

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.2
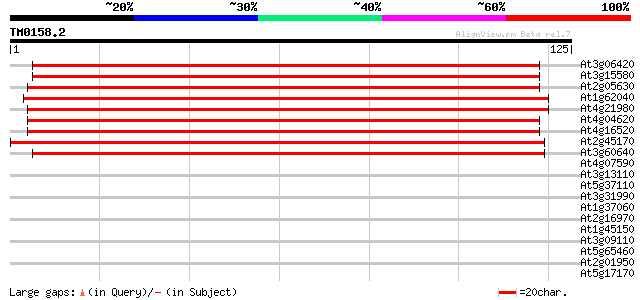
(125 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g06420 unknown protein 175 6e-45
At3g15580 unknown protein 173 2e-44
At2g05630 putative microtubule-associated protein 137 1e-33
At1g62040 unknown protein 137 2e-33
At4g21980 symbiosis-related like protein 136 3e-33
At4g04620 putative symbiosis-related protein 133 3e-32
At4g16520 symbiosis-related like protein 127 1e-30
At2g45170 putative microtubule-associated protein 127 2e-30
At3g60640 unknown protein 125 5e-30
At4g07590 30 0.31
At3g13110 serine acetyltransferase (Sat-1) 28 0.91
At5g37110 putative helicase 28 1.5
At3g31990 hypothetical protein 28 1.5
At1g37060 Athila retroelment ORF 1, putative 27 2.0
At2g16970 putative tetracycline transporter protein 27 2.6
At1g45150 unknown protein 27 2.6
At3g09110 hypothetical protein 27 3.4
At5g65460 putative protein 26 4.5
At2g01950 putative receptor protein kinase 26 4.5
At5g17170 unknown protein 26 5.9
>At3g06420 unknown protein
Length = 119
Score = 175 bits (443), Expect = 6e-45
Identities = 80/113 (70%), Positives = 98/113 (85%)
Query: 6 SFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHI 65
SFKD+F+ D+RL+ES IIAKYPDR+PVI+E+Y +DLP++ K KYLVPRD++VGHFIH+
Sbjct: 7 SFKDQFSSDERLKESNNIIAKYPDRIPVIIEKYSNADLPDMEKNKYLVPRDMTVGHFIHM 66
Query: 66 LSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
LS R+ L P KALFVFV NTLPQTAS MDS+Y +F++EDGFLYM YSTEKTFG
Sbjct: 67 LSKRMQLDPSKALFVFVHNTLPQTASRMDSLYNTFKEEDGFLYMCYSTEKTFG 119
>At3g15580 unknown protein
Length = 115
Score = 173 bits (439), Expect = 2e-44
Identities = 80/113 (70%), Positives = 100/113 (87%)
Query: 6 SFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHI 65
SFK+++T D+RL ESREIIAKYP R+PVI E+Y K+DLP + K+K+LVPRD+SVG FI+I
Sbjct: 3 SFKEQYTLDERLAESREIIAKYPTRIPVIAEKYCKTDLPAIEKKKFLVPRDMSVGQFIYI 62
Query: 66 LSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
LS+RL L PGKALFVFV NTLPQTA++MDSVY S++D+DGF+YM YS+EKTFG
Sbjct: 63 LSARLHLSPGKALFVFVNNTLPQTAALMDSVYESYKDDDGFVYMCYSSEKTFG 115
>At2g05630 putative microtubule-associated protein
Length = 120
Score = 137 bits (346), Expect = 1e-33
Identities = 63/114 (55%), Positives = 85/114 (74%)
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYPDR+PVIVER KSD+P++ ++KYLVP DL+VG F++
Sbjct: 4 SSFKHEHPLEKRQAEAARIREKYPDRIPVIVERAEKSDVPDIDRKKYLVPADLTVGQFVY 63
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
++ R+ L P KA+F+FVKN LP TA++M ++Y +DEDGFLYM YS E TFG
Sbjct: 64 VVRKRIKLSPEKAIFIFVKNILPPTAAIMSAIYEEHKDEDGFLYMSYSGENTFG 117
>At1g62040 unknown protein
Length = 119
Score = 137 bits (344), Expect = 2e-33
Identities = 67/117 (57%), Positives = 86/117 (73%)
Query: 4 SSSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFI 63
+SSFK E ++R ES I KYPDR+PVIVER +SD+P + K+KYLVP DL+VG F+
Sbjct: 3 NSSFKLEHPLERRQIESSRIREKYPDRIPVIVERAERSDVPNIDKKKYLVPADLTVGQFV 62
Query: 64 HILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
+++ R+ L KA+FVFVKNTLP TA+MM ++Y +DEDGFLYM YS E TFG V
Sbjct: 63 YVVRKRIKLSAEKAIFVFVKNTLPPTAAMMSAIYDENKDEDGFLYMTYSGENTFGLV 119
>At4g21980 symbiosis-related like protein
Length = 122
Score = 136 bits (342), Expect = 3e-33
Identities = 64/116 (55%), Positives = 86/116 (73%)
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK + R+ ES I KYPDR+PVIVE+ +SD+P++ K+KYLVP DL+VG F++
Sbjct: 4 SSFKISNPLEARMSESSRIREKYPDRIPVIVEKAGQSDVPDIDKKKYLVPADLTVGQFVY 63
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L KA+FVFVKNTLP TA++M ++Y +DEDGFLYM YS E TFGS+
Sbjct: 64 VVRKRIKLGAEKAIFVFVKNTLPPTAALMSAIYEEHKDEDGFLYMTYSGENTFGSL 119
>At4g04620 putative symbiosis-related protein
Length = 122
Score = 133 bits (334), Expect = 3e-32
Identities = 62/114 (54%), Positives = 85/114 (74%)
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
+SFK + R+ ES I AKYP+RVPVIVE+ +SD+P++ K+KYLVP DL++G F++
Sbjct: 4 NSFKLSNPLEMRMAESTRIRAKYPERVPVIVEKAGQSDVPDIDKKKYLVPADLTIGQFVY 63
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
++ R+ L KA+FVFVKNTLP TA++M ++Y +DEDGFLYM YS E TFG
Sbjct: 64 VVRKRIKLGAEKAIFVFVKNTLPPTAALMSAIYEEHKDEDGFLYMTYSGENTFG 117
>At4g16520 symbiosis-related like protein
Length = 121
Score = 127 bits (320), Expect = 1e-30
Identities = 59/114 (51%), Positives = 80/114 (69%)
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYPDR+PVIVE+ KSD+P + K+KYLVP DL+VG F++
Sbjct: 4 SSFKQEHDLEKRRAEAARIREKYPDRIPVIVEKAEKSDIPTIDKKKYLVPADLTVGQFVY 63
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
++ R+ L KA+F+FV N LP ++M SVY +D+DGFLY+ YS E TFG
Sbjct: 64 VIRKRIKLSAEKAIFIFVDNVLPPAGALMSSVYEEKKDDDGFLYVTYSGENTFG 117
>At2g45170 putative microtubule-associated protein
Length = 122
Score = 127 bits (318), Expect = 2e-30
Identities = 60/119 (50%), Positives = 82/119 (68%)
Query: 1 MGGSSSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVG 60
M S FK + F++R E+ I KYPDR+PVIVE+ KS++P + K+KYLVP DL+VG
Sbjct: 1 MNKGSIFKMDDDFEKRKAEAGRIREKYPDRIPVIVEKAEKSEVPNIDKKKYLVPSDLTVG 60
Query: 61 HFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGS 119
F++++ R+ L KA+F+FV N LP T +M SVY +DEDGFLY+ YS E TFG+
Sbjct: 61 QFVYVIRKRIKLSAEKAIFIFVDNVLPPTGELMSSVYEDKKDEDGFLYITYSGENTFGA 119
>At3g60640 unknown protein
Length = 121
Score = 125 bits (314), Expect = 5e-30
Identities = 58/114 (50%), Positives = 81/114 (70%)
Query: 6 SFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHI 65
SF+ + F++R E+ I KY DRVPVIVE+ KSD+P + K+KYLVP DL+VG F+++
Sbjct: 5 SFRQDHDFEKRKAEALRIREKYSDRVPVIVEKSEKSDIPNIDKKKYLVPADLTVGQFVYV 64
Query: 66 LSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGS 119
+ R+ L KA+F+FV N LP T +MM ++Y ++EDGFLY+ YS E TFGS
Sbjct: 65 IRKRIQLSAEKAIFIFVDNVLPPTGAMMSTIYDENKEEDGFLYVTYSGENTFGS 118
>At4g07590
Length = 860
Score = 30.0 bits (66), Expect = 0.31
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 5/68 (7%)
Query: 13 FDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKR----KYLVPRDLSVGHFIHILSS 68
FD + E + +I + D + +E Y +S + + G + K ++ +D VG + + +S
Sbjct: 93 FDIKTAEEKRLI-QLRDLDEIRLEAYERSKIYKEGTKLFHDKKIITKDFQVGDQVLLFNS 151
Query: 69 RLSLPPGK 76
RL L PGK
Sbjct: 152 RLKLFPGK 159
>At3g13110 serine acetyltransferase (Sat-1)
Length = 391
Score = 28.5 bits (62), Expect = 0.91
Identities = 22/76 (28%), Positives = 32/76 (41%), Gaps = 5/76 (6%)
Query: 39 LKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFV---KNTLPQTASMMDS 95
L+S P + +L+P +S H LS S PP + + + PQ + S
Sbjct: 17 LRSSSPHINHHSFLLPSFVSSKFKHHTLSPPPSPPPPPPMAACIDTCRTGKPQISPRDSS 76
Query: 96 VYRSFRDEDGFLYMYY 111
+ DE GF YM Y
Sbjct: 77 KHHD--DESGFRYMNY 90
>At5g37110 putative helicase
Length = 1307
Score = 27.7 bits (60), Expect = 1.5
Identities = 15/27 (55%), Positives = 17/27 (62%)
Query: 24 IAKYPDRVPVIVERYLKSDLPELGKRK 50
I K PDRV V+VE L S E GK+K
Sbjct: 623 IHKGPDRVTVVVESSLNSKNKENGKQK 649
Score = 25.8 bits (55), Expect = 5.9
Identities = 16/57 (28%), Positives = 31/57 (54%), Gaps = 11/57 (19%)
Query: 46 LGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRD 102
+G+ Y VPRD+ G+++ IL L++ PG F + ++ D +Y+ ++D
Sbjct: 731 IGRINY-VPRDIEDGYYLRIL---LNVQPGPRCF-------EELRTVNDVLYKEWKD 776
>At3g31990 hypothetical protein
Length = 450
Score = 27.7 bits (60), Expect = 1.5
Identities = 18/82 (21%), Positives = 39/82 (46%), Gaps = 8/82 (9%)
Query: 39 LKSDLPELGKRKYLVPRDLSVGHFIHILSSR-----LSLPPGKALFVFVKNTLPQTASMM 93
+ +D+PE+ + K +P+D G + ++ S+ LP G F K T+ + + M
Sbjct: 166 INTDIPEILEFKDALPKD---GLALTLIESKPKSEVAELPTGDFYLQFAKKTIKEVSEMF 222
Query: 94 DSVYRSFRDEDGFLYMYYSTEK 115
D + + + + + Y + +K
Sbjct: 223 DVLCTIYDIDRDWSWYYIACKK 244
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 27.3 bits (59), Expect = 2.0
Identities = 20/68 (29%), Positives = 34/68 (49%), Gaps = 5/68 (7%)
Query: 13 FDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKY----LVPRDLSVGHFIHILSS 68
FD + E + +I + D + +E Y S + + + + +V RD VG + + +S
Sbjct: 1609 FDIKTAEEKRLI-QLNDLNKIRLEAYESSKIYKERTKSFHDKKIVSRDFKVGDQVLLFNS 1667
Query: 69 RLSLPPGK 76
RL L PGK
Sbjct: 1668 RLRLFPGK 1675
>At2g16970 putative tetracycline transporter protein
Length = 414
Score = 26.9 bits (58), Expect = 2.6
Identities = 11/39 (28%), Positives = 19/39 (48%)
Query: 85 TLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSVHNI 123
TLP S++ ++R + F Y +Y T+ F N+
Sbjct: 85 TLPMCLSILPPAILAYRRDTNFFYAFYITKILFDMAKNV 123
>At1g45150 unknown protein
Length = 643
Score = 26.9 bits (58), Expect = 2.6
Identities = 21/59 (35%), Positives = 29/59 (48%), Gaps = 8/59 (13%)
Query: 29 DRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLP 87
DR+ V L+ +P LG LVP DL V S+LSL PG+ ++V +P
Sbjct: 113 DRLVVGQSLKLRRVVPVLGVPDALVPLDLPV--------SQLSLFPGETSVIWVSIDVP 163
>At3g09110 hypothetical protein
Length = 452
Score = 26.6 bits (57), Expect = 3.4
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query: 62 FIHILSSRLSLPPGKALFVFVKNTLPQTA--SMMDSVYRSFRDED 104
F+ +L S L+LP G + + K+ PQ++ + ++Y+S D D
Sbjct: 32 FVDVLCSLLTLPMGTIVRLLEKHQNPQSSVVGCLHNLYKSVADMD 76
>At5g65460 putative protein
Length = 1264
Score = 26.2 bits (56), Expect = 4.5
Identities = 28/103 (27%), Positives = 43/103 (41%), Gaps = 9/103 (8%)
Query: 11 FTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYL-VPRDLSVGHFIHILSSR 69
F+F +++E R + RV ++ R L + PEL K V R L + SS
Sbjct: 761 FSFIRKMEPRRVMDTMLVSRVRILYIRSLLARSPELQSIKVSPVERFLEKPYTGRTRSSS 820
Query: 70 LSLPPGKA--------LFVFVKNTLPQTASMMDSVYRSFRDED 104
S PG++ ++ F N P+ S + SV R D
Sbjct: 821 GSSSPGRSPVRYYDEQIYGFKVNLKPEKKSKLVSVVSRIRGHD 863
>At2g01950 putative receptor protein kinase
Length = 1143
Score = 26.2 bits (56), Expect = 4.5
Identities = 17/36 (47%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query: 44 PELGKRKYLVPRDLSVGHFIHILSSRLSLPPG-KAL 78
PELGK LV DL+ H + RL PG KAL
Sbjct: 513 PELGKCTTLVWLDLNTNHLTGEIPPRLGRQPGSKAL 548
>At5g17170 unknown protein
Length = 271
Score = 25.8 bits (55), Expect = 5.9
Identities = 16/46 (34%), Positives = 23/46 (49%)
Query: 4 SSSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKR 49
SS F DE +L ++ I PD V +V R + D+ +L KR
Sbjct: 132 SSFFGDELWPADKLGFTKTAIQAKPDSVYFVVSRGAEVDVKKLNKR 177
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,972,522
Number of Sequences: 26719
Number of extensions: 118281
Number of successful extensions: 275
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 253
Number of HSP's gapped (non-prelim): 30
length of query: 125
length of database: 11,318,596
effective HSP length: 87
effective length of query: 38
effective length of database: 8,994,043
effective search space: 341773634
effective search space used: 341773634
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0158.2


