

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0154.9
(839 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
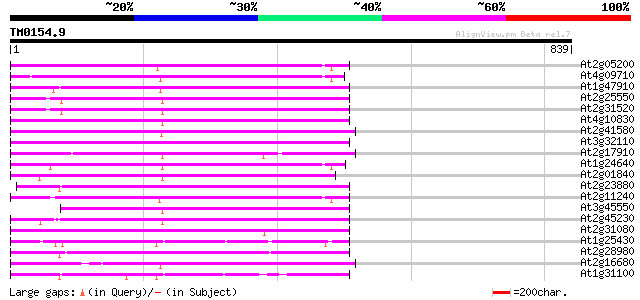
Score E
Sequences producing significant alignments: (bits) Value
At2g05200 putative non-LTR retroelement reverse transcriptase 304 2e-82
At4g09710 RNA-directed DNA polymerase -like protein 302 4e-82
At1g47910 reverse transcriptase, putative 302 6e-82
At2g25550 putative non-LTR retroelement reverse transcriptase 301 8e-82
At2g31520 putative non-LTR retroelement reverse transcriptase 300 2e-81
At4g10830 putative protein 295 9e-80
At2g41580 putative non-LTR retroelement reverse transcriptase 292 6e-79
At3g32110 non-LTR reverse transcriptase, putative 286 3e-77
At2g17910 putative non-LTR retroelement reverse transcriptase 279 5e-75
At1g24640 hypothetical protein 274 1e-73
At2g01840 putative non-LTR retroelement reverse transcriptase 274 2e-73
At2g23880 putative non-LTR retroelement reverse transcriptase 273 3e-73
At2g11240 pseudogene 271 8e-73
At3g45550 putative protein 270 3e-72
At2g45230 putative non-LTR retroelement reverse transcriptase 269 4e-72
At2g31080 putative non-LTR retroelement reverse transcriptase 269 6e-72
At1g25430 hypothetical protein 262 5e-70
At2g28980 putative non-LTR retroelement reverse transcriptase 259 6e-69
At2g16680 putative non-LTR retroelement reverse transcriptase 257 2e-68
At1g31100 hypothetical protein 254 1e-67
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 304 bits (778), Expect = 2e-82
Identities = 177/517 (34%), Positives = 281/517 (54%), Gaps = 12/517 (2%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLV-VDGGWVEDPKVVKSKMKEFFEARF 59
Q+SR W+ GD NT +FH RR N + + ++G + + + +F+ F
Sbjct: 233 QRSRVLWLHSGDRNTGYFHAVTRNRRTQNRLTVMEDINGVAQHEEHQISQIISGYFQQIF 292
Query: 60 TNIS-GDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIK 118
T+ S GD +++ VS+ DN LTR N EE++ AV+ K+PGPDG+ F
Sbjct: 293 TSESDGDFSVVDEAIEPMVSQGDNDFLTRIPNDEEVKDAVFSINASKAPGPDGFTAGFYH 352
Query: 119 SFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIV 178
S+WH++ D+ R + F T+ +P+ N + I LIPK P + DY PI+L YKIV
Sbjct: 353 SYWHIISTDVGREIRLFFTSKNFPRRMNETHIRLIPKDLGPRKVADYRPIALCNIFYKIV 412
Query: 179 SKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVID---EAKVKKRGFLVFKVDY 235
+K++T R+ ++ +I ENQSAF+ GR + D+VL+ +EV+ + KK + K D
Sbjct: 413 AKIMTKRMQLILPKLISENQSAFVPGRVISDNVLITHEVLHFLRTSSAKKHCSMAVKTDM 472
Query: 236 EKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGD 295
K YD V W+FL +L+R GF++ WI W+ C+ S S S L+NG+P + + L GD
Sbjct: 473 SKAYDRVEWDFLKKVLQRFGFHSIWIDWVLECVTSVSYSFLINGTPQGKVVPTRGLRQGD 532
Query: 296 PLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVL 355
PL+P LF++ E LSG+ +A+RL G +V G V+ L FADDT+F + +
Sbjct: 533 PLSPCLFILCTEVLSGLCTRAQRLRQLPGVRVSINGPRVNHLLFADDTMFFSKSDPESCN 592
Query: 356 AMKSMLRCFELMSGLKVNFFKSKLAGVS-VAEDVLLRYANLLHCKTTEIPFVYLGIHVGA 414
+ +L + SG +NF KS + S V + +L + YLG+
Sbjct: 593 KLSEILSRYGKASGQSINFHKSSVTFSSKTPRSVKGQVKRILKIRKEGGTGKYLGLPEHF 652
Query: 415 NPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASL 474
RK+ + ++ K+R++ W + LS GK ++++VL+S+PL+ +S FKLP ++L
Sbjct: 653 GRRKRDIFGAIIDKIRQKSHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKLP---SAL 709
Query: 475 CNRIEK---QFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
C +I+ +F W + RK +WV WSK+ + G
Sbjct: 710 CRKIQSLLTRFWWDTKPDVRKTSWVAWSKLTNPKNAG 746
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 302 bits (774), Expect = 4e-82
Identities = 173/511 (33%), Positives = 281/511 (54%), Gaps = 14/511 (2%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVE--DPKVVKSKMKEFFEAR 58
Q SR +W+ GD N +FH + RR N + ++ DG E + + + S + +F+
Sbjct: 279 QWSRVQWLNSGDRNKGYFHATTRTRRMLNNL-SVIEDGSGQEFHEEEQIASTISSYFQNI 337
Query: 59 FTNISGDGV-LLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFI 117
FT + + +++ +S N L + +L EI+ A++ DK+PGPDG++ F
Sbjct: 338 FTTSNNSDLQVVQEALSPIISSHCNEELIKISSLLEIKEALFSISADKAPGPDGFSASFF 397
Query: 118 KSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKI 177
++W +++ D+ R + F + N + +TLIPK++ P ++DY PI+L YKI
Sbjct: 398 HAYWDIIEADVSRDIRSFFVDSCLSPRLNETHVTLIPKISAPRKVSDYRPIALCNVQYKI 457
Query: 178 VSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKV---KKRGFLVFKVD 234
V+K+LT RL + +I +QSAF+ GR + D+VL+ +E++ +V KK + K D
Sbjct: 458 VAKILTRRLQPWLSELISLHQSAFVPGRAIADNVLITHEILHFLRVSGAKKYCSMAIKTD 517
Query: 235 YEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLG 294
K YD + WNFL +L RLGF+ KWI W+ C+ + S S L+NGSP + L G
Sbjct: 518 MSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGSPQGSVVPSRGLRQG 577
Query: 295 DPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNV 354
DPL+P+LF++ E LSG+ R+A+ + G +V R +V+ L FADDT+F + +
Sbjct: 578 DPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVARGSPQVNHLLFADDTMFFCKTNPTCC 637
Query: 355 LAMKSMLRCFELMSGLKVNFFKSKLAGVS-VAEDVLLRYANLLHCKTTEIPFVYLGIHVG 413
A+ ++L+ +EL SG +N KS + S +D+ R L YLG+
Sbjct: 638 GALSNILKKYELASGQSINLAKSAITFSSKTPQDIKRRVKLSLRIDNEGGIGKYLGLPEH 697
Query: 414 ANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVAS 473
RK+ + ++ ++R+R W + LS GK L+++VLSS+P + + FKLP AS
Sbjct: 698 FGRRKRDIFSSIVDRIRQRSHSWSIRFLSSAGKQILLKAVLSSMPSYAMMCFKLP---AS 754
Query: 474 LCNRIEK---QFLWGGEEGTRKIAWVKWSKV 501
LC +I+ +F W + RK+AWV W K+
Sbjct: 755 LCKQIQSVLTRFWWDSKPDKRKMAWVSWDKL 785
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 302 bits (773), Expect = 6e-82
Identities = 175/516 (33%), Positives = 271/516 (51%), Gaps = 10/516 (1%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGG-WVEDPKVVKSKMKEFFEARF 59
QKSR W+K GD+N+KFFH RR N I GL + G W + +++ +F+ F
Sbjct: 122 QKSRSLWMKLGDNNSKFFHALTKQRRARNRITGLHDENGIWSIEDDDIQNIAVSYFQNLF 181
Query: 60 TNISG---DGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHF 116
T + D L E +++ N LT E+R A++ +K+PGPDG F
Sbjct: 182 TTANPQVFDEALGEVQVL--ITDRINDLLTADATECEVRAALFMIHPEKAPGPDGMTALF 239
Query: 117 IKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYK 176
+ W ++K D++ ++ F GV+ K N++ I LIPK P + + PISL YK
Sbjct: 240 FQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLCNVGYK 299
Query: 177 IVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVK---KRGFLVFKV 233
++SK+L RL V+ ++I E QSAF++GR + D++L+A E+ + K F+ K
Sbjct: 300 VISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKFMAIKT 359
Query: 234 DYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLML 293
D K YD V WNF+ +LR++GF KWI WI CI + VL+NG P E+ L
Sbjct: 360 DMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERGLRQ 419
Query: 294 GDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHN 353
GDPL+P+LF++ E L +R+A R NL G KV VS L FADD+LF + +
Sbjct: 420 GDPLSPYLFILCTEVLIANIRKAERQNLITGIKVATPSPAVSHLLFADDSLFFCKANKEQ 479
Query: 354 VLAMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGIHV 412
+ +L+ +E +SG ++NF KS + G V + + +L YLG+
Sbjct: 480 CGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSYLGLPE 539
Query: 413 GANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVA 472
K + + +L+ R++ W K LS GGK +I+SV +++P + +S F+LPK +
Sbjct: 540 SLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPKAIT 599
Query: 473 SLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
S +F W +R + W+ W K+C + +G
Sbjct: 600 SKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDG 635
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 301 bits (772), Expect = 8e-82
Identities = 173/518 (33%), Positives = 276/518 (52%), Gaps = 14/518 (2%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWV-EDPKVVKSKMKEFFEARF 59
QKSR++W+K+GD NT +FH R N + ++ D G + K + + ++FF
Sbjct: 720 QKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFF---- 775
Query: 60 TNI-SGDGVLLEGTTFR----SVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNF 114
TNI S +G+ + F +V+ NL LT+ F+ EI A+ DK+PGPDG
Sbjct: 776 TNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTA 835
Query: 115 HFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCM 174
F K+ W ++ D++ ++ F N + I +IPK+ NP L+DY PI+L +
Sbjct: 836 RFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIALCNVL 895
Query: 175 YKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVF 231
YK++SK L RL + I+ ++Q+AF+ GR + D+V++A+EV+ KV+KR ++
Sbjct: 896 YKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAV 955
Query: 232 KVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRL 291
K D K YD V W+FL +R GF KWIGWI ++S SVL+NGSP + +
Sbjct: 956 KTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGI 1015
Query: 292 MLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPST 351
GDPL+P+LF++ + LS ++ +G ++G ++ LQFADD+LF + +
Sbjct: 1016 RQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFADDSLFFCQANV 1075
Query: 352 HNVLAMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGI 410
N A+K + +E SG K+N KS + G V R +L YLG+
Sbjct: 1076 RNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSRLKQILEIPNQGGGGKYLGL 1135
Query: 411 HVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKG 470
+KK ++ ++ +++KR S W + LS GK +++SV ++P++ +S FKLPKG
Sbjct: 1136 PEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKG 1195
Query: 471 VASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
+ S + F W R I WV W ++ + EG
Sbjct: 1196 IVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEG 1233
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 300 bits (769), Expect = 2e-81
Identities = 172/518 (33%), Positives = 276/518 (53%), Gaps = 14/518 (2%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWV-EDPKVVKSKMKEFFEARF 59
QKSR++W+K+GD NT +FH R N + ++ D G + K + + ++FF
Sbjct: 494 QKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQDFF---- 549
Query: 60 TNI-SGDGVLLEGTTFR----SVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNF 114
TNI S +G+ + F +V+ NL LT+ F+ EI A+ DK+PGPDG
Sbjct: 550 TNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDGLTA 609
Query: 115 HFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCM 174
F K+ W ++ D++ ++ F N + I +IPK+ NP L+DY PI+L +
Sbjct: 610 RFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIALCNVL 669
Query: 175 YKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVF 231
YK++SK L RL + I+ ++Q+AF+ GR + D+V++A+EV+ KV+KR ++
Sbjct: 670 YKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAV 729
Query: 232 KVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRL 291
K D K YD V W+FL +R GF KWIGWI ++S SVL+NGSP + +
Sbjct: 730 KTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGI 789
Query: 292 MLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPST 351
GDPL+P+LF++ + LS ++ +G ++G ++ LQFADD+LF + +
Sbjct: 790 RQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFADDSLFFCQANV 849
Query: 352 HNVLAMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGI 410
N A+K + +E SG K+N KS + G V + +L YLG+
Sbjct: 850 RNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSKLKQILEIPNQGGGGKYLGL 909
Query: 411 HVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKG 470
+KK ++ ++ +++KR S W + LS GK +++SV ++P++ +S FKLPKG
Sbjct: 910 PEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKG 969
Query: 471 VASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
+ S + F W R I WV W ++ + EG
Sbjct: 970 IVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEG 1007
>At4g10830 putative protein
Length = 1294
Score = 295 bits (754), Expect = 9e-80
Identities = 168/514 (32%), Positives = 277/514 (53%), Gaps = 6/514 (1%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWV-EDPKVVKSKMKEFFEARF 59
QKSR++W+K+GD NT+FFH R N + + + G + K + +EFF +
Sbjct: 700 QKSRNQWMKEGDRNTEFFHACTKTRFSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVY 759
Query: 60 TNISGDGVLLEGTTFRS-VSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIK 118
+ +++ F+ V+E+ N LT+ + EI A+ DK+PGPDG F K
Sbjct: 760 ESNGRPVSIIDFAGFKPIVTEQINDDLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYK 819
Query: 119 SFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIV 178
S W ++ D+++ ++ F + N + I +IPK+ NP L+DY PI+L +YKI+
Sbjct: 820 SCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSDYRPIALCNVLYKII 879
Query: 179 SKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVFKVDY 235
SK L RL + I+ ++Q+AF+ GR + D+V++A+E++ K +KR ++ K D
Sbjct: 880 SKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDV 939
Query: 236 EKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGD 295
K YD V WNFL +R GF WI WI G ++S + SVLVNG P ++ + GD
Sbjct: 940 SKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGD 999
Query: 296 PLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVL 355
PL+P+LF++ A+ L+ +++ +G ++G V+ LQFADD+LF + + N
Sbjct: 1000 PLSPYLFILCADILNHLIKNRVAEGDIRGIRIGNGVPGVTHLQFADDSLFFCQSNVRNCQ 1059
Query: 356 AMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGA 414
A+K + +E SG K+N KS + G V R N+L ++ YLG+
Sbjct: 1060 ALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQF 1119
Query: 415 NPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASL 474
+K+ ++ ++ +++KR S W K LS GK +++SV S+P++ +S FKLP + S
Sbjct: 1120 GRKKRDMFNYIIERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSE 1179
Query: 475 CNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
+ F W R+I W+ W ++ + EG
Sbjct: 1180 IEALLMNFWWEKNAKKREIPWIAWKRLQYSKKEG 1213
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 292 bits (747), Expect = 6e-79
Identities = 176/523 (33%), Positives = 276/523 (52%), Gaps = 8/523 (1%)
Query: 2 KSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKVVKSKMK-EFFEARFT 60
KSRD+W+ GD N+KFF ++ R +N++R LV + G + K K+ FFE F+
Sbjct: 68 KSRDKWMVGGDKNSKFFQATVKANRVSNSLRFLVDENGNEQTVNREKGKIAVTFFEDLFS 127
Query: 61 NI--SGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIK 118
+ S +LEG R V+E+ N LT+ N +EI AV+ + +PGPDG+ F +
Sbjct: 128 SSYPSSMDSVLEGFNKR-VTEDMNQDLTKKVNEQEIYKAVFSINAESAPGPDGFTALFFQ 186
Query: 119 SFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIV 178
W ++K+ I+ +E F G+ P++ N + + LIPK+ P + D PISL MYKI+
Sbjct: 187 RQWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPISLCSVMYKII 246
Query: 179 SKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKK---RGFLVFKVDY 235
SK+L+ RL K + I+ QSAF+ R + D++++A+E++ + + + F+VFK D
Sbjct: 247 SKILSARLKKYLPVIVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDM 306
Query: 236 EKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGD 295
K YD V W FL +L LGF + WI W+ C+ S S SVL+NG P + L GD
Sbjct: 307 SKAYDRVEWPFLKGILLALGFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLRQGD 366
Query: 296 PLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVL 355
PL+PFLF++ E L ++ QA ++ G + G V+ L FADDTL I + S
Sbjct: 367 PLSPFLFVLCTEALIHILNQAEKIGKISGIQFNGTGPSVNHLLFADDTLLICKASQLECA 426
Query: 356 AMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGA 414
+ L + +SG +N KS + G V E+ N +T YLG+
Sbjct: 427 EIMHCLSQYGHISGQMINSEKSAITFGAKVNEETKQWIMNRSGIQTEGGTGKYLGLPECF 486
Query: 415 NPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASL 474
K+ + + KL+ RLS W KTLS GGK L++S+ + P++ ++ F+L K + +
Sbjct: 487 QGSKQVLFGFIKEKLQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLSKTLCTK 546
Query: 475 CNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEGSWESRT*SC 517
+ F W + +KI W+ K+ + G + + C
Sbjct: 547 LTSVMMDFWWNSVQDKKKIHWIGAQKLMLPKFLGGFGFKDLQC 589
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 286 bits (732), Expect = 3e-77
Identities = 166/514 (32%), Positives = 278/514 (53%), Gaps = 6/514 (1%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVV-DGGWVEDPKVVKSKMKEFFEARF 59
QKSR++W GD NTKFFH S RRR N I L DG W+ + + +++ ++++ +
Sbjct: 877 QKSREKWFVHGDRNTKFFHTSTIIRRRRNQIEMLQDNDGRWLSNAQELETHAIDYYKRLY 936
Query: 60 TNISGDGVL--LEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFI 117
+ D V+ L F ++SE D +LT+ F+ E+ A+ K+PGPDG+ F
Sbjct: 937 SLDDLDAVVEQLPQEGFTALSEADFSSLTKPFSPLEVEGAIRSMGKYKAPGPDGFQPVFY 996
Query: 118 KSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKI 177
+ W V+ + + + + DF ++G +P+E N + LI KV P + + PISL ++K
Sbjct: 997 QQGWEVVGESVTKFVMDFFSSGSFPQETNDVLVVLIAKVLKPEKITQFRPISLCNVLFKT 1056
Query: 178 VSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKK--RGFLVFKVDY 235
++KV+ RL V+ +I Q++F+ GR D+++V EV+ + KK +G+++ K+D
Sbjct: 1057 ITKVMVGRLKGVINKLIGPAQTSFIPGRLSTDNIVVVQEVVHSMRRKKGVKGWMLLKLDL 1116
Query: 236 EKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGD 295
EK YD + W+ L L+ G W+ WI C++ S+ +L NG TD F + L GD
Sbjct: 1117 EKAYDRIRWDLLEDTLKAAGLPGTWVQWIMKCVEGPSMRLLWNGEKTDAFKPLRGLRQGD 1176
Query: 296 PLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVL 355
PL+P+LF++ E L ++ + +K K+ + G +S + FADD + E S +
Sbjct: 1177 PLSPYLFVLCIERLCHLIESSIAAKKWKPIKISQSGPRLSHICFADDLILFAEASIDQIR 1236
Query: 356 AMKSMLRCFELMSGLKVNFFKSKL-AGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGA 414
++ +L F SG KV+ KSK+ +V D+ R + K T+ YLG+ +
Sbjct: 1237 VLRGVLEKFCGASGQKVSLEKSKIYFSKNVLRDLGKRISEESGMKATKDLGKYLGVPILQ 1296
Query: 415 NPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASL 474
K T+ ++ + RL+ WK + LSF G++ L ++VL+SI + +S KLP+
Sbjct: 1297 KRINKETFGEVIKRFSSRLAGWKGRMLSFAGRLTLTKAVLTSILVHTMSTIKLPQSTLDG 1356
Query: 475 CNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
+++ + FLWG +K V W++VC R EG
Sbjct: 1357 LDKVSRAFLWGSTLEKKKQHLVAWTRVCLPRREG 1390
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 279 bits (713), Expect = 5e-75
Identities = 172/528 (32%), Positives = 268/528 (50%), Gaps = 16/528 (3%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKVVKSKM-KEFFEARF 59
QKSR +W+ GD NT FFH +++ R N + L+ + K K+ FFE F
Sbjct: 316 QKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSFFENLF 375
Query: 60 TN--ISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFI 117
T+ I LEG + SE ++ + LE + AV+ + +PGPDG+ F
Sbjct: 376 TSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELE-VYNAVFSINKESAPGPDGFTALFF 434
Query: 118 KSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKI 177
+ W ++K I+ + F GV P++ N + I LIPK+ +P ++D PISL +YKI
Sbjct: 435 QQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSVLYKI 494
Query: 178 VSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVFKVD 234
+SK+LT RL K + I+ QSAF+ R + D++LVA+E+I + R + FK D
Sbjct: 495 ISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMAFKTD 554
Query: 235 YEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLG 294
K YD V W FL M+ LGF KWI WI C+ S S SVL+NG P + + G
Sbjct: 555 MSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQG 614
Query: 295 DPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNV 354
DPL+P LF++ E L ++ +A + G + + V V+ L FADDTL + + +
Sbjct: 615 DPLSPALFVLCTEALIHILNKAEQAGKITGIQFQDKKVSVNHLLFADDTLLMCKATKQEC 674
Query: 355 LAMKSMLRCFELMSGLKVNFFKS-----KLAGVSVAEDVLLRYANLLHCKTTEIPFVYLG 409
+ L + +SG +N KS K + + + + R L T + YLG
Sbjct: 675 EELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQIKDWIKSRSGISLEGGTGK----YLG 730
Query: 410 IHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPK 469
+ + K+ + + KL+ RL+ W KTLS GGK L++S+ ++P++ +S FKLPK
Sbjct: 731 LPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPK 790
Query: 470 GVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEGSWESRT*SC 517
+ + F W + RKI W+ W ++ + +G + + C
Sbjct: 791 NLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQC 838
>At1g24640 hypothetical protein
Length = 1270
Score = 274 bits (701), Expect = 1e-73
Identities = 163/512 (31%), Positives = 268/512 (51%), Gaps = 14/512 (2%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKVVKSKMKEFFEARF- 59
QKSR +W++ G+ N+K+FH ++ R+ I L G ++ + K ++ +
Sbjct: 301 QKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLF 360
Query: 60 --TNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFI 117
+N SG G R VSE N +L + +EI+ AV+ + +PGPDG + F
Sbjct: 361 KSSNPSGFTDWFSGLVPR-VSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFF 419
Query: 118 KSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKI 177
+ +W + + + ++ F +G+ P E N + + LIPK +P + D PISL +YKI
Sbjct: 420 QHYWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKI 479
Query: 178 VSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVFKVD 234
+SK++ RL + I+ + QSAF+ R + D++LVA+E++ KV R F+ K D
Sbjct: 480 ISKIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSD 539
Query: 235 YEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLG 294
K YD V W++L +L LGF+ KW+ WI C+ S + SVL+N P +++ L G
Sbjct: 540 MSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQG 599
Query: 295 DPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNV 354
DPL+PFLF++ E L+ ++ +A+ +G + G V L FADD+LF+ + S
Sbjct: 600 DPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASREQS 659
Query: 355 LAMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVG 413
L ++ +L+ + +G +N KS + G V E + L T YLG+
Sbjct: 660 LVLQKILKVYGNATGQTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPEC 719
Query: 414 ANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVAS 473
+ K L +L+++L +W + LS GGK L++SV ++P+F +S FKLP +
Sbjct: 720 FSGSKVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLP---IT 776
Query: 474 LCNRIEK---QFLWGGEEGTRKIAWVKWSKVC 502
C +E F W + +RKI W W ++C
Sbjct: 777 TCENLESAMASFWWDSCDHSRKIHWQSWERLC 808
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 274 bits (700), Expect = 2e-73
Identities = 160/493 (32%), Positives = 264/493 (53%), Gaps = 8/493 (1%)
Query: 2 KSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVE---DPKVVKSKMKEFFEAR 58
KSR+RW+ GD NT FF+ S R+ N I+ + D +E D + K F +
Sbjct: 701 KSRNRWMLLGDRNTMFFYASTKLRKSRNRIKA-ITDAQGIENFRDDTIGKVAENYFADLF 759
Query: 59 FTNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIK 118
T + D + V+E+ N L ++ +E+R AV+ D++PG DG+ F
Sbjct: 760 TTTQTSDWEEIISGIAPKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAAFYH 819
Query: 119 SFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIV 178
W ++ +D+ ++ F + V + N + I LIPK+ +P ++DY PISL YKI+
Sbjct: 820 HLWDLIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASYKII 879
Query: 179 SKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVFKVDY 235
SK+L RL + +G +I ++Q+AF+ G+ + D+VLVA+E++ K ++ G++ K D
Sbjct: 880 SKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKTDI 939
Query: 236 EKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGD 295
K YD V WNFL ++ +LGF +W+ WI C+ S S VL+NGSP + + + GD
Sbjct: 940 SKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQGD 999
Query: 296 PLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVL 355
PL+P+LFL AE LS M+R+A G K+ ++ + +S L FADD+LF S N+
Sbjct: 1000 PLSPYLFLFCAEVLSNMLRKAEVNKQIHGMKITKDCLAISHLLFADDSLFFCRASNQNIE 1059
Query: 356 AMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGA 414
+ + + +E SG K+N+ KS + G + R LL YLG+
Sbjct: 1060 QLALIFKKYEEASGQKINYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGGKYLGLPEQL 1119
Query: 415 NPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASL 474
RK ++ +++K+++R W LS GK +I+++ ++P++ ++ F LP + +
Sbjct: 1120 GRRKVELFEYIVTKVKERTEGWAYNYLSPAGKEIVIKAIAMALPVYSMNCFLLPTLICNE 1179
Query: 475 CNRIEKQFLWGGE 487
N + F WG E
Sbjct: 1180 INSLITAFWWGKE 1192
>At2g23880 putative non-LTR retroelement reverse transcriptase
Length = 1216
Score = 273 bits (698), Expect = 3e-73
Identities = 165/506 (32%), Positives = 269/506 (52%), Gaps = 10/506 (1%)
Query: 11 GDSNTKFFHNSINWRRRTNAIRGLVV-DGGWVEDPKVVKSKMKEFFEARFTNISGD--GV 67
GD N K FH +I R N+IR +V DG V + ++++ +F+ I D G+
Sbjct: 90 GDRNNKTFHRAITTREAVNSIREIVTRDGLVVTSQQDIQTEAVNYFQDFLQTIPADYEGM 149
Query: 68 LLEGTT----FRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHV 123
+E FR SE+D+ LTR EEI+ ++ DKSPGPDGY F K+ W +
Sbjct: 150 CVEELENLLPFRC-SEDDHRLLTRVVTGEEIKKVIFSMPKDKSPGPDGYTSEFYKASWEI 208
Query: 124 LKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLT 183
+ D+++ ++ F G PK NS+ + LIPK + DY PIS +YK +SK+L
Sbjct: 209 IGDEVIIAIQSFFAKGFLPKGVNSTILALIPKKKEAREIKDYRPISCCNVLYKAISKILA 268
Query: 184 TRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVI-DEAKVKKRGFLVFKVDYEKVYDSV 242
RL +++ I NQSAF++ R L+++VL+A E++ D K K+D K +DS+
Sbjct: 269 NRLKRILPKFIVGNQSAFVKDRLLIENVLLATELVKDYHKDSISTRCAMKIDISKAFDSL 328
Query: 243 NWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLF 302
W+FL ++L + F ++I WI C+ +AS S+ VNG F + L G L+P+LF
Sbjct: 329 QWSFLTHVLAAMNFPGEFIHWISLCMSTASFSIQVNGELAGYFRSARGLRQGCSLSPYLF 388
Query: 303 LIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLR 362
+I + LS M+ +A + G+ + + ++ L FADD + + + +V + +L
Sbjct: 389 VISMDVLSRMLDKAAGAREF-GYHPRCKTLGLTHLCFADDLMILTDGKIRSVDGIVKVLN 447
Query: 363 CFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPRKKTTW 422
F GLK+ K+ L V++ ++ ++P YLG+ + + +
Sbjct: 448 QFAAKLGLKICMEKTTLYLAGVSDHSRQLMSSRYSFGVGKLPVRYLGLPLVTKRLTTSDY 507
Query: 423 DPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASLCNRIEKQF 482
PL+ ++R+R+ +W + LSF G++ LI SVL SI F+++ F+LP+ + NRI
Sbjct: 508 SPLIDQIRRRIGMWTSRYLSFAGRLSLINSVLWSITNFWMNAFRLPRECINEINRISSAL 567
Query: 483 LWGGEEGTRKIAWVKWSKVCRLRMEG 508
LW G E K A V W ++C+ + EG
Sbjct: 568 LWSGPELNPKKAKVSWDEICKPKKEG 593
>At2g11240 pseudogene
Length = 1044
Score = 271 bits (694), Expect = 8e-73
Identities = 165/516 (31%), Positives = 269/516 (51%), Gaps = 18/516 (3%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKV-VKSKMKEFFEARF 59
Q+SR W+ GD N+ +FH R N + + G E + + + + E+F+ F
Sbjct: 172 QRSRQLWLALGDKNSGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISEYFQKLF 231
Query: 60 TNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKS 119
+ EG ++ E ++ N EEI++A + DK+PGPDG++ F +S
Sbjct: 232 S-------ANEGARAATIKEAIKPFISPEQNPEEIKSACFSIHADKAPGPDGFSASFFQS 284
Query: 120 FWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVS 179
W + +IV ++ F ++ N + ITLIPK+ + + DY PI+L YKI+S
Sbjct: 285 NWMTVGPNIVLEIQSFFSSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVFYKIIS 344
Query: 180 KVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAK---VKKRGFLVFKVDYE 236
K+L+ RL ++ II ENQSAF+ R D+VL+ +E + K +KR F+ K +
Sbjct: 345 KLLSRRLQPILQEIISENQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMS 404
Query: 237 KVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGDP 296
K YD + W+F+ +++ +GF+ WI WI CI + S S L+NGS E+ L GDP
Sbjct: 405 KAYDRIEWDFIKLVMQEMGFHQTWISWILQCITTVSYSFLLNGSAQGAVTPERGLRQGDP 464
Query: 297 LAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVLA 356
L+PFLF+I +E LSG+ R+A+ G +V + V+ L FADDT+F +
Sbjct: 465 LSPFLFIICSEVLSGLCRKAQLDGSLLGLRVSKGNPRVNHLLFADDTIFFCRSDLKSCKT 524
Query: 357 MKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYA-NLLHCKTTEIPFVYLGIHVGAN 415
+L+ +E SG +N KS + D + A +L + YLG+
Sbjct: 525 FLCILKKYEEASGQMINKSKSAITFSRKTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFG 584
Query: 416 PRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASLC 475
+K+ ++ ++ ++R+R W + LS GK +++SVL+S+P + +S FKL + SLC
Sbjct: 585 RKKRDLFNQIVDRIRQRSLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKL---LVSLC 641
Query: 476 NRIEK---QFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
RI+ F W +K+ W+ WSK+ + + EG
Sbjct: 642 KRIQSALTHFWWDSSADKKKMCWIAWSKMAKNKKEG 677
>At3g45550 putative protein
Length = 851
Score = 270 bits (689), Expect = 3e-72
Identities = 146/437 (33%), Positives = 237/437 (53%), Gaps = 4/437 (0%)
Query: 76 SVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDF 135
SV+ N LT+ F EI A+ DK+PGPDG F K W ++ +D+++ ++ F
Sbjct: 36 SVTNIINSELTQDFRDSEIFEAICQIGDDKAPGPDGLTARFYKQCWDIVGNDVIKEVKLF 95
Query: 136 HTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIID 195
+ N + I +IPK+ NP L+DY PI+L +YK++SK + RL + I+
Sbjct: 96 FESSHMKTSVNHTNICMIPKIQNPQTLSDYRPIALCNVLYKVISKCMVNRLKAHLNSIVS 155
Query: 196 ENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVFKVDYEKVYDSVNWNFLFYMLR 252
++Q+AF+ GR + D+V++A+E++ KV+KR ++ K D K YD V W+FL +R
Sbjct: 156 DSQAAFIPGRIINDNVMIAHEIMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMR 215
Query: 253 RLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGM 312
GF KWIGWI ++S SVL+NGSP + + GDPL+P+LF++ + LS +
Sbjct: 216 LFGFCDKWIGWIMAAVKSVHYSVLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSHL 275
Query: 313 MRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKV 372
++ +G ++G ++ LQFADD+LF + + N A+K + +E SG K+
Sbjct: 276 IKVKASSGDIRGVRIGNGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKI 335
Query: 373 NFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSKLRK 431
N KS + G V R LL+ YLG+ +KK ++ ++ ++++
Sbjct: 336 NVQKSLITFGSRVYGSTQTRLKTLLNIPNQGGGGKYLGLPEQFGRKKKEMFNYIIDRVKE 395
Query: 432 RLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGTR 491
R + W K LS GK L++SV ++P++ +S FKLP+G+ S + F W R
Sbjct: 396 RTASWSAKFLSPAGKEILLKSVALAMPVYAMSCFKLPQGIVSEIESLLMNFWWEKASNKR 455
Query: 492 KIAWVKWSKVCRLRMEG 508
I WV W ++ + EG
Sbjct: 456 GIPWVAWKRLQYSKKEG 472
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 269 bits (688), Expect = 4e-72
Identities = 163/517 (31%), Positives = 265/517 (50%), Gaps = 12/517 (2%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGG--WVEDP---KVVKSKMKEFF 55
+KSR W+++GD NTK+FH + RR N I+ L+ + G W D +V ++ K+ F
Sbjct: 338 EKSRIMWMRNGDRNTKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYFKKLF 397
Query: 56 EARFTNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFH 115
+ + + LE T VS++ N L EE++ A + K PGPDG N
Sbjct: 398 ASEDVGYTVEE--LENLT-PLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGF 454
Query: 116 FIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMY 175
+ FW + D I +++ F +G + N + I LIPK+ + D+ PISL +Y
Sbjct: 455 LYQQFWETMGDQITEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIY 514
Query: 176 KIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKR---GFLVFK 232
K++ K++ RL K++ +I E Q+AF++GR + D++L+A+E++ + F+ K
Sbjct: 515 KVIGKLMANRLKKILPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIK 574
Query: 233 VDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLM 292
D K YD V W FL +R LGF WI I C++S VL+NG+P E + L
Sbjct: 575 TDISKAYDRVEWPFLEKAMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLR 634
Query: 293 LGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTH 352
GDPL+P+LF+I E L M++ A + N G KV R +S L FADD++F + +
Sbjct: 635 QGDPLSPYLFVICTEMLVKMLQSAEQKNQITGLKVARGAPPISHLLFADDSMFYCKVNDE 694
Query: 353 NVLAMKSMLRCFELMSGLKVNFFKSKL-AGVSVAEDVLLRYANLLHCKTTEIPFVYLGIH 411
+ + ++ + L SG +VN+ KS + G ++E+ L + VYLG+
Sbjct: 695 ALGQIIRIIEEYSLASGQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLP 754
Query: 412 VGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGV 471
K T L +L K++ W+ LS GGK L+++V ++P + +S FK+PK +
Sbjct: 755 ESFQGSKVATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTI 814
Query: 472 ASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
+ +F W ++ R + W W + R + G
Sbjct: 815 CQQIESVMAEFWWKNKKEGRGLHWKAWCHLSRPKAVG 851
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 269 bits (687), Expect = 6e-72
Identities = 166/516 (32%), Positives = 270/516 (52%), Gaps = 10/516 (1%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGG-WVEDPKVVKSKMKEFFEARF 59
QKSR+++++ GD NT FFH S RRR N I L D WV D +++ +++ +
Sbjct: 201 QKSREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMALTYYKRLY 260
Query: 60 T--NISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFI 117
+ ++S +L F S+SE + AL + F E+ +AV K+PGPDGY F
Sbjct: 261 SLEDVSEVRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPDGYQPVFY 320
Query: 118 KSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKI 177
+ W + + R + +F GV P N + + LI KV P + + P+SL ++KI
Sbjct: 321 QQCWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQFRPVSLCNVLFKI 380
Query: 178 VSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKK--RGFLVFKVDY 235
++K++ TRL V+ +I Q++F+ GR +D++++ E + + KK +G+++ K+D
Sbjct: 381 ITKMMVTRLKNVISKLIGPAQASFIPGRLSIDNIVLVQEAVHSMRRKKGRKGWMLLKLDL 440
Query: 236 EKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGD 295
EK YD V W+FL L G W I + S+SVL NG TD F + L GD
Sbjct: 441 EKAYDRVRWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSVLWNGERTDSFVPARGLRQGD 500
Query: 296 PLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVL 355
PL+P+LF++ E L ++ + +K V G ++S + FADD + E S +
Sbjct: 501 PLSPYLFVLCLERLCHLIEASVGKREWKPIAVSCGGSKLSHVCFADDLILFAEASVAQIR 560
Query: 356 AMKSMLRCFELMSGLKVNFFKSKL---AGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHV 412
++ +L F SG KV+ KSK+ VS + L+ + + C T E+ YLG+ +
Sbjct: 561 IIRRVLERFCEASGQKVSLEKSKIFFSHNVSREMEQLISEESGIGC-TKELG-KYLGMPI 618
Query: 413 GANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVA 472
K T+ +L ++ RL+ WK ++LS G++ L ++VLSSIP+ +S LP
Sbjct: 619 LQKRMNKETFGEVLERVSARLAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPVSTL 678
Query: 473 SLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
+R + FLWG +K + W K+C+ + EG
Sbjct: 679 DTLDRYSRTFLWGSTMEKKKQHLLSWRKICKPKAEG 714
>At1g25430 hypothetical protein
Length = 1213
Score = 262 bits (670), Expect = 5e-70
Identities = 172/526 (32%), Positives = 271/526 (50%), Gaps = 32/526 (6%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKVVKSKMKEFFEARFT 60
QKSR W +GD NTK+FH + R +N+I L G + D + + + + F
Sbjct: 354 QKSRISWFAEGDGNTKYFHRMADARNSSNSISALYDGNGKLVDSQ---EGILDLCASYFG 410
Query: 61 NISGDGV---LLEGTTFRSV-----SEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGY 112
++ GD V L+E + S L TF+ E+IR A++ +KS GPDG+
Sbjct: 411 SLLGDEVDPYLMEQNDMNLLLSYRCSPAQVCELESTFSNEDIRAALFSLPRNKSCGPDGF 470
Query: 113 NFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIG 172
F W ++ ++ +++F ++G K+ N++ I LIPK+ NP +D+ PIS +
Sbjct: 471 TAEFFIDSWSIVGAEVTDAIKEFFSSGCLLKQWNATTIVLIPKIVNPTCTSDFRPISCLN 530
Query: 173 CMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVI---DEAKVKKRGFL 229
+YK+++++LT RL +++ +I QSAFL GR L ++VL+A +++ + + + RG L
Sbjct: 531 TLYKVIARLLTDRLQRLLSGVISSAQSAFLPGRSLAENVLLATDLVHGYNWSNISPRGML 590
Query: 230 VFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEK 289
KVD +K +DSV W F+ LR L K+I WI CI + + +V +NG F K
Sbjct: 591 --KVDLKKAFDSVRWEFVIAALRALAIPEKFINWISQCISTPTFTVSINGGNGGFFKSTK 648
Query: 290 RLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEP 349
L GDPL+P+LF++ E S ++ L + + +S L FADD + ++
Sbjct: 649 GLRQGDPLSPYLFVLAMEAFSNLLHSRYESGLIH-YHPKASNLSISHLMFADDVMIFFDG 707
Query: 350 STHNVLAMKSMLRCFELMSGLKVNFFKSK--LAGVSVAEDVLLRYANLLH-CKTTEIPFV 406
+ ++ + L F SGLKVN KS LAG++ E AN + +P
Sbjct: 708 GSFSLHGICETLDDFASWSGLKVNKDKSHLYLAGLNQLES----NANAAYGFPIGTLPIR 763
Query: 407 YLGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFK 466
YLG+ + + ++PLL K+ R W K LSF G++ LI SV+ F++S F
Sbjct: 764 YLGLPLMNRKLRIAEYEPLLEKITARFRSWVNKCLSFAGRIQLISSVIFGSINFWMSTFL 823
Query: 467 LPKG----VASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
LPKG + SLC+R FLW G K V W+ +C + EG
Sbjct: 824 LPKGCIKRIESLCSR----FLWSGNIEQAKGIKVSWAALCLPKSEG 865
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 259 bits (661), Expect = 6e-69
Identities = 162/518 (31%), Positives = 271/518 (52%), Gaps = 14/518 (2%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLV-VDGGWVEDPKVVKSKMKEFFEARF 59
QKS+ W+ GD N +FH + R+ N+IR + + ++ + +K + + FF
Sbjct: 656 QKSKLHWMNVGDGNNSYFHKAAQVRKMRNSIREIRGPNAETLQTSEEIKGEAERFFNEFL 715
Query: 60 TNISGD--GVLLEGT----TFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYN 113
SGD G+ +E ++R + N+ LTR EEI+ ++ +KSPGPDGY
Sbjct: 716 NRQSGDFHGISVEDLRNLMSYRCSVTDQNI-LTREVTGEEIQKVLFAMPNNKSPGPDGYT 774
Query: 114 FHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGC 173
F K+ W + D + ++ F G PK N++ + LIPK + + + DY PIS
Sbjct: 775 SEFFKATWSLTGPDFIAAIQSFFVKGFLPKGLNATILALIPKKDEAIEMKDYRPISCCNV 834
Query: 174 MYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVI-DEAKVKKRGFLVFK 232
+YK++SK+L RL ++ I +NQSAF++ R L+++VL+A E++ D K K
Sbjct: 835 LYKVISKILANRLKLLLPSFILQNQSAFVKERLLMENVLLATELVKDYHKESVTPRCAMK 894
Query: 233 VDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLM 292
+D K +DSV W FL L L F + WIK CI +A+ SV VNG F + L
Sbjct: 895 IDISKAFDSVQWQFLLNTLEALNFPETFRHWIKLCISTATFSVQVNGELAGFFGSSRGLR 954
Query: 293 LGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTH 352
G L+P+LF+I LS M+ +A ++ G+ E + ++ L FADD + +
Sbjct: 955 QGCALSPYLFVICMNVLSHMIDEA-AVHRNIGYHPKCEKIGLTHLCFADDLMVFVDGHQW 1013
Query: 353 NVLAMKSMLRCFELMSGLKVNFFKSK--LAGVSVAEDVLLRYANLLHCKTTEIPFVYLGI 410
++ + ++ + F SGL+++ KS LAGVS ++ V + + ++P YLG+
Sbjct: 1014 SIEGVINVFKEFAGRSGLQISLEKSTIYLAGVSASDRV--QTLSSFPFANGQLPVRYLGL 1071
Query: 411 HVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKG 470
+ + PL+ ++ ++S W ++LS+ G++ L+ SV+ SI F++S ++LP G
Sbjct: 1072 PLLTKQMTTADYSPLIEAVKTKISSWTARSLSYAGRLALLNSVIVSIANFWMSAYRLPAG 1131
Query: 471 VASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
++ FLW G K A + WS +C+ + EG
Sbjct: 1132 CIREIEKLCSAFLWSGPVLNPKKAKIAWSSICQPKKEG 1169
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 257 bits (656), Expect = 2e-68
Identities = 168/523 (32%), Positives = 260/523 (49%), Gaps = 19/523 (3%)
Query: 2 KSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKVVKSKMKE-FFEARFT 60
KS+D+W+ GD N+KFF + R N++R LV + G K + +FE F
Sbjct: 302 KSKDKWLFGGDRNSKFFQAMVKANRTKNSLRFLVDENGNEHTLNREKGNIASVYFENLFM 361
Query: 61 NI--SGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIK 118
+ + L+G R VSEE N LT+ EI +AV+ + +P +
Sbjct: 362 SSYPANSQSALDGFKTR-VSEEMNQELTQAVTELEIHSAVFSINVESAP----------E 410
Query: 119 SFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIV 178
D + +L F T GV P+E N + + LIPK NP ++D PISL +YKI+
Sbjct: 411 KLECCQGSDYIEILGFFET-GVLPQEWNHTHLYLIPKFTNPQRMSDIRPISLCSVLYKII 469
Query: 179 SKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVK---KRGFLVFKVDY 235
SK+L+ +L K + I+ +QSAF R + D++L+A+E++ + + F+VFK D
Sbjct: 470 SKILSFKLKKHLPSIVSPSQSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMVFKTDM 529
Query: 236 EKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGD 295
K YD V W+FL +L LGF KWI WI GC+ S + SVL+NG E+ + GD
Sbjct: 530 SKAYDRVEWSFLQEILVALGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIRQGD 589
Query: 296 PLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVL 355
P++PFLF++ E L +++QA G + G V+ L F DDT + + +
Sbjct: 590 PISPFLFVLCTEALIHILQQAENSKKVSGIQFNGSGPSVNHLLFVDDTQLVCRATKSDCE 649
Query: 356 AMKSMLRCFELMSGLKVNFFKSKLA-GVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGA 414
M L + +SG +N KS + GV V ED N YLG+
Sbjct: 650 QMMLCLSQYGHISGQLINVEKSSITFGVKVDEDTKRWIKNRSGIHLEGGTGKYLGLPENL 709
Query: 415 NPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASL 474
+ K+ + + KL+ LS W KTLS GGK L++S+ ++P++ ++ F+LPKG+ +
Sbjct: 710 SGSKQDLFGYIKEKLQSHLSGWYDKTLSQGGKEILLKSIALALPVYIMTCFRLPKGLCTK 769
Query: 475 CNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEGSWESRT*SC 517
+ F W E + KI W+ K+ + G + + C
Sbjct: 770 LTSVMMDFWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQC 812
>At1g31100 hypothetical protein
Length = 1090
Score = 254 bits (649), Expect = 1e-67
Identities = 164/521 (31%), Positives = 263/521 (50%), Gaps = 38/521 (7%)
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKV-VKSKMKEFFEARF 59
Q+SR W+ +GDSNT +FH + R+ N I ++ D G D ++ +K E+F
Sbjct: 265 QRSRVTWMGEGDSNTSYFHRMADSRKAVNTIHIIIDDNGVKIDTQLGIKEHCIEYFSNLL 324
Query: 60 TNISGDGVLLEGT-----TFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNF 114
G +L++ FR S + L +F+ ++I++A + +K+ GPDG+
Sbjct: 325 GGEVGPPMLIQEDFDLLLPFRC-SHDQKKELAMSFSRQDIKSAFFSFPSNKTSGPDGFPV 383
Query: 115 HFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIG-- 172
F K W V+ ++ + +F T+ V K+ N++ + LIPK+ N +ND+ PIS
Sbjct: 384 EFFKETWSVIGTEVTDAVSEFFTSSVLLKQWNATTLVLIPKITNASKMNDFRPISCNDFG 443
Query: 173 --CMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVI---DEAKVKKRG 227
+YK+++++LT RL ++ +I QSAFL GR L ++VL+A E++ + + RG
Sbjct: 444 PITLYKVIARLLTNRLQCLLSQVISPFQSAFLPGRFLAENVLLATELVQGYNRQNIDPRG 503
Query: 228 FLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*M 287
L KVD K +DS+ W+F+ L+ +G +++ WI CI + + SV VNG+ F
Sbjct: 504 ML--KVDLRKAFDSIRWDFIISALKAIGIPDRFVYWITQCISTPTFSVCVNGNTGGFFKS 561
Query: 288 EKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIY 347
+ L G+PL+PFLF++ E S ++ + Y + + +S L FADD + +
Sbjct: 562 TRGLRQGNPLSPFLFVLAMEVFSSLLNSRFQAG-YIHYHPKTSPLSISHLMFADDIMVFF 620
Query: 348 EPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVY 407
+ + ++ + L F SGL +N E L A L + + I
Sbjct: 621 DGGSSSLHGISEALEDFAFWSGLVLN-----------REKTHLYLAGLDRIEASTI---- 665
Query: 408 LGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKL 467
A + + PLL KL KR W K LSF G+V LI SV+S I F++S F L
Sbjct: 666 ------ARKLRIAEYGPLLEKLAKRFRSWSVKCLSFAGRVQLIASVISGIINFWISTFIL 719
Query: 468 PKGVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
PKG + +FLW G +K A V WS+VC + EG
Sbjct: 720 PKGCVKRIEALCARFLWSGNIDVKKGAKVAWSEVCLPKEEG 760
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.148 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,391,168
Number of Sequences: 26719
Number of extensions: 772651
Number of successful extensions: 2548
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 2294
Number of HSP's gapped (non-prelim): 119
length of query: 839
length of database: 11,318,596
effective HSP length: 108
effective length of query: 731
effective length of database: 8,432,944
effective search space: 6164482064
effective search space used: 6164482064
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0154.9


