

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0157b.6
(293 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
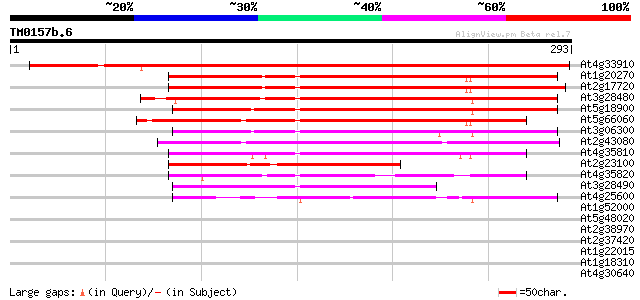
Score E
Sequences producing significant alignments: (bits) Value
At4g33910 unknown protein 385 e-107
At1g20270 unknown protein 179 2e-45
At2g17720 similar to prolyl 4-hydroxylase alpha subunit 164 7e-41
At3g28480 prolyl 4-hydroxylase like protein 160 1e-39
At5g18900 unknown protein 159 2e-39
At5g66060 prolyl 4-hydroxylase, alpha subunit-like protein 153 9e-38
At3g06300 unknown protein 149 2e-36
At2g43080 unknown protein 146 1e-35
At4g35810 putative protein 127 5e-30
At2g23100 hypothetical protein 121 4e-28
At4g35820 putative protein 120 9e-28
At3g28490 prolyl 4-hydroxylase, putative 102 2e-22
At4g25600 unknown protein 78 6e-15
At1g52000 30 1.5
At5g48020 unknown protein 29 2.6
At2g38970 putative retroelement pol polyprotein 29 2.6
At2g37420 putative kinesin heavy chain 29 3.4
At1g22015 unknown protein 29 3.4
At1g18310 beta-glucan-elicitor receptor, putative 29 3.4
At4g30640 putative protein 28 5.8
>At4g33910 unknown protein
Length = 288
Score = 385 bits (989), Expect = e-107
Identities = 188/284 (66%), Positives = 223/284 (78%), Gaps = 5/284 (1%)
Query: 11 SSRSNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNL--M 68
S R KL + + C FL GF+GSTL S + ++PR R+L+ E E M
Sbjct: 7 SYRRKKLGLATVIVFCSLCFLFGFYGSTLLSQNVPR---VKPRLRMLDMVENGEEEASSM 63
Query: 69 TAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETE 128
G G++SI SIPFQVLSW+PRA+YFPNFATAEQC++I+ AK LKPSALALR+GET
Sbjct: 64 PHGVTGEESIGSIPFQVLSWRPRAIYFPNFATAEQCQAIIERAKVNLKPSALALRKGETA 123
Query: 129 QNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHY 188
+NTKG RTSSG F+S+SE+ TG L +E KIARATMIPRSHGE+FNILRYE+ Q+Y+ HY
Sbjct: 124 ENTKGTRTSSGTFISASEESTGALDFVERKIARATMIPRSHGESFNILRYELGQKYDSHY 183
Query: 189 DSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQ 248
D FNP EYGPQ SQR+ASFLLYL+DV+EGGETMFPFENG NM + Y Y+ CIGL+V+PR+
Sbjct: 184 DVFNPTEYGPQSSQRIASFLLYLSDVEEGGETMFPFENGSNMGIGYDYKQCIGLKVKPRK 243
Query: 249 GDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQD 292
GDGLLFYS+FPNGTID TSLHGSCPV KGEKWVATKWIRD DQ+
Sbjct: 244 GDGLLFYSVFPNGTIDQTSLHGSCPVTKGEKWVATKWIRDQDQE 287
>At1g20270 unknown protein
Length = 287
Score = 179 bits (454), Expect = 2e-45
Identities = 93/207 (44%), Positives = 130/207 (61%), Gaps = 7/207 (3%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
+VLSW+PRA + NF + E+C+ ++ +AK + S + E ++++ +RTSSG F+
Sbjct: 77 EVLSWEPRAFVYHNFLSKEECEYLISLAKPHMVKSTVVDSETGKSKDSR-VRTSSGTFLR 135
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
DK + IE++IA T IP HGE +L YE Q+Y PHYD F QR
Sbjct: 136 RGRDKI--IKTIEKRIADYTFIPADHGEGLQVLHYEAGQKYEPHYDYFVDEFNTKNGGQR 193
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFP 259
MA+ L+YL+DV+EGGET+FP N V + E +C GL V+PR GD LLF+S+ P
Sbjct: 194 MATMLMYLSDVEEGGETVFPAANMNFSSVPWYNELSECGKKGLSVKPRMGDALLFWSMRP 253
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
+ T+DPTSLHG CPVI+G KW +TKW+
Sbjct: 254 DATLDPTSLHGGCPVIRGNKWSSTKWM 280
>At2g17720 similar to prolyl 4-hydroxylase alpha subunit
Length = 291
Score = 164 bits (414), Expect = 7e-41
Identities = 83/211 (39%), Positives = 131/211 (61%), Gaps = 7/211 (3%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
+V+SW+PRA+ + NF T E+C+ ++ +AK + S + + ++++ +RTSSG F+
Sbjct: 81 EVISWEPRAVVYHNFLTNEECEHLISLAKPSMVKSTVVDEKTGGSKDSR-VRTSSGTFLR 139
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
D+ + +IE++I+ T IP +GE +L Y+V Q+Y PHYD F QR
Sbjct: 140 RGHDEV--VEVIEKRISDFTFIPVENGEGLQVLHYQVGQKYEPHYDYFLDEFNTKNGGQR 197
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFP 259
+A+ L+YL+DV +GGET+FP G V + E C GL V P++ D LLF+++ P
Sbjct: 198 IATVLMYLSDVDDGGETVFPAARGNISAVPWWNELSKCGKEGLSVLPKKRDALLFWNMRP 257
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWIRDHD 290
+ ++DP+SLHG CPV+KG KW +TKW H+
Sbjct: 258 DASLDPSSLHGGCPVVKGNKWSSTKWFHVHE 288
>At3g28480 prolyl 4-hydroxylase like protein
Length = 316
Score = 160 bits (404), Expect = 1e-39
Identities = 90/224 (40%), Positives = 135/224 (60%), Gaps = 15/224 (6%)
Query: 69 TAGEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALRE-G 125
+A FG D P +V LSW PR + F + E+C + +AK L+ S +A + G
Sbjct: 45 SASSFGFD-----PTRVTQLSWTPRVFLYEGFLSDEECDHFIKLAKGKLEKSMVADNDSG 99
Query: 126 ETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYN 185
E+ ++ +RTSSG+F+S +D ++ +E K+A T +P +GE+ IL YE Q+Y
Sbjct: 100 ESVESE--VRTSSGMFLSKRQDDI--VSNVEAKLAAWTFLPEENGESMQILHYENGQKYE 155
Query: 186 PHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSY-RYEDCI--GL 242
PH+D F+ R+A+ L+YL++V++GGET+FP G + + +C G
Sbjct: 156 PHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLKDDSWTECAKQGY 215
Query: 243 RVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
V+PR+GD LLF++L PN T D SLHGSCPV++GEKW AT+WI
Sbjct: 216 AVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWI 259
>At5g18900 unknown protein
Length = 298
Score = 159 bits (402), Expect = 2e-39
Identities = 90/207 (43%), Positives = 127/207 (60%), Gaps = 9/207 (4%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+S KPRA + F T +C +V +AKA LK SA+A + E +RTSSG F+S
Sbjct: 40 VSSKPRAFVYEGFLTELECDHMVSLAKASLKRSAVADNDSG-ESKFSEVRTSSGTFISKG 98
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+D ++ IE+KI+ T +P+ +GE +LRYE Q+Y+ H+D F+ + RMA
Sbjct: 99 KDPI--VSGIEDKISTWTFLPKENGEDIQVLRYEHGQKYDAHFDYFHDKVNIVRGGHRMA 156
Query: 206 SFLLYLTDVQEGGETMFP-FENGLNMDVSYRYEDCI-----GLRVRPRQGDGLLFYSLFP 259
+ L+YL++V +GGET+FP E +S ED G+ V+PR+GD LLF++L P
Sbjct: 157 TILMYLSNVTKGGETVFPDAEIPSRRVLSENKEDLSDCAKRGIAVKPRKGDALLFFNLHP 216
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
+ DP SLHG CPVI+GEKW ATKWI
Sbjct: 217 DAIPDPLSLHGGCPVIEGEKWSATKWI 243
>At5g66060 prolyl 4-hydroxylase, alpha subunit-like protein
Length = 267
Score = 153 bits (387), Expect = 9e-38
Identities = 87/209 (41%), Positives = 129/209 (61%), Gaps = 11/209 (5%)
Query: 67 LMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGE 126
L +GE DDS +++SW+PRA + NF T E+CK ++ +AK ++ S + + +
Sbjct: 64 LQRSGE--DDSKNERWVEIISWEPRASVYHNFLTKEECKYLIELAKPHMEKSTVV--DEK 119
Query: 127 TEQNTKG-IRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYN 185
T ++T +RTSSG F++ DKT + IE++I+ T IP HGE +L YE+ Q+Y
Sbjct: 120 TGKSTDSRVRTSSGTFLARGRDKT--IREIEKRISDFTFIPVEHGEGLQVLHYEIGQKYE 177
Query: 186 PHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IG 241
PHYD F QR+A+ L+YL+DV+EGGET+FP G V + E +C G
Sbjct: 178 PHYDYFMDEYNTRNGGQRIATVLMYLSDVEEGGETVFPAAKGNYSAVPWWNELSECGKGG 237
Query: 242 LRVRPRQGDGLLFYSLFPNGTIDPTSLHG 270
L V+P+ GD LLF+S+ P+ T+DP+SLHG
Sbjct: 238 LSVKPKMGDALLFWSMTPDATLDPSSLHG 266
>At3g06300 unknown protein
Length = 299
Score = 149 bits (376), Expect = 2e-36
Identities = 82/207 (39%), Positives = 124/207 (59%), Gaps = 9/207 (4%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+S KPRA + F T +C ++ +AK L+ SA+A + E +RTSSG F+S
Sbjct: 41 VSSKPRAFVYEGFLTDLECDHLISLAKENLQRSAVADNDNG-ESQVSDVRTSSGTFISKG 99
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+D ++ IE+K++ T +P+ +GE +LRYE Q+Y+ H+D F+ + R+A
Sbjct: 100 KDPI--VSGIEDKLSTWTFLPKENGEDLQVLRYEHGQKYDAHFDYFHDKVNIARGGHRIA 157
Query: 206 SFLLYLTDVQEGGETMFP----FENGLNMDVSYRYEDCI--GLRVRPRQGDGLLFYSLFP 259
+ LLYL++V +GGET+FP F + DC G+ V+P++G+ LLF++L
Sbjct: 158 TVLLYLSNVTKGGETVFPDAQEFSRRSLSENKDDLSDCAKKGIAVKPKKGNALLFFNLQQ 217
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
+ DP SLHG CPVI+GEKW ATKWI
Sbjct: 218 DAIPDPFSLHGGCPVIEGEKWSATKWI 244
>At2g43080 unknown protein
Length = 283
Score = 146 bits (369), Expect = 1e-35
Identities = 79/211 (37%), Positives = 125/211 (58%), Gaps = 5/211 (2%)
Query: 78 ITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKG-IRT 136
I ++ +V+SW PR + +F + E+C+ + +A+ L+ S + + +T + K +RT
Sbjct: 72 IGNVKPEVVSWSPRIIVLHDFLSPEECEYLKAIARPRLQVSTVV--DVKTGKGVKSDVRT 129
Query: 137 SSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEY 196
SSG+F++ E + IE++IA + +P +GE +LRYE Q Y PH+D F
Sbjct: 130 SSGMFLTHVERSYPIIQAIEKRIAVFSQVPAENGELIQVLRYEPQQFYKPHHDYFADTFN 189
Query: 197 GPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYS 256
+ QR+A+ L+YLTD EGGET FP + D + + G+ V+P +GD +LF+S
Sbjct: 190 LKRGGQRVATMLMYLTDDVEGGETYFPLAG--DGDCTCGGKIMKGISVKPTKGDAVLFWS 247
Query: 257 LFPNGTIDPTSLHGSCPVIKGEKWVATKWIR 287
+ +G DP S+HG C V+ GEKW ATKW+R
Sbjct: 248 MGLDGQSDPRSIHGGCEVLSGEKWSATKWMR 278
>At4g35810 putative protein
Length = 307
Score = 127 bits (320), Expect = 5e-30
Identities = 79/221 (35%), Positives = 119/221 (53%), Gaps = 36/221 (16%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGL-KPSALALREG----------------- 125
+V+SW+PRA + NF T E+C+ ++ +AK + K + ++ G
Sbjct: 81 EVISWEPRAFVYHNFLTNEECEHLISLAKPSMMKSKVVDVKTGKSIDSRFCTLTSVVVFT 140
Query: 126 -----ETEQNTK-------GIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAF 173
E +N+K +RTSSG F++ D+ + IE +I+ T IP +GE
Sbjct: 141 FQLNLERFENSKFANPSLCRVRTSSGTFLNRGHDEI--VEEIENRISDFTFIPPENGEGL 198
Query: 174 NILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVS 233
+L YEV QRY PH+D F + QR+A+ L+YL+DV EGGET+FP G DV
Sbjct: 199 QVLHYEVGQRYEPHHDYFFDEFNVRKGGQRIATVLMYLSDVDEGGETVFPAAKGNVSDVP 258
Query: 234 Y--RYEDC--IGLRVRPRQGDGLLFYSLFPNGTIDPTSLHG 270
+ C GL V P++ D LLF+S+ P+ ++DP+SLHG
Sbjct: 259 WWDELSQCGKEGLSVLPKKRDALLFWSMKPDASLDPSSLHG 299
>At2g23100 hypothetical protein
Length = 1036
Score = 121 bits (304), Expect = 4e-28
Identities = 62/121 (51%), Positives = 85/121 (70%), Gaps = 4/121 (3%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
Q LSW PR Y PNFAT +QC++++ +AK LKPS LALR+ ET+ R+ +
Sbjct: 798 QGLSWNPRVFYLPNFATKQQCEAVIDMAKPKLKPSTLALRK-ETKHFQMQYRS---LHQH 853
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
+ ED++G LA IEEKIA AT P+ + E+FNILRY++ Q+Y+ HYD+F+ AEYGP SQR
Sbjct: 854 TDEDESGVLAAIEEKIALATRFPKDYYESFNILRYQLGQKYDSHYDAFHSAEYGPLISQR 913
Query: 204 M 204
+
Sbjct: 914 V 914
>At4g35820 putative protein
Length = 272
Score = 120 bits (301), Expect = 9e-28
Identities = 71/196 (36%), Positives = 112/196 (56%), Gaps = 31/196 (15%)
Query: 84 QVLSWKPRALYFPNFA--------TAEQCKSIVGVAKAGLKPSALA-LREGETEQNTKGI 134
+V++ +PRA + NF T E+C ++ +AK + S + G E+++
Sbjct: 89 EVITKEPRAFVYHNFLALFFKICKTNEECDHLISLAKPSMARSKVRNALTGLGEESSS-- 146
Query: 135 RTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPA 194
RTSSG F+ S DK + IE++I+ T IP+ +GE ++ YEV Q++ PH+D F
Sbjct: 147 RTSSGTFIRSGHDKI--VKEIEKRISEFTFIPQENGETLQVINYEVGQKFEPHFDGF--- 201
Query: 195 EYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLF 254
QR+A+ L+YL+DV +GGET+FP G+ G+ VRP++GD LLF
Sbjct: 202 -------QRIATVLMYLSDVDKGGETVFPEAKGIKSKK--------GVSVRPKKGDALLF 246
Query: 255 YSLFPNGTIDPTSLHG 270
+S+ P+G+ DP+S HG
Sbjct: 247 WSMRPDGSRDPSSKHG 262
>At3g28490 prolyl 4-hydroxylase, putative
Length = 213
Score = 102 bits (254), Expect = 2e-22
Identities = 52/138 (37%), Positives = 81/138 (58%), Gaps = 2/138 (1%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
LSW PRA + F + E+C ++ +AK L+ S + E +RTSSG+F++
Sbjct: 35 LSWTPRAFLYKGFLSDEECDHLIKLAKGKLEKSMVVADVDSGESEDSEVRTSSGMFLTKR 94
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+D +A +E K+A T +P +GEA IL YE Q+Y+PH+D F + R+A
Sbjct: 95 QDDI--VANVEAKLAAWTFLPEENGEALQILHYENGQKYDPHFDYFYDKKALELGGHRIA 152
Query: 206 SFLLYLTDVQEGGETMFP 223
+ L+YL++V +GGET+FP
Sbjct: 153 TVLMYLSNVTKGGETVFP 170
>At4g25600 unknown protein
Length = 291
Score = 77.8 bits (190), Expect = 6e-15
Identities = 58/206 (28%), Positives = 97/206 (46%), Gaps = 35/206 (16%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
LSW PR + F + E+C ++ +LR+ TE V+ +
Sbjct: 62 LSWLPRVFLYRGFLSEEECDHLI------------SLRKETTE-----------VYSVDA 98
Query: 146 EDKTG---TLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQ 202
+ KT +A IEEK++ T +P +G + + Y ++ D F
Sbjct: 99 DGKTQLDPVVAGIEEKVSAWTFLPGENGGSIKVRSY-TSEKSGKKLDYFGEEPSSVLHES 157
Query: 203 RMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCI--GLRVRPRQGDGLLFYSLFPN 260
+A+ +LYL++ +GGE +FP N ++ + C+ G +RP +G+ +LF++ N
Sbjct: 158 LLATVVLYLSNTTQGGELLFP-----NSEMKPK-NSCLEGGNILRPVKGNAILFFTRLLN 211
Query: 261 GTIDPTSLHGSCPVIKGEKWVATKWI 286
++D S H CPV+KGE VATK I
Sbjct: 212 ASLDGKSTHLRCPVVKGELLVATKLI 237
>At1g52000
Length = 730
Score = 30.0 bits (66), Expect = 1.5
Identities = 14/27 (51%), Positives = 17/27 (62%)
Query: 113 AGLKPSALALREGETEQNTKGIRTSSG 139
AG P A A+ GETE+N G + SSG
Sbjct: 178 AGTNPGASAVGNGETEKNAGGSKPSSG 204
Score = 28.1 bits (61), Expect = 5.8
Identities = 14/27 (51%), Positives = 16/27 (58%)
Query: 113 AGLKPSALALREGETEQNTKGIRTSSG 139
AG P A A GETE+N G + SSG
Sbjct: 206 AGTNPGASAGGNGETEKNVGGSKPSSG 232
Score = 27.7 bits (60), Expect = 7.6
Identities = 14/28 (50%), Positives = 17/28 (60%)
Query: 112 KAGLKPSALALREGETEQNTKGIRTSSG 139
KAG P A A G TE+N G ++SSG
Sbjct: 233 KAGTNPGANAGGNGGTEKNAGGSKSSSG 260
>At5g48020 unknown protein
Length = 355
Score = 29.3 bits (64), Expect = 2.6
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query: 37 STLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITSI 81
STLF+H D L+P ESS ++Y + AGE+ + ++ I
Sbjct: 303 STLFAHIASDAE-LKPLGHFAESSLASKYPAIPAGEYVEQELSVI 346
>At2g38970 putative retroelement pol polyprotein
Length = 692
Score = 29.3 bits (64), Expect = 2.6
Identities = 18/74 (24%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query: 45 DDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSI-TSIPFQVLSWK--PRALYFPNFATA 101
+ GRGL P P + + E+ E L+ +G+ D++ + P +++ K P P +
Sbjct: 159 NQGRGLAPEPSMFDDDERLEQQLVFSGKSYSDALENNHPVRMMDLKIYPEVSAVPRADSR 218
Query: 102 EQCKSIVGVAKAGL 115
E+ +V + A +
Sbjct: 219 EKFDVLVHLRAAAM 232
>At2g37420 putative kinesin heavy chain
Length = 1022
Score = 28.9 bits (63), Expect = 3.4
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 12/59 (20%)
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILR----------YEVDQRYN--PHYDSFN 192
E+K T AI+E + + T++ +HG+A + +R Y+VDQ N P S N
Sbjct: 915 ENKETTEAIVETCMNQVTLLQENHGQAVSNIRNKAEQSLIKDYQVDQHKNETPKKQSIN 973
>At1g22015 unknown protein
Length = 398
Score = 28.9 bits (63), Expect = 3.4
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Query: 14 SNKLTFPYIFLICIF-FFLAGFFGSTLFSHSQDDGRGL 50
S +LT ++ L+CI FFL F S L S S D G L
Sbjct: 8 SKRLTMTWVPLLCISCFFLGAIFTSKLRSASSDSGSQL 45
>At1g18310 beta-glucan-elicitor receptor, putative
Length = 649
Score = 28.9 bits (63), Expect = 3.4
Identities = 26/111 (23%), Positives = 45/111 (40%), Gaps = 6/111 (5%)
Query: 106 SIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMI 165
S+ GV K K AL + N+ TSS F + A+I E++ +I
Sbjct: 300 SMKGVKKDSYKEIISALGKDVNGLNSSAEVTSSSYFYGKLIARAARFALIAEEVCYLDVI 359
Query: 166 PRSHGEAFNILRYEVDQRYNPHYDSFNP------AEYGPQKSQRMASFLLY 210
P+ N++ +D + P+ ++P + G + SQ F +Y
Sbjct: 360 PKIVTYLKNMIEPWLDGSFKPNGFLYDPKWGGLITKQGSKDSQADFGFGIY 410
>At4g30640 putative protein
Length = 301
Score = 28.1 bits (61), Expect = 5.8
Identities = 18/67 (26%), Positives = 31/67 (45%), Gaps = 2/67 (2%)
Query: 178 YEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE 237
++++ R+ +S N + P+ ++ SFL + D EGG T + +SY E
Sbjct: 64 FDLETRFLSFPESIN--WWTPEFEDKVDSFLRSVVDRSEGGLTEIRIRHCTERSLSYAAE 121
Query: 238 DCIGLRV 244
C L V
Sbjct: 122 RCPNLEV 128
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,868,211
Number of Sequences: 26719
Number of extensions: 296272
Number of successful extensions: 731
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 678
Number of HSP's gapped (non-prelim): 29
length of query: 293
length of database: 11,318,596
effective HSP length: 99
effective length of query: 194
effective length of database: 8,673,415
effective search space: 1682642510
effective search space used: 1682642510
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0157b.6


