

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.11
(361 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
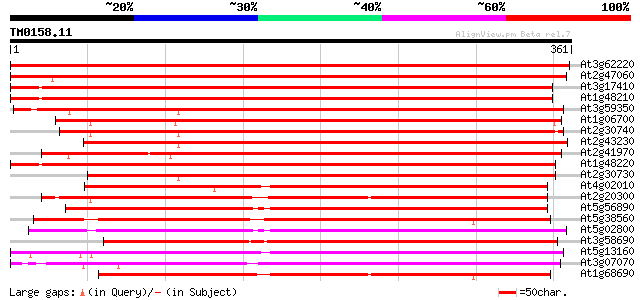
Score E
Sequences producing significant alignments: (bits) Value
At3g62220 serine/threonine protein kinase-like protein 604 e-173
At2g47060 putative protein kinase 596 e-171
At3g17410 protein kinase like protein 554 e-158
At1g48210 Pto kinase interactor 1, putative 530 e-151
At3g59350 protein kinase-like protein 491 e-139
At1g06700 protein kinase interactor, putative 486 e-138
At2g30740 putative protein kinase 482 e-136
At2g43230 protein kinase like protein 477 e-135
At2g41970 putative protein kinase 470 e-133
At1g48220 Pto kinase interactor 1, putative 463 e-131
At2g30730 putative protein kinase 444 e-125
At4g02010 putative NAK-like ser/thr protein kinase 286 2e-77
At2g20300 protein kinase like protein 271 3e-73
At5g56890 unknown protein 271 4e-73
At5g38560 putative protein 269 2e-72
At5g02800 protein kinase - like 267 8e-72
At3g58690 serine/threonine-specific protein kinase -like protein 266 1e-71
At5g13160 protein kinase-like 266 2e-71
At3g07070 putative protein kinase 265 3e-71
At1g68690 protein kinase, putative 262 2e-70
>At3g62220 serine/threonine protein kinase-like protein
Length = 361
Score = 604 bits (1557), Expect = e-173
Identities = 300/360 (83%), Positives = 326/360 (90%)
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
MSCF CC EDD + GG + K GNDG + SETA++G Q+VK QPIEV I D
Sbjct: 1 MSCFGCCREDDLPGANDYGGHNMTKQSGGNDGRRNGSETAQKGAQSVKVQPIEVAAILAD 60
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
EL E T++FG +SLIGEGSY RVY+GVLKNGQ AAIKKLD++KQP+EEFLAQVSMVSRLK
Sbjct: 61 ELIEATNDFGTNSLIGEGSYARVYHGVLKNGQRAAIKKLDSNKQPNEEFLAQVSMVSRLK 120
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
H NFV+LLGYSVDGNSRILV+EFA NGSLHDILHGRKGVKGA+PGP+L+W QRVKIAVGA
Sbjct: 121 HVNFVELLGYSVDGNSRILVFEFAQNGSLHDILHGRKGVKGAKPGPLLSWHQRVKIAVGA 180
Query: 181 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 240
ARGLEYLHEKA+PH+IHRDIKSSNVLIFD+DVAKIADFDLSNQAPDMAARLHSTRVLGTF
Sbjct: 181 ARGLEYLHEKANPHVIHRDIKSSNVLIFDNDVAKIADFDLSNQAPDMAARLHSTRVLGTF 240
Query: 241 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 300
GYHAPEYAMTGQL+AKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV
Sbjct: 241 GYHAPEYAMTGQLSAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 300
Query: 301 RQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPAGETA 360
+QCVD+RLGG+YPPKAVAK+AAVAALCVQYEADFRPNMSIVVKALQPLL AR GPAGE A
Sbjct: 301 KQCVDSRLGGDYPPKAVAKLAAVAALCVQYEADFRPNMSIVVKALQPLLNARTGPAGEGA 360
>At2g47060 putative protein kinase
Length = 365
Score = 596 bits (1537), Expect = e-171
Identities = 303/362 (83%), Positives = 322/362 (88%), Gaps = 4/362 (1%)
Query: 1 MSCFNCC-EEDDFHKTAESGGPYVVKN--PSGNDGNHH-ASETAKQGGQTVKPQPIEVPN 56
MSCF CC E+DD HKTA+ GG + P GND HH ASETA++G VK QPIEVP
Sbjct: 1 MSCFGCCGEDDDMHKTADYGGRHNQAKHFPPGNDARHHQASETAQKGPPVVKLQPIEVPI 60
Query: 57 ISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMV 116
I ELKE TD+FG +SLIGEGSYGRVYYGVL N +AIKKLD++KQPD EFLAQVSMV
Sbjct: 61 IPFSELKEATDDFGSNSLIGEGSYGRVYYGVLNNDLPSAIKKLDSNKQPDNEFLAQVSMV 120
Query: 117 SRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKI 176
SRLKH+NFVQLLGY VDGNSRIL YEFA+NGSLHDILHGRKGVKGAQPGPVL+W QRVKI
Sbjct: 121 SRLKHDNFVQLLGYCVDGNSRILSYEFANNGSLHDILHGRKGVKGAQPGPVLSWYQRVKI 180
Query: 177 AVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRV 236
AVGAARGLEYLHEKA+PHIIHRDIKSSNVL+F+DDVAKIADFDLSNQAPDMAARLHSTRV
Sbjct: 181 AVGAARGLEYLHEKANPHIIHRDIKSSNVLLFEDDVAKIADFDLSNQAPDMAARLHSTRV 240
Query: 237 LGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLS 296
LGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDH LPRGQQSLVTWATPKLS
Sbjct: 241 LGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHRLPRGQQSLVTWATPKLS 300
Query: 297 EDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPA 356
EDKV+QCVD RLGG+YPPKAVAK+AAVAALCVQYEADFRPNMSIVVKALQPLL AR
Sbjct: 301 EDKVKQCVDARLGGDYPPKAVAKLAAVAALCVQYEADFRPNMSIVVKALQPLLNARAVAP 360
Query: 357 GE 358
GE
Sbjct: 361 GE 362
>At3g17410 protein kinase like protein
Length = 364
Score = 554 bits (1427), Expect = e-158
Identities = 275/350 (78%), Positives = 304/350 (86%), Gaps = 2/350 (0%)
Query: 1 MSCFNCCEE-DDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISE 59
M CF CC +DF + +E+G P V N G +G HH + ++ QPI V I
Sbjct: 1 MGCFGCCGGGEDFRRVSETG-PKPVHNTGGYNGGHHQRADPPKNLPVIQMQPISVAAIPA 59
Query: 60 DELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRL 119
DEL+++TDN+G SLIGEGSYGRV+YG+LK+G+AAAIKKLD+SKQPD+EFLAQVSMVSRL
Sbjct: 60 DELRDITDNYGSKSLIGEGSYGRVFYGILKSGKAAAIKKLDSSKQPDQEFLAQVSMVSRL 119
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
+ EN V LLGY VDG R+L YE+A NGSLHDILHGRKGVKGAQPGPVL+W QRVKIAVG
Sbjct: 120 RQENVVALLGYCVDGPLRVLAYEYAPNGSLHDILHGRKGVKGAQPGPVLSWHQRVKIAVG 179
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
AARGLEYLHEKA+PH+IHRDIKSSNVL+FDDDVAKIADFDLSNQAPDMAARLHSTRVLGT
Sbjct: 180 AARGLEYLHEKANPHVIHRDIKSSNVLLFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDK 299
FGYHAPEYAMTG L+ KSDVYSFGVVLLELLTGRKPVDHTLPRGQQS+VTWATPKLSEDK
Sbjct: 240 FGYHAPEYAMTGTLSTKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSVVTWATPKLSEDK 299
Query: 300 VRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLL 349
V+QCVD RL GEYPPKAVAK+AAVAALCVQYEADFRPNMSIVVKALQPLL
Sbjct: 300 VKQCVDARLNGEYPPKAVAKLAAVAALCVQYEADFRPNMSIVVKALQPLL 349
>At1g48210 Pto kinase interactor 1, putative
Length = 363
Score = 530 bits (1364), Expect = e-151
Identities = 260/349 (74%), Positives = 292/349 (83%), Gaps = 1/349 (0%)
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
MSCF C +DF ++G P NP+G +G H+ + QPI VP I D
Sbjct: 1 MSCFGWCGSEDFRNATDTG-PRPAHNPAGYNGGHYQRADPPMNQPVIPMQPISVPAIPVD 59
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
EL+++TDN+G +LIGEGSYGRV+YGVLK+G AAAIKKLD+SKQPD+EFL+Q+SMVSRL+
Sbjct: 60 ELRDITDNYGSKTLIGEGSYGRVFYGVLKSGGAAAIKKLDSSKQPDQEFLSQISMVSRLR 119
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
H+N L+GY VDG R+L YEFA GSLHD LHG+KG KGA GPV+TW QRVKIAVGA
Sbjct: 120 HDNVTALMGYCVDGPLRVLAYEFAPKGSLHDTLHGKKGAKGALRGPVMTWQQRVKIAVGA 179
Query: 181 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 240
ARGLEYLHEK P +IHRDIKSSNVL+FDDDVAKI DFDLS+QAPDMAARLHSTRVLGTF
Sbjct: 180 ARGLEYLHEKVSPQVIHRDIKSSNVLLFDDDVAKIGDFDLSDQAPDMAARLHSTRVLGTF 239
Query: 241 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 300
GYHAPEYAMTG L++KSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV
Sbjct: 240 GYHAPEYAMTGTLSSKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 299
Query: 301 RQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLL 349
+QCVD RL GEYPPKAV K+AAVAALCVQYEA+FRPNMSIVVKALQPLL
Sbjct: 300 KQCVDARLLGEYPPKAVGKLAAVAALCVQYEANFRPNMSIVVKALQPLL 348
>At3g59350 protein kinase-like protein
Length = 408
Score = 491 bits (1264), Expect = e-139
Identities = 250/358 (69%), Positives = 286/358 (79%), Gaps = 7/358 (1%)
Query: 3 CFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHAS--ETAKQGGQTVKPQPIEVPNISED 60
C C E+ +H S ++ ND HH + A + +P I+VP +S D
Sbjct: 48 CCACHVEEPYHS---SENEHLRSPKHHNDFGHHTRKPQAAVKPDALKEPPSIDVPALSLD 104
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE--EFLAQVSMVSR 118
ELKE TDNFG SLIGEGSYGR YY LK+G+A A+KKLD + +P+ EFL QVS VS+
Sbjct: 105 ELKEKTDNFGSKSLIGEGSYGRAYYATLKDGKAVAVKKLDNAAEPESNVEFLTQVSRVSK 164
Query: 119 LKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAV 178
LKH+NFV+L GY V+GN RIL YEFA+ GSLHDILHGRKGV+GAQPGP L W QRV+IAV
Sbjct: 165 LKHDNFVELFGYCVEGNFRILAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAV 224
Query: 179 GAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLG 238
AARGLEYLHEK P +IHRDI+SSNVL+F+D AKIADF+LSNQ+PDMAARLHSTRVLG
Sbjct: 225 DAARGLEYLHEKVQPAVIHRDIRSSNVLLFEDFKAKIADFNLSNQSPDMAARLHSTRVLG 284
Query: 239 TFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSED 298
TFGYHAPEYAMTGQL KSDVYSFGVVLLELLTGRKPVDHT+PRGQQSLVTWATP+LSED
Sbjct: 285 TFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSED 344
Query: 299 KVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPA 356
KV+QCVD +L GEYPPKAVAK+AAVAALCVQYE++FRPNMSIVVKALQPLL + A
Sbjct: 345 KVKQCVDPKLKGEYPPKAVAKLAAVAALCVQYESEFRPNMSIVVKALQPLLRSSTAAA 402
>At1g06700 protein kinase interactor, putative
Length = 361
Score = 486 bits (1251), Expect = e-138
Identities = 247/333 (74%), Positives = 278/333 (83%), Gaps = 7/333 (2%)
Query: 30 NDGNHHASETAKQGGQTVKPQ--PIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGV 87
+D NH S+ A VK + PIEVP +S DE+KE T+NFG +LIGEGSYGRVYY
Sbjct: 27 SDANHKNSKPAPVAKHEVKKEALPIEVPPLSLDEVKEKTENFGSKALIGEGSYGRVYYAT 86
Query: 88 LKNGQAAAIKKLDASKQP--DEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFAS 145
L +G A A+KKLD + + D EFL+QVSMVSRLKHEN +QLLG+ VDGN R+L YEFA+
Sbjct: 87 LNDGVAVALKKLDVAPEAETDTEFLSQVSMVSRLKHENLIQLLGFCVDGNLRVLAYEFAT 146
Query: 146 NGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNV 205
GSLHDILHGRKGV+GAQPGP L W RVKIAV AARGLEYLHEK+ P +IHRDI+SSNV
Sbjct: 147 MGSLHDILHGRKGVQGAQPGPTLDWITRVKIAVEAARGLEYLHEKSQPPVIHRDIRSSNV 206
Query: 206 LIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVV 265
L+F+D AKIADF+LSNQAPD AARLHSTRVLGTFGYHAPEYAMTGQL KSDVYSFGVV
Sbjct: 207 LLFEDYKAKIADFNLSNQAPDNAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVV 266
Query: 266 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 325
LLELLTGRKPVDHT+PRGQQSLVTWATP+LSEDKV+QC+D +L +YPPKAVAK+AAVAA
Sbjct: 267 LLELLTGRKPVDHTMPRGQQSLVTWATPRLSEDKVKQCIDPKLKADYPPKAVAKLAAVAA 326
Query: 326 LCVQYEADFRPNMSIVVKALQPLL---TARPGP 355
LCVQYEA+FRPNMSIVVKALQPLL A P P
Sbjct: 327 LCVQYEAEFRPNMSIVVKALQPLLKPPAAAPAP 359
>At2g30740 putative protein kinase
Length = 366
Score = 482 bits (1241), Expect = e-136
Identities = 245/332 (73%), Positives = 277/332 (82%), Gaps = 9/332 (2%)
Query: 33 NHHASETAKQGGQTVKPQ------PIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYG 86
N A++ ++ VKP+ PIEVP +S DE+KE TDNFG SLIGEGSYGRVYY
Sbjct: 29 NSEANQKNQKPQAVVKPEAQKEALPIEVPPLSVDEVKEKTDNFGSKSLIGEGSYGRVYYA 88
Query: 87 VLKNGQAAAIKKLDASKQPDE--EFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFA 144
L +G+A A+KKLD + + + EFL QVSMVSRLKHEN +QL+GY VD N R+L YEFA
Sbjct: 89 TLNDGKAVALKKLDVAPEAETNTEFLNQVSMVSRLKHENLIQLVGYCVDENLRVLAYEFA 148
Query: 145 SNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSN 204
+ GSLHDILHGRKGV+GAQPGP L W RVKIAV AARGLEYLHEK P +IHRDI+SSN
Sbjct: 149 TMGSLHDILHGRKGVQGAQPGPTLDWLTRVKIAVEAARGLEYLHEKVQPPVIHRDIRSSN 208
Query: 205 VLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGV 264
VL+F+D AK+ADF+LSNQAPD AARLHSTRVLGTFGYHAPEYAMTGQL KSDVYSFGV
Sbjct: 209 VLLFEDYQAKVADFNLSNQAPDNAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGV 268
Query: 265 VLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVA 324
VLLELLTGRKPVDHT+PRGQQSLVTWATP+LSEDKV+QCVD +L GEYPPK+VAK+AAVA
Sbjct: 269 VLLELLTGRKPVDHTMPRGQQSLVTWATPRLSEDKVKQCVDPKLKGEYPPKSVAKLAAVA 328
Query: 325 ALCVQYEADFRPNMSIVVKALQPLLTARPGPA 356
ALCVQYE++FRPNMSIVVKALQPLL P PA
Sbjct: 329 ALCVQYESEFRPNMSIVVKALQPLLKP-PAPA 359
>At2g43230 protein kinase like protein
Length = 406
Score = 477 bits (1227), Expect = e-135
Identities = 239/314 (76%), Positives = 268/314 (85%), Gaps = 2/314 (0%)
Query: 48 KPQPIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE 107
+P PI+VP +S ELKE T NFG +LIGEGSYGRVYY +G+A A+KKLD + +P+
Sbjct: 90 EPPPIDVPAMSLVELKEKTQNFGSKALIGEGSYGRVYYANFNDGKAVAVKKLDNASEPET 149
Query: 108 --EFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPG 165
EFL QVS VSRLK +NFVQLLGY V+GN R+L YEFA+ SLHDILHGRKGV+GAQPG
Sbjct: 150 NVEFLTQVSKVSRLKSDNFVQLLGYCVEGNLRVLAYEFATMRSLHDILHGRKGVQGAQPG 209
Query: 166 PVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAP 225
P L W QRV++AV AA+GLEYLHEK P +IHRDI+SSNVLIF+D AKIADF+LSNQAP
Sbjct: 210 PTLEWMQRVRVAVDAAKGLEYLHEKVQPAVIHRDIRSSNVLIFEDFKAKIADFNLSNQAP 269
Query: 226 DMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQ 285
DMAARLHSTRVLGTFGYHAPEYAMTGQL KSDVYSFGVVLLELLTGRKPVDHT+PRGQQ
Sbjct: 270 DMAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQ 329
Query: 286 SLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKAL 345
SLVTWATP+LSEDKV+QCVD +L GEYPPKAVAK+AAVAALCVQYEA+FRPNMSIVVKAL
Sbjct: 330 SLVTWATPRLSEDKVKQCVDPKLKGEYPPKAVAKLAAVAALCVQYEAEFRPNMSIVVKAL 389
Query: 346 QPLLTARPGPAGET 359
QPLL + A T
Sbjct: 390 QPLLRSATAAAPPT 403
>At2g41970 putative protein kinase
Length = 365
Score = 470 bits (1209), Expect = e-133
Identities = 231/340 (67%), Positives = 283/340 (82%), Gaps = 6/340 (1%)
Query: 21 PYVVKNPSGNDGNHHA--SETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQDSLIGEG 78
P NP+ GN + A + G K PIE+P+++ DEL + NFG +LIGEG
Sbjct: 23 PNKAGNPNFGGGNRGEPRNPNAPRSGAPAKVLPIEIPSVALDELNRMAGNFGNKALIGEG 82
Query: 79 SYGRVYYGVLKNGQAAAIKKLDAS--KQPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNS 136
SYGRV+ G K G+A AIKKLDAS ++PD +F +Q+S+VSRLKH++FV+LLGY ++ N+
Sbjct: 83 SYGRVFCGKFK-GEAVAIKKLDASSSEEPDSDFTSQLSVVSRLKHDHFVELLGYCLEANN 141
Query: 137 RILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHII 196
RIL+Y+FA+ GSLHD+LHGRKGV+GA+PGPVL W QRVKIA GAA+GLE+LHEK P I+
Sbjct: 142 RILIYQFATKGSLHDVLHGRKGVQGAEPGPVLNWNQRVKIAYGAAKGLEFLHEKVQPPIV 201
Query: 197 HRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAK 256
HRD++SSNVL+FDD VAK+ADF+L+N + D AARLHSTRVLGTFGYHAPEYAMTGQ+ K
Sbjct: 202 HRDVRSSNVLLFDDFVAKMADFNLTNASSDTAARLHSTRVLGTFGYHAPEYAMTGQITQK 261
Query: 257 SDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKA 316
SDVYSFGVVLLELLTGRKPVDHT+P+GQQSLVTWATP+LSEDKV+QC+D +L ++PPKA
Sbjct: 262 SDVYSFGVVLLELLTGRKPVDHTMPKGQQSLVTWATPRLSEDKVKQCIDPKLNNDFPPKA 321
Query: 317 VAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARP-GP 355
VAK+AAVAALCVQYEADFRPNM+IVVKALQPLL ++P GP
Sbjct: 322 VAKLAAVAALCVQYEADFRPNMTIVVKALQPLLNSKPAGP 361
>At1g48220 Pto kinase interactor 1, putative
Length = 364
Score = 463 bits (1191), Expect = e-131
Identities = 231/352 (65%), Positives = 276/352 (77%), Gaps = 2/352 (0%)
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
MSCF C +D A++G P N G +G HH V QPI VP I D
Sbjct: 1 MSCFGWCGSEDVRNPADTG-PSQAHNSIGYNGRHHQRADPPMNQPVVNMQPIAVPAIPVD 59
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
EL+++T+NF + L+G+GSYGRV+YGVLK+G+ AAIKKL +KQPD+EFL+QVSMVSRL
Sbjct: 60 ELEDITENFSSEVLVGKGSYGRVFYGVLKSGKEAAIKKLYPTKQPDQEFLSQVSMVSRLH 119
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
HEN V L+ Y VDG R+L YEFA+ G+LHD+LHG+ GV GA GPV+TW +RVKIA+GA
Sbjct: 120 HENVVALMAYCVDGPLRVLAYEFATYGTLHDVLHGQTGVIGALQGPVMTWQRRVKIALGA 179
Query: 181 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRV-LGT 239
ARGLEYLH+K +P +IHRDIK+SN+L+FDDD+AKI DFDL +QAP+MA RLHS R+ LG
Sbjct: 180 ARGLEYLHKKVNPQVIHRDIKASNILLFDDDIAKIGDFDLYDQAPNMAGRLHSCRMALGA 239
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDK 299
H PE+AMTG L KSDVYSFGVVLLELLTGRKPVD TLPRGQQ+LVTWATPKLS+DK
Sbjct: 240 SRSHCPEHAMTGILTTKSDVYSFGVVLLELLTGRKPVDRTLPRGQQNLVTWATPKLSKDK 299
Query: 300 VRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTA 351
V+QCVD RL GEYPPKAVAK+AAV+A CV Y+ DFRP+MSIVVKALQPLL +
Sbjct: 300 VKQCVDARLLGEYPPKAVAKLAAVSARCVHYDPDFRPDMSIVVKALQPLLNS 351
>At2g30730 putative protein kinase
Length = 338
Score = 444 bits (1142), Expect = e-125
Identities = 222/303 (73%), Positives = 255/303 (83%), Gaps = 2/303 (0%)
Query: 51 PIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE--E 108
PI VP++S DE+ E TDNFG +SLIGEGSYGRVYY L +G+A A+KKLD + + + E
Sbjct: 29 PIIVPSLSVDEVNEQTDNFGPNSLIGEGSYGRVYYATLNDGKAVALKKLDLAPEDETNTE 88
Query: 109 FLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVL 168
FL+QVSMVSRLKHEN +QL+GY VD N R+L YEFA+ GSLHDILHGRKGV+ A PGP L
Sbjct: 89 FLSQVSMVSRLKHENLIQLVGYCVDENLRVLAYEFATMGSLHDILHGRKGVQDALPGPTL 148
Query: 169 TWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMA 228
W RVKIAV AARGLEYLHEK P +IHRDI+SSN+L+FDD AKIADF+LSNQ+PD A
Sbjct: 149 DWITRVKIAVEAARGLEYLHEKVQPQVIHRDIRSSNILLFDDYQAKIADFNLSNQSPDNA 208
Query: 229 ARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLV 288
ARL STRVLG+FGY++PEYAMTG+L KSDVY FGVVLLELLTGRKPVDHT+PRGQQSLV
Sbjct: 209 ARLQSTRVLGSFGYYSPEYAMTGELTHKSDVYGFGVVLLELLTGRKPVDHTMPRGQQSLV 268
Query: 289 TWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPL 348
TWATPKLSED V +CVD +L GEY PK+VAK+AAVAALCVQYE++ RP MS VVKALQ L
Sbjct: 269 TWATPKLSEDTVEECVDPKLKGEYSPKSVAKLAAVAALCVQYESNCRPKMSTVVKALQQL 328
Query: 349 LTA 351
L A
Sbjct: 329 LIA 331
>At4g02010 putative NAK-like ser/thr protein kinase
Length = 725
Score = 286 bits (731), Expect = 2e-77
Identities = 154/302 (50%), Positives = 202/302 (65%), Gaps = 9/302 (2%)
Query: 49 PQPIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS-KQPDE 107
P P +S +ELKE T NF S++GEG +G+VY G+L +G A AIKKL + Q D+
Sbjct: 360 PHPASTRFLSYEELKEATSNFESASILGEGGFGKVYRGILADGTAVAIKKLTSGGPQGDK 419
Query: 108 EFLAQVSMVSRLKHENFVQLLGY--SVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPG 165
EF ++ M+SRL H N V+L+GY S D + +L YE NGSL LHG G+
Sbjct: 420 EFQVEIDMLSRLHHRNLVKLVGYYSSRDSSQHLLCYELVPNGSLEAWLHGPLGLNCP--- 476
Query: 166 PVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAP 225
L W R+KIA+ AARGL YLHE + P +IHRD K+SN+L+ ++ AK+ADF L+ QAP
Sbjct: 477 --LDWDTRMKIALDAARGLAYLHEDSQPSVIHRDFKASNILLENNFNAKVADFGLAKQAP 534
Query: 226 DMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQ 285
+ STRV+GTFGY APEYAMTG L KSDVYS+GVVLLELLTGRKPVD + P GQ+
Sbjct: 535 EGRGNHLSTRVMGTFGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRKPVDMSQPSGQE 594
Query: 286 SLVTWATPKL-SEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKA 344
+LVTW P L +D++ + VD+RL G+YP + ++ +AA CV EA RP M VV++
Sbjct: 595 NLVTWTRPVLRDKDRLEELVDSRLEGKYPKEDFIRVCTIAAACVAPEASQRPTMGEVVQS 654
Query: 345 LQ 346
L+
Sbjct: 655 LK 656
>At2g20300 protein kinase like protein
Length = 744
Score = 271 bits (694), Expect = 3e-73
Identities = 150/332 (45%), Positives = 207/332 (62%), Gaps = 19/332 (5%)
Query: 21 PYVVKNPSGNDGNHHASETAKQGGQTVKPQ----PIEVPNISEDELKEVTDNFGQDSLIG 76
P + K P G+ +S G ++ + V + EL++ TD F ++G
Sbjct: 299 PSINKRPGA--GSMFSSSARSSGSDSLMSSMATCALSVKTFTLSELEKATDRFSAKRVLG 356
Query: 77 EGSYGRVYYGVLKNGQAAAIKKLDASKQP-DEEFLAQVSMVSRLKHENFVQLLGYSVDGN 135
EG +GRVY G +++G A+K L Q D EF+A+V M+SRL H N V+L+G ++G
Sbjct: 357 EGGFGRVYQGSMEDGTEVAVKLLTRDNQNRDREFIAEVEMLSRLHHRNLVKLIGICIEGR 416
Query: 136 SRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHI 195
+R L+YE NGS+ LH L W R+KIA+GAARGL YLHE ++P +
Sbjct: 417 TRCLIYELVHNGSVESHLHEG----------TLDWDARLKIALGAARGLAYLHEDSNPRV 466
Query: 196 IHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNA 255
IHRD K+SNVL+ DD K++DF L+ +A + + + STRV+GTFGY APEYAMTG L
Sbjct: 467 IHRDFKASNVLLEDDFTPKVSDFGLAREATEGSQHI-STRVMGTFGYVAPEYAMTGHLLV 525
Query: 256 KSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKL-SEDKVRQCVDTRLGGEYPP 314
KSDVYS+GVVLLELLTGR+PVD + P G+++LVTWA P L + + + Q VD L G Y
Sbjct: 526 KSDVYSYGVVLLELLTGRRPVDMSQPSGEENLVTWARPLLANREGLEQLVDPALAGTYNF 585
Query: 315 KAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
+AK+AA+A++CV E RP M VV+AL+
Sbjct: 586 DDMAKVAAIASMCVHQEVSHRPFMGEVVQALK 617
>At5g56890 unknown protein
Length = 1113
Score = 271 bits (693), Expect = 4e-73
Identities = 150/313 (47%), Positives = 200/313 (62%), Gaps = 8/313 (2%)
Query: 37 SETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAI 96
S T+ ++ P + + E+ + T+NF + ++GEG +GRVY GV +G A+
Sbjct: 691 SSTSLSFESSIAPFTLSAKTFTASEIMKATNNFDESRVLGEGGFGRVYEGVFDDGTKVAV 750
Query: 97 KKLDAS-KQPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHG 155
K L +Q EFLA+V M+SRL H N V L+G ++ +R LVYE NGS+ LHG
Sbjct: 751 KVLKRDDQQGSREFLAEVEMLSRLHHRNLVNLIGICIEDRNRSLVYELIPNGSVESHLHG 810
Query: 156 RKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKI 215
K + P L W R+KIA+GAARGL YLHE + P +IHRD KSSN+L+ +D K+
Sbjct: 811 ID--KASSP---LDWDARLKIALGAARGLAYLHEDSSPRVIHRDFKSSNILLENDFTPKV 865
Query: 216 ADFDLSNQAPDMAARLH-STRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRK 274
+DF L+ A D H STRV+GTFGY APEYAMTG L KSDVYS+GVVLLELLTGRK
Sbjct: 866 SDFGLARNALDDEDNRHISTRVMGTFGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRK 925
Query: 275 PVDHTLPRGQQSLVTWATPKL-SEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEAD 333
PVD + P GQ++LV+W P L S + + +D LG E ++AK+AA+A++CVQ E
Sbjct: 926 PVDMSQPPGQENLVSWTRPFLTSAEGLAAIIDQSLGPEISFDSIAKVAAIASMCVQPEVS 985
Query: 334 FRPNMSIVVKALQ 346
RP M VV+AL+
Sbjct: 986 HRPFMGEVVQALK 998
>At5g38560 putative protein
Length = 681
Score = 269 bits (687), Expect = 2e-72
Identities = 150/340 (44%), Positives = 211/340 (61%), Gaps = 23/340 (6%)
Query: 16 AESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQDSLI 75
+ S P +++ SG+D + +S++ Q S DEL +VT F + +L+
Sbjct: 294 SRSSAPPKMRSHSGSDYMYASSDSGMVSNQRSW--------FSYDELSQVTSGFSEKNLL 345
Query: 76 GEGSYGRVYYGVLKNGQAAAIKKLD-ASKQPDEEFLAQVSMVSRLKHENFVQLLGYSVDG 134
GEG +G VY GVL +G+ A+K+L Q + EF A+V ++SR+ H + V L+GY +
Sbjct: 346 GEGGFGCVYKGVLSDGREVAVKQLKIGGSQGEREFKAEVEIISRVHHRHLVTLVGYCISE 405
Query: 135 NSRILVYEFASNGSLHDILHGRKGVKGAQPG-PVLTWAQRVKIAVGAARGLEYLHEKADP 193
R+LVY++ N +LH LH PG PV+TW RV++A GAARG+ YLHE P
Sbjct: 406 QHRLLVYDYVPNNTLHYHLHA--------PGRPVMTWETRVRVAAGAARGIAYLHEDCHP 457
Query: 194 HIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLH-STRVLGTFGYHAPEYAMTGQ 252
IIHRDIKSSN+L+ + A +ADF L+ A ++ H STRV+GTFGY APEYA +G+
Sbjct: 458 RIIHRDIKSSNILLDNSFEALVADFGLAKIAQELDLNTHVSTRVMGTFGYMAPEYATSGK 517
Query: 253 LNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSE----DKVRQCVDTRL 308
L+ K+DVYS+GV+LLEL+TGRKPVD + P G +SLV WA P L + ++ + VD RL
Sbjct: 518 LSEKADVYSYGVILLELITGRKPVDTSQPLGDESLVEWARPLLGQAIENEEFDELVDPRL 577
Query: 309 GGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPL 348
G + P + +M AA CV++ A RP MS VV+AL L
Sbjct: 578 GKNFIPGEMFRMVEAAAACVRHSAAKRPKMSQVVRALDTL 617
>At5g02800 protein kinase - like
Length = 395
Score = 267 bits (682), Expect = 8e-72
Identities = 157/349 (44%), Positives = 207/349 (58%), Gaps = 13/349 (3%)
Query: 13 HKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQD 72
HK V + SE+ +G + Q + EL T NF ++
Sbjct: 22 HKLDRKSSDCSVSTSEKSRAKSSLSESKSKGSDHIVAQ-----TFTFSELATATRNFRKE 76
Query: 73 SLIGEGSYGRVYYGVLKN-GQAAAIKKLDASK-QPDEEFLAQVSMVSRLKHENFVQLLGY 130
LIGEG +GRVY G L + Q AAIK+LD + Q + EFL +V M+S L H N V L+GY
Sbjct: 77 CLIGEGGFGRVYKGYLASTSQTAAIKQLDHNGLQGNREFLVEVLMLSLLHHPNLVNLIGY 136
Query: 131 SVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEK 190
DG+ R+LVYE+ GSL D LH G QP L W R+KIA GAA+GLEYLH+K
Sbjct: 137 CADGDQRLLVYEYMPLGSLEDHLHDIS--PGKQP---LDWNTRMKIAAGAAKGLEYLHDK 191
Query: 191 ADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMT 250
P +I+RD+K SN+L+ DD K++DF L+ P STRV+GT+GY APEYAMT
Sbjct: 192 TMPPVIYRDLKCSNILLDDDYFPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMT 251
Query: 251 GQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSE-DKVRQCVDTRLG 309
GQL KSDVYSFGVVLLE++TGRK +D + G+Q+LV WA P + K Q D L
Sbjct: 252 GQLTLKSDVYSFGVVLLEIITGRKAIDSSRSTGEQNLVAWARPLFKDRRKFSQMADPMLQ 311
Query: 310 GEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPAGE 358
G+YPP+ + + AVAA+CVQ + + RP ++ VV AL L + + P +
Sbjct: 312 GQYPPRGLYQALAVAAMCVQEQPNLRPLIADVVTALSYLASQKFDPLAQ 360
>At3g58690 serine/threonine-specific protein kinase -like protein
Length = 400
Score = 266 bits (681), Expect = 1e-71
Identities = 147/294 (50%), Positives = 198/294 (67%), Gaps = 4/294 (1%)
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLD-ASKQPDEEFLAQVSMVSRL 119
+L T F + +++G G +G VY GVL +G+ AIK +D A KQ +EEF +V ++SRL
Sbjct: 79 QLHSATGGFSKSNVVGNGGFGLVYRGVLNDGRKVAIKLMDHAGKQGEEEFKMEVELLSRL 138
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
+ + LLGY D + ++LVYEF +NG L + L+ G+ P P L W R++IAV
Sbjct: 139 RSPYLLALLGYCSDNSHKLLVYEFMANGGLQEHLY-LPNRSGSVP-PRLDWETRMRIAVE 196
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
AA+GLEYLHE+ P +IHRD KSSN+L+ + AK++DF L+ D A STRVLGT
Sbjct: 197 AAKGLEYLHEQVSPPVIHRDFKSSNILLDRNFNAKVSDFGLAKVGSDKAGGHVSTRVLGT 256
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSE-D 298
GY APEYA+TG L KSDVYS+GVVLLELLTGR PVD G+ LV+WA P+L++ D
Sbjct: 257 QGYVAPEYALTGHLTTKSDVYSYGVVLLELLTGRVPVDMKRATGEGVLVSWALPQLADRD 316
Query: 299 KVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTAR 352
KV +D L G+Y K V ++AA+AA+CVQ EAD+RP M+ VV++L PL+ R
Sbjct: 317 KVVDIMDPTLEGQYSTKEVVQVAAIAAMCVQAEADYRPLMADVVQSLVPLVRNR 370
>At5g13160 protein kinase-like
Length = 456
Score = 266 bits (679), Expect = 2e-71
Identities = 161/376 (42%), Positives = 216/376 (56%), Gaps = 25/376 (6%)
Query: 1 MSCFNCCEEDDF----------HKTAESGGPYVVKNPSGNDGNHHASETAKQGG---QTV 47
M CF+C + D H + P V N SG + GG + +
Sbjct: 1 MGCFSCFDSSDDEKLNPVDESNHGQKKQSQPTVSNNISGLPSGGEKLSSKTNGGSKRELL 60
Query: 48 KPQP----IEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKN-GQAAAIKKLDAS 102
P+ I + EL T NF D+ +GEG +GRVY G L + GQ A+K+LD +
Sbjct: 61 LPRDGLGQIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRN 120
Query: 103 K-QPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKG 161
Q + EFL +V M+S L H N V L+GY DG+ R+LVYEF GSL D LH K
Sbjct: 121 GLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKE 180
Query: 162 AQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLS 221
A L W R+KIA GAA+GLE+LH+KA+P +I+RD KSSN+L+ + K++DF L+
Sbjct: 181 A-----LDWNMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLA 235
Query: 222 NQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLP 281
P STRV+GT+GY APEYAMTGQL KSDVYSFGVV LEL+TGRK +D +P
Sbjct: 236 KLGPTGDKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMP 295
Query: 282 RGQQSLVTWATPKLSE-DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSI 340
G+Q+LV WA P ++ K + D RL G +P +A+ + AVA++C+Q +A RP ++
Sbjct: 296 HGEQNLVAWARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIAD 355
Query: 341 VVKALQPLLTARPGPA 356
VV AL L P+
Sbjct: 356 VVTALSYLANQAYDPS 371
>At3g07070 putative protein kinase
Length = 414
Score = 265 bits (677), Expect = 3e-71
Identities = 163/377 (43%), Positives = 224/377 (59%), Gaps = 38/377 (10%)
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQT--VKPQPIEVPNIS 58
M+CF+C FH+ + V S N + T + +T P+ + N +
Sbjct: 1 MNCFSCFY---FHEKKK------VPRDSDNSYRRNGEVTGRDNNKTHPENPKTVNEQNKN 51
Query: 59 EDELKEVTDN-----------------FGQDSLIGEGSYGRVYYGVL-KNGQAAAIKKLD 100
DE KEVT+N F Q+ LIGEG +GRVY G L K G A+K+LD
Sbjct: 52 NDEDKEVTNNIAAQTFSFRELATATKNFRQECLIGEGGFGRVYKGKLEKTGMIVAVKQLD 111
Query: 101 ASK-QPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGV 159
+ Q ++EF+ +V M+S L H++ V L+GY DG+ R+LVYE+ S GSL D L
Sbjct: 112 RNGLQGNKEFIVEVLMLSLLHHKHLVNLIGYCADGDQRLLVYEYMSRGSLEDHL------ 165
Query: 160 KGAQPGPV-LTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADF 218
P + L W R++IA+GAA GLEYLH+KA+P +I+RD+K++N+L+ + AK++DF
Sbjct: 166 LDLTPDQIPLDWDTRIRIALGAAMGLEYLHDKANPPVIYRDLKAANILLDGEFNAKLSDF 225
Query: 219 DLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDH 278
L+ P + S+RV+GT+GY APEY TGQL KSDVYSFGVVLLEL+TGR+ +D
Sbjct: 226 GLAKLGPVGDKQHVSSRVMGTYGYCAPEYQRTGQLTTKSDVYSFGVVLLELITGRRVIDT 285
Query: 279 TLPRGQQSLVTWATPKLSE-DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPN 337
T P+ +Q+LVTWA P E + + D L G +P KA+ + AVAA+C+Q EA RP
Sbjct: 286 TRPKDEQNLVTWAQPVFKEPSRFPELADPSLEGVFPEKALNQAVAVAAMCLQEEATVRPL 345
Query: 338 MSIVVKALQPLLTARPG 354
MS VV AL L TA G
Sbjct: 346 MSDVVTALGFLGTAPDG 362
>At1g68690 protein kinase, putative
Length = 708
Score = 262 bits (670), Expect = 2e-70
Identities = 139/296 (46%), Positives = 192/296 (63%), Gaps = 14/296 (4%)
Query: 58 SEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLD-ASKQPDEEFLAQVSMV 116
S +EL + T+ F Q++L+GEG +G VY G+L +G+ A+K+L Q D EF A+V +
Sbjct: 366 SYEELVKATNGFSQENLLGEGGFGCVYKGILPDGRVVAVKQLKIGGGQGDREFKAEVETL 425
Query: 117 SRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKI 176
SR+ H + V ++G+ + G+ R+L+Y++ SN L+ LHG K V L WA RVKI
Sbjct: 426 SRIHHRHLVSIVGHCISGDRRLLIYDYVSNNDLYFHLHGEKSV--------LDWATRVKI 477
Query: 177 AVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRV 236
A GAARGL YLHE P IIHRDIKSSN+L+ D+ A+++DF L+ A D + +TRV
Sbjct: 478 AAGAARGLAYLHEDCHPRIIHRDIKSSNILLEDNFDARVSDFGLARLALDCNTHI-TTRV 536
Query: 237 LGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLS 296
+GTFGY APEYA +G+L KSDV+SFGVVLLEL+TGRKPVD + P G +SLV WA P +S
Sbjct: 537 IGTFGYMAPEYASSGKLTEKSDVFSFGVVLLELITGRKPVDTSQPLGDESLVEWARPLIS 596
Query: 297 E----DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPL 348
++ D +LGG Y + +M A CV++ A RP M +V+A + L
Sbjct: 597 HAIETEEFDSLADPKLGGNYVESEMFRMIEAAGACVRHLATKRPRMGQIVRAFESL 652
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,593,079
Number of Sequences: 26719
Number of extensions: 373520
Number of successful extensions: 4450
Number of sequences better than 10.0: 1012
Number of HSP's better than 10.0 without gapping: 859
Number of HSP's successfully gapped in prelim test: 153
Number of HSP's that attempted gapping in prelim test: 1031
Number of HSP's gapped (non-prelim): 1113
length of query: 361
length of database: 11,318,596
effective HSP length: 101
effective length of query: 260
effective length of database: 8,619,977
effective search space: 2241194020
effective search space used: 2241194020
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0158.11


