQRRVSGSQRLFPTP 994
Query: 962 AMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
+ SM+AVA VCVCV + + P ++ FSW LD+G+
Sbjct: 995 IIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSWGGLDYGS 1034
Score = 152 bits (384), Expect = 9e-37
Identities = 95/219 (43%), Positives = 114/219 (51%), Gaps = 44/219 (20%)
Query: 26 KRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQQLFPLVSGIPASNSSSSCSEEVDLRDP 85
+R EW+ W WD F A V+ V Q F L
Sbjct: 38 QRRDEWNSKMWDWDSRRFEAKPVD------VEVQEFDLT--------------------- 70
Query: 86 MGSREGERKRRVIVLEDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKKSRVAGGGASN 145
+ +R GE E G L+L G A + + K +G N
Sbjct: 71 LRNRSGE--------------ERGLDLNLGSGLTAVEETTTTTQNVRPNKKVRSGSPGGN 116
Query: 146 RAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEG 205
+CQV++C DLS AKDYHRRHKVCE+HSKAT+ALVG MQRFCQQCSRFH+L EFDEG
Sbjct: 117 YPMCQVDNCTEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEG 176
Query: 206 KRSCRRRLAGHNKRRRKTNN-EAVPNG--NPLNDDQTSS 241
KRSCRRRLAGHN+RRRKT E V +G P N D T++
Sbjct: 177 KRSCRRRLAGHNRRRRKTTQPEEVASGVVVPGNHDTTNN 215
>At2g33810 squamosa-promoter binding protein-like 3
Length = 131
Score = 126 bits (317), Expect = 5e-29
Identities = 55/87 (63%), Positives = 65/87 (74%)
Query: 140 GGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHML 199
G S+ VCQVE C AD+S+AK YH+RHKVC+ H+KA + QRFCQQCSRFH L
Sbjct: 45 GKATSSSGVCQVESCTADMSKAKQYHKRHKVCQFHAKAPHVRISGLHQRFCQQCSRFHAL 104
Query: 200 QEFDEGKRSCRRRLAGHNKRRRKTNNE 226
EFDE KRSCRRRLAGHN+RRRK+ +
Sbjct: 105 SEFDEAKRSCRRRLAGHNERRRKSTTD 131
>At1g53160 squamosa promoter binding protein-like 4 (spl4)
Length = 174
Score = 125 bits (314), Expect = 1e-28
Identities = 59/106 (55%), Positives = 74/106 (69%), Gaps = 2/106 (1%)
Query: 122 DREVASWDGGNGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRAL 181
+ E D G + + GG+ +CQV+ C AD+ AK YHRRHKVCE+H+KA+
Sbjct: 29 EEEEVGRDRVRGSRGSINRGGSLR--LCQVDRCTADMKEAKLYHRRHKVCEVHAKASSVF 86
Query: 182 VGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEA 227
+ QRFCQQCSRFH LQEFDE KRSCRRRLAGHN+RRRK++ E+
Sbjct: 87 LSGLNQRFCQQCSRFHDLQEFDEAKRSCRRRLAGHNERRRKSSGES 132
>At1g02065 squamosa promoter binding protein-like 8 (SPL8)
Length = 333
Score = 123 bits (309), Expect = 5e-28
Identities = 66/123 (53%), Positives = 77/123 (61%), Gaps = 10/123 (8%)
Query: 112 LSLKLGGH----AADREVASWDGGNGKKSRVAGGGASNRAV---CQVEDCCADLSRAKDY 164
+ L LGG AAD + S ++SR G +N CQ E C ADLS AK Y
Sbjct: 147 IGLNLGGRTYFSAADDDFVS---RLYRRSRPGESGMANSLSTPRCQAEGCNADLSHAKHY 203
Query: 165 HRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTN 224
HRRHKVCE HSKA+ + QRFCQQCSRFH+L EFD GKRSCR+RLA HN+RRRK +
Sbjct: 204 HRRHKVCEFHSKASTVVAAGLSQRFCQQCSRFHLLSEFDNGKRSCRKRLADHNRRRRKCH 263
Query: 225 NEA 227
A
Sbjct: 264 QSA 266
>At1g76580 unknown protein
Length = 488
Score = 123 bits (308), Expect = 6e-28
Identities = 76/214 (35%), Positives = 114/214 (52%), Gaps = 6/214 (2%)
Query: 435 SESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIESYIRP 494
+ SGS +SP S AQ RT +I KLF K+PS P LR +I WLS P+D+ES+IRP
Sbjct: 272 ASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRP 331
Query: 495 GCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFIFNGQV 554
GC++L++Y+ + + WE++ +L + L V D FW + Q+A +G++
Sbjct: 332 GCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWSNSRFLVNAGRQLASHKHGRI 389
Query: 555 VIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCE-D 613
+ S +R+ N +++TVSP+A A + V+G N+ RL CA G Y E
Sbjct: 390 RLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVT 447
Query: 614 AHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIE 647
E +EL+ + Q A V+ GR FIE
Sbjct: 448 GREHRLTKVDELN-VSSFQVQSASSVSLGRCFIE 480
>At1g69170 squamosa promoter binding protein-like 6 (spl6)
Length = 405
Score = 123 bits (308), Expect = 6e-28
Identities = 58/89 (65%), Positives = 66/89 (73%), Gaps = 1/89 (1%)
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
KKSR A S +CQV C DLS +KDYH+RH+VCE HSK + +V QRFCQQC
Sbjct: 110 KKSR-ASNLCSQNPLCQVYGCSKDLSSSKDYHKRHRVCEAHSKTSVVIVNGLEQRFCQQC 168
Query: 194 SRFHMLQEFDEGKRSCRRRLAGHNKRRRK 222
SRFH L EFD+GKRSCRRRLAGHN+RRRK
Sbjct: 169 SRFHFLSEFDDGKRSCRRRLAGHNERRRK 197
>At5g50670 putative protein
Length = 359
Score = 121 bits (303), Expect = 2e-27
Identities = 54/104 (51%), Positives = 71/104 (67%), Gaps = 2/104 (1%)
Query: 132 NGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQ 191
+ K++R G G + +C V+ C +D S ++YH+RHKVC++HSK + QRFCQ
Sbjct: 84 SSKRTRGNGVGTNQMPICLVDGCDSDFSNCREYHKRHKVCDVHSKTPVVTINGHKQRFCQ 143
Query: 192 QCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLN 235
QCSRFH L+EFDEGKRSCR+RL GHN+RRRK E + G P N
Sbjct: 144 QCSRFHALEEFDEGKRSCRKRLDGHNRRRRKPQPEHI--GRPAN 185
>At5g50570 putative protein
Length = 359
Score = 121 bits (303), Expect = 2e-27
Identities = 54/104 (51%), Positives = 71/104 (67%), Gaps = 2/104 (1%)
Query: 132 NGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQ 191
+ K++R G G + +C V+ C +D S ++YH+RHKVC++HSK + QRFCQ
Sbjct: 84 SSKRTRGNGVGTNQMPICLVDGCDSDFSNCREYHKRHKVCDVHSKTPVVTINGHKQRFCQ 143
Query: 192 QCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLN 235
QCSRFH L+EFDEGKRSCR+RL GHN+RRRK E + G P N
Sbjct: 144 QCSRFHALEEFDEGKRSCRKRLDGHNRRRRKPQPEHI--GRPAN 185
>At5g43270 squamosa promoter binding protein-like 2 (emb|CAB56576.1)
Length = 419
Score = 120 bits (302), Expect = 3e-27
Identities = 87/257 (33%), Positives = 119/257 (45%), Gaps = 22/257 (8%)
Query: 149 CQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRS 208
CQVE C DLS AKDYHR+H++CE HSK + +V +RFCQQCSRFH L EFDE KRS
Sbjct: 169 CQVEGCNLDLSSAKDYHRKHRICENHSKFPKVVVSGVERRFCQQCSRFHCLSEFDEKKRS 228
Query: 209 CRRRLAGHNKRRRKTNNEAVPNGNPLNDDQTSSYLLI-----------SLLKILSNMNSD 257
CRRRL+ HN RRRK N +G P D + + LI S + S +
Sbjct: 229 CRRRLSDHNARRRKPNPGRTYDGKPQVDFVWNRFALIHPRSEEKFIWPSSKHVPSRVLMP 288
Query: 258 RSDQTTDQDLLTHLLRSLASRNGEQGGNNLSNILREPENLLREGGSSRESEMVPTLLSNG 317
+ +T D + L + S +E + G+S++ + +LLSN
Sbjct: 289 QPAKTEISDTEHNRFGLLDPKTKTARAELFS---KEKVTISSHMGASQDLDGALSLLSNS 345
Query: 318 SQG-SPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIKPSISNTPPAY---AEARD 373
+ S +D + T+ + AH + A + Y +P PPA
Sbjct: 346 TTWVSSSDQPRRFTLDHHPSSNLQPVAHRSAAQLNSVSGYWQPD----PPAVEGPTALHR 401
Query: 374 STAGQIKMNNFDLNDIY 390
+ GQ N F LN Y
Sbjct: 402 NGVGQFNENYFSLNQFY 418
>At3g15270 squamosa promoter binding protein-like 5
Length = 181
Score = 120 bits (301), Expect = 4e-27
Identities = 63/145 (43%), Positives = 84/145 (57%), Gaps = 2/145 (1%)
Query: 90 EGERKRRVIVLEDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKKSRVAGGGASN--RA 147
EG+R +R L+D ++ G + + D RV G
Sbjct: 2 EGQRTQRRGYLKDKATVSNLVEEEMENGMDGEEEDGGDEDKRKKVMERVRGPSTDRVPSR 61
Query: 148 VCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKR 207
+CQV+ C +L+ AK Y+RRH+VCE+H+KA+ A V QRFCQQCSRFH L EFDE KR
Sbjct: 62 LCQVDRCTVNLTEAKQYYRRHRVCEVHAKASAATVAGVRQRFCQQCSRFHELPEFDEAKR 121
Query: 208 SCRRRLAGHNKRRRKTNNEAVPNGN 232
SCRRRLAGHN+RRRK + ++ G+
Sbjct: 122 SCRRRLAGHNERRRKISGDSFGEGS 146
>At2g42200 squamosa promoter binding protein-like 9
Length = 375
Score = 117 bits (294), Expect = 2e-26
Identities = 66/135 (48%), Positives = 78/135 (56%), Gaps = 17/135 (12%)
Query: 101 EDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKKSRVAGGGASNRAV------------ 148
E G + E + S GG +++ DGG G S + GG SNR V
Sbjct: 17 ESGGSSTESSSFS---GGLMFGQKIYFEDGGGGSGSS-SSGGRSNRRVRGGGSGQSGQIP 72
Query: 149 -CQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKR 207
CQVE C DL+ AK Y+ RH+VC +HSK + V QRFCQQCSRFH L EFD KR
Sbjct: 73 RCQVEGCGMDLTNAKGYYSRHRVCGVHSKTPKVTVAGIEQRFCQQCSRFHQLPEFDLEKR 132
Query: 208 SCRRRLAGHNKRRRK 222
SCRRRLAGHN+RRRK
Sbjct: 133 SCRRRLAGHNERRRK 147
>At3g57920 squamosa promoter-binding protein homolog
Length = 354
Score = 112 bits (279), Expect = 1e-24
Identities = 64/125 (51%), Positives = 79/125 (63%), Gaps = 6/125 (4%)
Query: 104 GLNEEGGTLSLKL----GGHAADREVASWDG-GNGKKSRV-AGGGASNRAVCQVEDCCAD 157
G E GG+ S + GG +++ DG G+ K+RV +S A CQVE C D
Sbjct: 8 GQAESGGSSSTESSSLSGGLRFGQKIYFEDGSGSRSKNRVNTVRKSSTTARCQVEGCRMD 67
Query: 158 LSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHN 217
LS K Y+ RHKVC +HSK+++ +V QRFCQQCSRFH L EFD KRSCRRRLA HN
Sbjct: 68 LSNVKAYYSRHKVCCIHSKSSKVIVSGLHQRFCQQCSRFHQLSEFDLEKRSCRRRLACHN 127
Query: 218 KRRRK 222
+RRRK
Sbjct: 128 ERRRK 132
>At5g18830 squamosa promoter binding protein-like 7
Length = 801
Score = 109 bits (272), Expect = 9e-24
Identities = 78/309 (25%), Positives = 136/309 (43%), Gaps = 31/309 (10%)
Query: 385 DLNDIYIDSDDGIEDLERLPGTTNHVTSSLDYPWTQQ----------DSHQSSPPQTSRN 434
D DI SD E+ L H+T+ P+T+ S ++ P + +
Sbjct: 239 DGKDITCSSDQRAEEEPSLIFEDRHITTQGSVPFTRSINADNFVSVTGSGEAQPDEGMND 298
Query: 435 S--ESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIESYI 492
+ E ++ + S C T RI KL+ P++FP LR QI WL++ P ++E YI
Sbjct: 299 TKFERSPSNGDNKSAYSTVCPTGRISFKLYDWNPAEFPRRLRHQIFQWLANMPVELEGYI 358
Query: 493 RPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFIFNG 552
RPGC +LT+++ E +W ++ D + L + + G + + + + + + G
Sbjct: 359 RPGCTILTVFIAMPEIMWAKLSKDPVAYLDEFILKPGKMLFGRGSMTVYLNNMIFRLIKG 418
Query: 553 QVV---IDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYL 609
+D L K+ V P A K ++ V G N+++P R + + GKYL
Sbjct: 419 GTTLKRVDVKL-----ESPKLQFVYPTCFEAGKPIELVVCGQNLLQPKCRFLVSFSGKYL 473
Query: 610 VCEDAHESMDQYSEELDG-------LHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIV 662
H + + DG + I + P G F+E+E++ S+F P I+
Sbjct: 474 ----PHNYSVVPAPDQDGKRSCNNKFYKINIVNSDPSLFGPAFVEVENESGLSNFIPLII 529
Query: 663 AEEDVCSDI 671
+ VCS++
Sbjct: 530 GDAAVCSEM 538
Score = 101 bits (251), Expect = 2e-21
Identities = 47/89 (52%), Positives = 62/89 (68%), Gaps = 2/89 (2%)
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
KK RV GG S A CQV DC AD+S K YH+RH+VC + A+ ++ +R+CQQC
Sbjct: 125 KKKRVRGG--SGVARCQVPDCEADISELKGYHKRHRVCLRCATASFVVLDGENKRYCQQC 182
Query: 194 SRFHMLQEFDEGKRSCRRRLAGHNKRRRK 222
+FH+L +FDEGKRSCRR+L HN RR++
Sbjct: 183 GKFHLLPDFDEGKRSCRRKLERHNNRRKR 211
>At1g27370 squamosa-promoter binding protein-like 10
Length = 396
Score = 108 bits (271), Expect = 1e-23
Identities = 51/91 (56%), Positives = 64/91 (70%), Gaps = 7/91 (7%)
Query: 149 CQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRS 208
CQ++ C DLS +KDYHR+H+VCE HSK + +V +RFCQQCSRFH + EFDE KRS
Sbjct: 176 CQIDGCELDLSSSKDYHRKHRVCETHSKCPKVVVSGLERRFCQQCSRFHAVSEFDEKKRS 235
Query: 209 CRRRLAGHNKRRRKTNNEAVPNG-NPLNDDQ 238
CR+RL+ HN RRRK P G PLN ++
Sbjct: 236 CRKRLSHHNARRRK------PQGVFPLNSER 260
>At1g27360 squamosa-promoter binding protein-like 11
Length = 393
Score = 107 bits (266), Expect = 4e-23
Identities = 47/74 (63%), Positives = 55/74 (73%)
Query: 149 CQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRS 208
CQ++ C DLS AK YHR+HKVCE HSK + V +RFCQQCSRFH + EFDE KRS
Sbjct: 175 CQIDGCELDLSSAKGYHRKHKVCEKHSKCPKVSVSGLERRFCQQCSRFHAVSEFDEKKRS 234
Query: 209 CRRRLAGHNKRRRK 222
CR+RL+ HN RRRK
Sbjct: 235 CRKRLSHHNARRRK 248
>At5g60070 ankyrin-like protein
Length = 548
Score = 38.1 bits (87), Expect = 0.025
Identities = 49/184 (26%), Positives = 72/184 (38%), Gaps = 24/184 (13%)
Query: 766 DHPSLDLA--LSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALV-------- 815
+HP L + LS LH A + ++VE LL +++ + GK +
Sbjct: 130 EHPELSMTVDLSNTTALHTAAAQGHVEVVEYLLEAAGSSLAAIAKSNGKTALHSAARNGH 189
Query: 816 -EGEKQSFLFRPDAA---GPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDS 871
E K PD A G TPLH+A K S DV+ L M G + N DS
Sbjct: 190 AEVVKAIVAVEPDTATRTDKKGQTPLHMAV-KGQSIDVVVEL-----MKGHRSSLNMADS 243
Query: 872 TGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEI 931
G+T A +G I +V+ ++ + +P T DT +K +
Sbjct: 244 KGNTALHVATRKGR---IKIVELLLDNNETSPSTKAINRAGETPLDTAEKTGHPQIA-AV 299
Query: 932 AKTR 935
KTR
Sbjct: 300 LKTR 303
>At5g59020 putative protein
Length = 780
Score = 35.4 bits (80), Expect = 0.16
Identities = 49/207 (23%), Positives = 82/207 (38%), Gaps = 21/207 (10%)
Query: 89 REGERKRRVIV-LEDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKKSRVAGGGASNRA 147
R RK RV+ +E D + L G A+ ++ S D K S
Sbjct: 224 RSSNRKVRVVHGVEGDYCTQHSCPLPCNADGCLAESKLGSTDADQKKVS----------- 272
Query: 148 VCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQ-QCSRFHMLQEFDEGK 206
++ C + L++A++ + K+ E + + + V + M CQ Q S+ H +
Sbjct: 273 -VELSQCVSLLTKARNKSSKGKISEDRASSLLS-VKHCMYEPCQRQDSKTHKVTSEKGRS 330
Query: 207 RSCRRRLAGHNKRRRKTNNEAVPNGNPLNDDQTSSYLL----ISLLKIL--SNMNSDRSD 260
S +RL+ + + KTN+E D T+S + ++LL + SN N
Sbjct: 331 ISPFQRLSFNMGKASKTNSEGGTVPTTQLDSMTNSTKIDSQNVALLSDVDGSNCNKPSKK 390
Query: 261 QTTDQDLLTHLLRSLASRNGEQGGNNL 287
TT L LL L GN++
Sbjct: 391 DTTTTSHLRRLLEPLLKPRAANSGNSV 417
>At4g22200 potassium channel (AKT2)
Length = 802
Score = 33.9 bits (76), Expect = 0.47
Identities = 29/97 (29%), Positives = 41/97 (41%), Gaps = 10/97 (10%)
Query: 799 VPENISNE---VRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTN 855
VP NI++ V G A + E PD G TPLH+AA + G ED + L
Sbjct: 537 VPPNIASNLIAVVTTGNAALLDELLKAKLSPDITDSKGKTPLHVAASR-GYEDCVLVLLK 595
Query: 856 DPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLV 892
C + I RD G++ A + HY ++
Sbjct: 596 HGCNIHI------RDVNGNSALWEAIISKHYEIFRIL 626
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,520,592
Number of Sequences: 26719
Number of extensions: 1066094
Number of successful extensions: 3075
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 2998
Number of HSP's gapped (non-prelim): 63
length of query: 1003
length of database: 11,318,596
effective HSP length: 109
effective length of query: 894
effective length of database: 8,406,225
effective search space: 7515165150
effective search space used: 7515165150
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0158.4


