

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0151.7
(1675 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
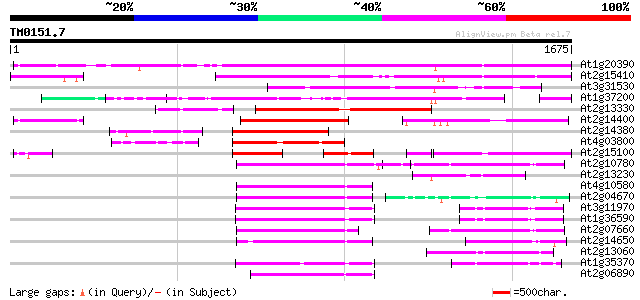
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 1058 0.0
At2g15410 putative retroelement pol polyprotein 826 0.0
At3g31530 hypothetical protein 570 e-162
At1g37200 hypothetical protein 558 e-158
At2g13330 F14O4.9 454 e-127
At2g14400 putative retroelement pol polyprotein 385 e-106
At2g14380 putative retroelement pol polyprotein 354 3e-97
At4g03800 322 9e-88
At2g15100 putative retroelement pol polyprotein 291 3e-78
At2g10780 pseudogene 231 3e-60
At2g13230 pseudogene 228 2e-59
At4g10580 putative reverse-transcriptase -like protein 223 7e-58
At2g04670 putative retroelement pol polyprotein 220 6e-57
At3g11970 hypothetical protein 214 4e-55
At1g36590 hypothetical protein 214 4e-55
At2g07660 putative retroelement pol polyprotein 213 6e-55
At2g14650 putative retroelement pol polyprotein 210 5e-54
At2g13060 pseudogene 181 4e-45
At1g35370 hypothetical protein 180 7e-45
At2g06890 putative retroelement integrase 179 2e-44
>At1g20390 hypothetical protein
Length = 1791
Score = 1058 bits (2737), Expect = 0.0
Identities = 648/1718 (37%), Positives = 934/1718 (53%), Gaps = 136/1718 (7%)
Query: 11 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKM----VIIAASDAVKCRMFPS 66
PF+ + +V+I K + L+SY+G +DPK+ L FN + + I DA +C++F
Sbjct: 156 PFTRRITNVSIRGAQK-IKLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIE 214
Query: 67 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 126
A WF+ L SI +F +S FL ++ + DL++I Q ESL+ ++
Sbjct: 215 HLTGPAHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFV 274
Query: 127 ARYS--AASVKVEDEEPRACALAFKNGL-LPGGLNSKLTR*PARSMEEMRARASTYILDE 183
R+ ++ V DE A +A +N + +T ++E+ RAS +I E
Sbjct: 275 DRFKLVVTNITVPDE---AAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELE 331
Query: 184 EDDAFKRKRAKLEKGDTSPKRAKKDKNGEDKGDGKQQRQDKGKAALRPTKEQLYPRRDDY 243
E+ ++ K K A K G D + +Q D+ +A
Sbjct: 332 EEKLILARKHNSTKTPAC-KDAVVIKVGPDDSNEPRQHLDRNPSA--------------- 375
Query: 244 EQRRPWQSKSHRQREEADMVMNTELTDMLRGARGANLVDEPEA-PKYQSRDANPKKWCEF 302
R+ +++TE D + D P A P Y CE+
Sbjct: 376 ------------GRKPTSFLVSTETPDAKPWNKYIRDADSPAAGPMY----------CEY 413
Query: 303 HRSAGHGTDDCWTLQREIDKLIRAG---YQGNRQGQWRNNGDHNKTHKREEERVDTKGKK 359
H+S H T++C LQ + ++G + +R N N+T R+ TK
Sbjct: 414 HKSRAHSTENCRFLQGLLMAKYKSGGITIECDRPPINNKNQRRNETTARQYLNDQTKPPT 473
Query: 360 KQESAAIATRGADDTFA--QHSGPPVGT-------INTIAGGFGGGGDTHAARKRHVRAV 410
E I + ADD A Q +G + ++ I GG D+ + K++ +
Sbjct: 474 PAEQGIITS--ADDPAAKRQRNGKAIAAEPVVVRQVHVIMGGLQNCSDSVRSIKQYRKKA 531
Query: 411 NSVHEVAFGFV-----HPDRTISMA----DFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQ 461
V VA+ +P+++ ++ D EG+ +DP+VV+L ++ V RVL+D
Sbjct: 532 EMV--VAWPSSTSTTRNPNQSAPISFTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDT 589
Query: 462 GSSADIIYGDAFDKLGLTDKDLTPDAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKV 521
GSS D+I+ D + +TD+ + P + L GF G+ VM G I L G + V
Sbjct: 590 GSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTIKLPIFVG----GLIAWV 645
Query: 522 RYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMARECYNNC 581
+++V+ A YNVI+G ++++ A+ ST H VK+P +N
Sbjct: 646 KFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPT------------------HNGI 687
Query: 582 LNLYGKKSALVGHRCYEIEASDENLDPRGEGRVNRPTPIEDTKALKFGDRTLKIGTRLTE 641
L K A R YE + L ++ P R + +G ++
Sbjct: 688 FTLRAPKEAKTPSRSYE----ESELCRTEMVNIDESDPT----------RCVGVGAEISP 733
Query: 642 EQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHRLALNPSVKPVSQLRRRLGGDKGKAV 701
L LL N FAWS +DM GIDP H L ++P+ KPV Q RR+LG ++ +AV
Sbjct: 734 SIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLGPERARAV 793
Query: 702 QQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKWLMCVDYTDLNKACPKDSYPLPSIDS 761
+EV+KLL A I EVKYP WLAN V+VKK NGKW +CVDYTDLNKACPKDSYPLP ID
Sbjct: 794 NEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPLPHIDR 853
Query: 762 LVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAFMTARVNYCYRTMPFGLKNAGATYQR 821
LV+ SGN LLS MDA+SGY+QI MH D++KT+F+T R YCY+ M FGLKNAGATYQR
Sbjct: 854 LVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQR 913
Query: 822 LMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCSFGVQG 881
++++ A Q+GR +EVY+DDM+VKS++ DH + L + F + + M+LNP KC+FGV
Sbjct: 914 FVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVTS 973
Query: 882 GKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFF 941
G+FLG+++T RGIE NP + +AI ++ +P N +EVQRLTGRIAAL+RF+ +S D+ PF+
Sbjct: 974 GEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDKCLPFY 1033
Query: 942 KCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKPMQGHPLHLYFAVSDSALSSVILQEG 1001
L++ A F+W + EEAF +LK+ LS+PPIL KP G L+LY AVSD A+SSV+++E
Sbjct: 1034 NLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHAVSSVLVRED 1093
Query: 1002 DGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTARRLRPYFQSFPVRVRTDLPLRQVLQ 1061
GE R I++ S +L AE RY IEKAALAV+ +AR+LRPYFQS + V TD PLR L
Sbjct: 1094 RGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLTDQPLRVALH 1153
Query: 1062 KPDMSGRLVAWSVELSEYGLQYDKRGKVGAQSLADFVVELTPDRFERV-----DTQWTLF 1116
P SGR+ W+VELSEY + + R + +Q LADF++EL ER +W+L+
Sbjct: 1154 SPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLADFLIELPLQSAERAVSGNRGEEWSLY 1213
Query: 1117 VDGSSNSSGSGTGVTLEGPGDLVLEQSLKFEFKATNNQAEYEALIAGLKLAREVKIGSLL 1176
VDGSS++ GSG G+ L P VLEQS + F ATNN AEYE LIAGL+LA ++I ++
Sbjct: 1214 VDGSSSARGSGIGIRLVSPTAEVLEQSFRLRFVATNNVAEYEVLIAGLRLAAGMQITTIH 1273
Query: 1177 IRTDSQLVENQVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEYVPRTENQRADALAKLA 1236
TDSQL+ Q+ G ++ K+ + YL+ V+ + F+ + +PR +N ADALA LA
Sbjct: 1274 AFTDSQLIAGQLSGEYEAKNEKMDAYLKIVQLMTKDFENFKLSKIPRGDNAPADALAALA 1333
Query: 1237 STRKPDNNRSVIQETLAFPSIEG----ELMACVDRG--------ATWMGPILSILAGDPA 1284
T D R + E++ PSI+ E++ + W I L+
Sbjct: 1334 LTSDSDLRRIIPVESIDKPSIDSTDAVEIVNTIRSSNAPDPADPTDWRVEIRDYLSDGTL 1393
Query: 1285 EVEQCTKEQRR-EASHYTLIDGHLYRRGFSAPLLKCVPPEKYEAIMSEVHEGMCASHIGG 1343
++ T + R +A+ YTL+ HL + +L C+ + IM E HEG +H GG
Sbjct: 1394 PSDKWTARRLRIKAAKYTLMKEHLLKVSAFGAMLNCLHGTEINEIMKETHEGAAGNHSGG 1453
Query: 1344 RSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMWGVD 1403
R+LA K+ + GFYWPT+ DC F KC++CQ A P + L AP+PF W +D
Sbjct: 1454 RALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTELLRAGVAPYPFMRWAMD 1513
Query: 1404 LVGPFPIARSQMKFILVAVDYFTKWIEAEPLAKITSAKIVNFYWKRIVCRFGIPRAIVSD 1463
+VGP P +R Q +FILV DYFTKW+EAE A I + + NF WK I+CR G+P I++D
Sbjct: 1514 IVGPMPASR-QKRFILVMTDYFTKWVEAESYATIRANDVQNFVWKFIICRHGLPYEIITD 1572
Query: 1464 NGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEE 1523
NG+QF S FC I++ ++ +PQ NGQ E N+ IL GL++RL E KGAW +E
Sbjct: 1573 NGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNKTILSGLKKRLDEKKGAWADE 1632
Query: 1524 LPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWRTEPDFEGE--NQANMAAEL 1581
L VLWSY TT +S T +TPF YG++AM PAE+ + R + N M L
Sbjct: 1633 LDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYSSLRRSMMVKNPELNDRMMLDRL 1692
Query: 1582 DLLSETRDEAHIRETAMKQRVAAKFNSRVRVRDMQVGDLVLKW----RSGASGNKLTPNW 1637
D L E R+ A R + A +N +V R VGDLVL+ + + KL NW
Sbjct: 1693 DDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGDLVLRKVFENTAEINAGKLGANW 1752
Query: 1638 EGPYRIIKVLGNEAYHLEELDGRRLPRSFNGLSLRYYY 1675
EG Y++ K++ Y L + G +PR++N + L+ YY
Sbjct: 1753 EGSYQVSKIVRPGDYELLTMSGTAVPRTWNSMHLKRYY 1790
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 826 bits (2134), Expect = 0.0
Identities = 470/1138 (41%), Positives = 641/1138 (56%), Gaps = 137/1138 (12%)
Query: 616 RPTPIEDTKA---LKFGDRTLKIGTR--LTEEQETRLTKLLGENLDLFAWSCKDMPGIDP 670
RP P + T + D + +G R L E + L L +N FAWS +DMPGID
Sbjct: 708 RPNPQKGTVVQVNIDESDPSRCVGIRIDLPSELQNELVNFLRQNAATFAWSVEDMPGIDS 767
Query: 671 NFICHRLALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVK 730
CH L ++P+ KP+ Q RR+LG D+ K V +EV KLL A I EV+YP WL N V+VK
Sbjct: 768 AVTCHELNVDPTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVK 827
Query: 731 KANGKWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPAD 790
K NGKW +C+D+TDLNKACPKDS+PLP ID LV+ +GNELLS MDA+SGY+QI MH D
Sbjct: 828 KKNGKWRVCIDFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQND 887
Query: 791 EDKTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGL 850
+KT F+T + YCY+ MPFGLKNAGATY RL++++F Q+ +MEVY+DDM+VKS+R
Sbjct: 888 REKTVFITDQGTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAE 947
Query: 851 DHHQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNP 910
+H L + F + ++NM+LNP KC+FGV G+FLG+++T RGIE NP + AI + +P
Sbjct: 948 EHITHLRQCFQVLNRYNMKLNPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSP 1007
Query: 911 SNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSP 970
N +EVQRL GRIAAL+RF+ +S D+ PF++ L+ N FEW +CEE
Sbjct: 1008 RNTREVQRLIGRIAALNRFISRSTDKCLPFYQLLRANKRFEWDEKCEE------------ 1055
Query: 971 PILSKPMQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAAL 1030
G L+LY AVS SA+S V+++E GE I++VS TL GAE+RY +EK A
Sbjct: 1056 --------GETLYLYIAVSTSAVSGVLVREDRGEQHPIFYVSKTLDGAELRYPTLEKLAF 1107
Query: 1031 AVLVTARRLRPYFQSFPVRVRTDLPLRQVLQKPDMSGRLVAWSVELSEYGLQYDKRGKVG 1090
AV+++AR+LRPYF+S+ V V T+ PLR +L P SGRL W+VELSEY + Y R
Sbjct: 1108 AVVISARKLRPYFKSYTVEVLTNQPLRTILHSPSQSGRLAKWAVELSEYDIAYKNR---- 1163
Query: 1091 AQSLADFVVELTPDRFERVDTQWTLFVDGSSNSSGSGTGVTLEGPGDLVLEQSLKFEFKA 1150
T + R T+ S V L F A
Sbjct: 1164 -----------TCAKSHRAPTR-------PYRRSIPCRKVDLA-------------RFPA 1192
Query: 1151 TNNQAEYEALIAGLKLAREVKIGSLLIRTDSQLVENQVKGTFQVKDPNLIKYLE*VRYLM 1210
+NN+AEYEALIAGL+LA +++ + DSQLV +Q G ++ K+ + YL+ VR L
Sbjct: 1193 SNNEAEYEALIAGLRLAHGIEVKKIQAYCDSQLVASQFSGNYEAKNERMDAYLKVVRELS 1252
Query: 1211 TLFQEVVVEYVPRTENQRADALAKLASTRKPDNNRSVIQETLAFPS--IEGEL-----MA 1263
F+ + +PR++N ADALA LAST PD R + E + PS I+G +
Sbjct: 1253 YNFEVFELTKIPRSDNAPADALAVLASTSDPDLRRVIPVECIDVPSIKIQGTITKPSEYT 1312
Query: 1264 CVDRGATWMGPILSIL------------------AGDPAEVEQCTKEQ------------ 1293
+D A + ++ IL + DP + C +
Sbjct: 1313 VIDLTAEQLSIVMVILPPATDRSEDAGPSGNLVCSSDPGQSSGCDRSDDSNRSADSDSPA 1372
Query: 1294 ---------------------------RREASHYTLIDGHLYRRGFSAPLLKCVPPEKYE 1326
R ++ YTL HL R + LL C+ E+ +
Sbjct: 1373 STNWIEEIRSYIADRIVPKDKWAARRLRARSAQYTLPHEHLLRWSATGVLLSCLDDEEAQ 1432
Query: 1327 AIMSEVHEGMCASHIGGRSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPK 1386
+M E+HEG +H GGR+LA K+ + G YWPT+ DC FV +C+KCQ A P +
Sbjct: 1433 QVMREIHEGAGGNHSGGRALALKIRKHGQYWPTMNADCEKFVARCEKCQGHAPFIHIPSE 1492
Query: 1387 ELVTMSAPWPFAMWGVDLVGPFPIARSQMKFILVAVDYFTKWIEAEPLAKITSAKIVNFY 1446
L T P+PF W +D++ PFP +R Q K+ILV DYFTKW EAE A+I S ++ NF
Sbjct: 1493 VLQTAMPPYPFMRWAMDIIRPFPASR-QKKYILVMTDYFTKWFEAESYARIQSKEVQNFV 1551
Query: 1447 WKRIVCRFGIPRAIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVIL 1506
WK I+C G+P IV+DNGTQF+S Q FC + I++ ++ +PQ NGQ E N+ IL
Sbjct: 1552 WKNIICHHGLPYEIVTDNGTQFTSLQFEGFCAKWKIRLSKSTPRYPQCNGQAEATNKTIL 1611
Query: 1507 RGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWR-- 1564
GL++RL E KGAW +EL VLWSY TT + +T TPF +TYG++A+ P E T R
Sbjct: 1612 DGLKKRLDEKKGAWADELDGVLWSYRTTPRRSTDRTPFSLTYGMEALAPCEAGLPTVRRS 1671
Query: 1565 ---TEPDFEGENQANMAAELDLLSETRDEAHIRETAMKQRVAAKFNSRVRVRDMQVGDLV 1621
+P N + LD L E RD+A +R +Q A +N +V+ R GDLV
Sbjct: 1672 MLINDPTL---NDQMLLDNLDTLEEQRDQALLRIQNYQQAAAKFYNKKVKNRLFAEGDLV 1728
Query: 1622 LKW----RSGASGNKLTPNWEGPYRIIKVLGNEAYHLEELDGRRLPRSFNGLSLRYYY 1675
L+ KL +WEGPY I KV+ Y L +DG +PRS+N +L+ YY
Sbjct: 1729 LRKVFENTVEQDAGKLGAHWEGPYLISKVVKPGVYELLTMDGTPIPRSWNSANLKRYY 1786
Score = 51.6 bits (122), Expect = 4e-06
Identities = 58/242 (23%), Positives = 102/242 (41%), Gaps = 27/242 (11%)
Query: 2 EADSVAEFR---PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASD- 57
E D V E PF+ + V I ++ + L +Y G DP+ L + + SD
Sbjct: 225 EVDKVLEETQRSPFTSCISDVRIR-HISKIKLANYEGLVDPRPFLTSVSIAIGRAHFSDE 283
Query: 58 ---AVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNI 114
A C++F A+ WF+ L SI + ++ FL + + + DL+ +
Sbjct: 284 DRDAGSCQLFVKHLSGAALTWFSRLEANSIDSVHALTTSFLKNYGVFMEKGASNVDLWTM 343
Query: 115 RQQEGESLKEYMARYSAASVKVEDEEPRACALAFKNGLLPGGLNSKLT-------R*PAR 167
Q ESL+ ++ R+ V + A A A +N L G + T + A+
Sbjct: 344 AQTAKESLRSFIGRFKEIVTSVATPDDAAIA-ALRNALWHGDSSRCTTLCKPFHRQGSAK 402
Query: 168 S-MEEMRARASTYILDEEDDAFKRKRAKLEK------GDTSPKRAKK----DKNGEDKGD 216
+ + + + + + + DA K KL+K GDT+P+ K +++ + KGD
Sbjct: 403 APIAKEKPKEGHHESRQHYDADYAKEEKLKKGTSYYVGDTAPRSDKPWNKWNRSADAKGD 462
Query: 217 GK 218
K
Sbjct: 463 QK 464
>At3g31530 hypothetical protein
Length = 831
Score = 570 bits (1468), Expect = e-162
Identities = 332/838 (39%), Positives = 474/838 (55%), Gaps = 103/838 (12%)
Query: 769 NELL--SLMDAYSGYHQIRMHPADEDKTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRV 826
N+LL S +++ + + M+P D++KTAF T + +CYR MPFGLKNAGATY+R ++++
Sbjct: 77 NQLLETSYCHSWTLFVVMMMNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKI 136
Query: 827 FAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLG 886
FA Q+G+ MEVY+DDM+VKS+ DH L E F ++ +N++LNP KC FGV+ G
Sbjct: 137 FALQIGKTMEVYIDDMLVKSMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRSG---- 192
Query: 887 FMITSRGIEINPDKCKAIQQMKNPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLQK 946
IE NP + +A+ M +P N +EVQ LTGR+AAL+RF+ +S ++ F+ L++
Sbjct: 193 -------IEANPKQIEALLGMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRR 245
Query: 947 NAAFEWTAECEEAFVRLKELLSSPPILSKPMQGHPLHLYFAVSDSALSSVILQEGDGEHR 1006
N FEWT CEEAF LK+ L++PPIL+KP+ G P +LY AVSD+A+S +++E GE +
Sbjct: 246 NKKFEWTTRCEEAFQELKKYLATPPILAKPVIGEPQYLYVAVSDTAVSGELVREDRGEQK 305
Query: 1007 VIYFVSHTLQGAEVRYKKIEKAALAVLVTARRLRPYFQSFPVRVRTDLPLRQVLQKPDMS 1066
+I++VS T AE RY ++EK ALAV+++A++LRPYFQS + V +PLR +L P S
Sbjct: 306 LIFYVSQTFTSAESRYPQMEKLALAVVMSAQKLRPYFQSHSIIVMGSMPLRVILHSPSQS 365
Query: 1067 GRLVAWSVELSEYGLQYDKRGKVGAQSLADFVVELTPD--RFERVDTQWTLFVDGSSNSS 1124
GRL W++ELSEY ++Y + +Q LADF+VEL R +DT W L VDGSS+
Sbjct: 366 GRLAKWTIELSEYDIEYQNKTCAKSQVLADFIVELPTKEARENPLDTTWLLHVDGSSSKQ 425
Query: 1125 GSGTGVTLEGPGDLVLEQSLKFEFKATNNQAEYEALIAGLKLAREVKIGSLLIRTDSQLV 1184
GSG G+ L P +ATNN AEYEAL+AGL LA +KIG DSQL+
Sbjct: 426 GSGVGIRLTSPTG-----------EATNNVAEYEALVAGLNLAWGLKIGKTRAFCDSQLI 474
Query: 1185 ENQVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEYVPRTENQRADALAKLASTRKPDNN 1244
NQ G + +D + YL V+ L F E + +PR EN ADALA LAST
Sbjct: 475 ANQFNGEYTTQDKKMEAYLIHVQNLAKNFDEFELTRIPRGENTSADALAALASTSDTSLK 534
Query: 1245 RSVIQETLAFPSIEGELMACV-------------DRGATWMGPILSILA-GDPAEVEQCT 1290
R + E + PSIE V D + WM PI+S ++ G +
Sbjct: 535 RVIPVEFIEKPSIELGKEEHVLPIQISADQDDPDDCNSEWMEPIISYISEGKLPSDKWKA 594
Query: 1291 KEQRREASHYTLIDGHLYRRGFSAPLLKCVPPEKYEAIMSEVHEGMCASHIGGRSLACKV 1350
++ + +A+ + L+D LY+ S PL+ CV E IM E+H G A
Sbjct: 595 RKLKAQAARFVLVDTKLYKWRLSGPLMTCVEAEAICKIMKEIHSGSHA------------ 642
Query: 1351 LRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMWGVDLVGPFPI 1410
PT+ + P + L ++++P+PF W +D++GP
Sbjct: 643 -------PTIHQ---------------------PAELLSSIASPYPFMRWSMDIIGPMHP 674
Query: 1411 ARSQMKFILVAVDYFTKWIEAEPLAKITSAKIVNFYWKRIVCRFGIPRAIVSDNGTQFSS 1470
++ Q K +LV DYF+KWIEAE A I A++ NF K I+CR GIP IV+DNG+QF S
Sbjct: 675 SK-QKKLVLVLTDYFSKWIEAESYASIKDAQVENFVLKHILCRHGIPYEIVTDNGSQFIS 733
Query: 1471 NQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLWS 1530
+ + FC + GI++ ++ +PQ NGQ E AN EL VLW
Sbjct: 734 TRFQGFCDKWGIRLSKSTPRYPQGNGQAEAAN--------------------ELEGVLWL 773
Query: 1531 YNTTVQSTTRETPFRMTYGVDAMLPAE--IDNFTWRTEPDFEGENQANMAAELDLLSE 1586
+ TT + TRETPF YG + ++PAE + + P+ + +N ELDL+ E
Sbjct: 774 HRTTPRRATRETPFASIYGTECVIPAEMIVPSLRRSLSPENDPDNTQTPLDELDLIDE 831
Score = 35.8 bits (81), Expect = 0.22
Identities = 21/51 (41%), Positives = 27/51 (52%)
Query: 532 YNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMARECYNNCL 582
YN IIG LN+ AV ST HL +K+P S KL ++ A + N L
Sbjct: 30 YNAIIGTPWLNQFRAVASTYHLCLKFPTSDSVWEKLGHERAEAVKAENQLL 80
>At1g37200 hypothetical protein
Length = 1564
Score = 558 bits (1438), Expect = e-158
Identities = 408/1268 (32%), Positives = 602/1268 (47%), Gaps = 206/1268 (16%)
Query: 285 EAPKYQSRDANPKKWCEFHRSAGHGTDDCWTLQREIDKLIRAGYQGNRQGQWRNNGDHNK 344
EA + +S + + K+C++H GH T++C R + KLI AG + + G + K
Sbjct: 309 EAAEDESPELDLGKYCKYHNKRGHSTEEC----RAVKKLIAAGGKTKK-------GSNPK 357
Query: 345 THKREEERVDTKGKKKQESAAIATRGADDTFAQHSGPPVGT--INTIAGGFGGGGDTHAA 402
+ + + KQ+ G D S PP I+ + GG T
Sbjct: 358 VETPPPDEQEEEQTPKQKKRERTLEGGD------SPPPARRERIDLVFTELDLGGKTG-- 409
Query: 403 RKRHVRAVNSV-HEVAFGFVHPDRTISMADFEGIKPHKDDPIVVQLRMNSFNVRRVLLDQ 461
R V S H+ F + E + P + D I+ ++ + ++ + Q
Sbjct: 410 --RSVTTPPSAPHKKNMRFPLRLSKFCRSATEHLSPKRIDFIMGGSQLCNDSINSIKTHQ 467
Query: 462 GSSADIIYGDAF-----DKLGL-----TDKDLTPDAGTLVGF-AGEQVMVRGYIDLDTIF 510
+ G + K+ L TD D D ++ G + R I +DT
Sbjct: 468 RKADSYTKGKSLMMGPDHKITLWESETTDLDKPHDDAIVIRIDVGNYKLSR--IMIDTGS 525
Query: 511 GEDECARVLKVRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVD 570
D+ A +N I+GR L+ + AV ST H +K+P G + +
Sbjct: 526 SVDK---------------APFNAILGRPWLHAMKAVPSTYHQCIKFPSEKG-IAVIYGS 569
Query: 571 QKMARECYNNCLNLYGKKSALVGHRCYEIEASDENLDPRGEGRVNRPT---PIEDTKALK 627
Q+ +R CY L KK+ LV + ++ L R + P+ P + +
Sbjct: 570 QRSSRRCYMGSYELI-KKAELV------VLMIEDKLAEMKTVRSSDPSQCGPQKSSITQV 622
Query: 628 FGDRT-----LKIGTRLTEEQETRLTKLLGENLDLFAWSCKDMPGIDPNFICHRLALNPS 682
+ D + + +G L L EN D FAWS ++ GI H L ++P+
Sbjct: 623 YIDESDPKQCVGVGQDLDPAIREDFITFLKENKDSFAWSSANLQGISLEVTSHELNVDPT 682
Query: 683 VKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKWLMCVDY 742
+P+ Q RR+LG ++ KAVQ EVD+LL IREVKYP WLAN V+VK NGKW +C+D+
Sbjct: 683 YRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPDWLANPVVVKNKNGKWRVCIDF 742
Query: 743 TDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAFMTARVN 802
DLNKACPKDS+PLP ID LV +G+ELLS MDA+SGY+QI M P D++KTAF+T
Sbjct: 743 MDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQILMRPDDQEKTAFITD--- 799
Query: 803 YCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGE 862
CY+ MPFGLKN GATYQRL++R+FA Q+G+ ME+Y+DDM+VKS DH L E F
Sbjct: 800 -CYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVKSAHEKDHLPQLRECFKI 858
Query: 863 IRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEVQRLTGR 922
+ K M+ +NP+KC
Sbjct: 859 LNKFEMK--------------------------LNPEKCS-------------------- 872
Query: 923 IAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKPMQGHPL 982
F SG+ F L E + AF+ ++ S +P +
Sbjct: 873 ------FGVPSGE----FLGYLVTQRGIEANPKQIAAFI---DMPSPKMARDRPYRRPQQ 919
Query: 983 HLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTARRLRPY 1042
+Y A D EG + IY+VS TL AE Y+ +EK ALAV+++AR+LR Y
Sbjct: 920 VIYRAPPD--------HEGQ---KPIYYVSKTLIDAETHYRAMEKLALAVVMSARKLRQY 968
Query: 1043 FQSFPVRVRTDLPLRQVLQKPDMSGRLVAWSVELSEYGLQYDKRGKVGAQSLADFVVELT 1102
QS P+ V T P+R +L S RL W++ELSEY ++Y R AQ LADFV+EL
Sbjct: 969 LQSHPIVVMTSQPIRTILHSTTQSSRLAKWAIELSEYDIEYRTRTSFKAQVLADFVIELP 1028
Query: 1103 PDRFERVDT--QWTLFVDGSSNSSGSGTGVTLEGPGDLVLEQSLKFEFKATNNQAEYEAL 1160
+ ++ +W L VDGSSN GSG G+ L P V+EQS + F A+NN++EYEAL
Sbjct: 1029 LADLDGTNSNKKWLLHVDGSSNRQGSGEGIQLTSPTGEVIEQSFRLGFNASNNESEYEAL 1088
Query: 1161 IAGLKLAREVKIGSLLIRTDSQLVENQVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEY 1220
I G+KLA+ ++I + +DSQLV +Q G ++ KD + YLE V+ L F+ +
Sbjct: 1089 IDGIKLAQGMRIRDIHAHSDSQLVTSQFHGEYEAKDERMEAYLELVKTLTQQFESFELTR 1148
Query: 1221 VPRTENQRADALAKLASTRKPDNNRSVIQETLAFPSIE---------------------- 1258
+PR EN DALA LAST P R + E + PSI+
Sbjct: 1149 IPRGENTSTDALAALASTLDPFVKRIIPVEGIEHPSIDLTVKHAGMETSATCNFTRVTRS 1208
Query: 1259 -----GELMACVDRGAT----------------------WMGPILS-ILAGDPAEVEQCT 1290
L A + GA W PI+ I G + T
Sbjct: 1209 TTAAARALAAAAEAGAAEEEAKLRTPEQLASYEPEPYNDWRIPIIDYIERGITPPDKWET 1268
Query: 1291 KEQRREASHYTLIDGHLYRRGFSAPLLKCVPPEKYEAIMSEVHEGMCASHIGGRSLACKV 1350
++ + +++ Y +++G L +R + P + C ++ + +M +HEG C SH GR+L C +
Sbjct: 1269 RKLKAQSARYCIMEGRLMKRSVAGPYMVCTYGQQTKDLMKSMHEGQCGSHYSGRTLRCIM 1328
Query: 1351 LRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMWGVDLVGPFPI 1410
L DC+ +C KCQ A PP+E+ ++S+P+PF W +D+V P
Sbjct: 1329 L----------ADCIAHSLRCDKCQRHAPTLHQPPEEMSSISSPYPFMKWSMDVVRPMEA 1378
Query: 1411 A--RSQMKFILVAVDYFTKWIEAEPLAKITSAKIVNFYWKRIVCRFGIPRAIVSDNGTQF 1468
+ + ++K +LV DY TKWIE + ++T ++ F + IV R GIP IV+DNG
Sbjct: 1379 SGGKKRLKNLLVLTDYSTKWIEGKAFQQVTEKQVEVFLLENIVYRHGIPYEIVTDNGLTS 1438
Query: 1469 SSNQTREF 1476
+++ F
Sbjct: 1439 PREKSKHF 1446
Score = 55.5 bits (132), Expect = 3e-07
Identities = 33/99 (33%), Positives = 54/99 (54%), Gaps = 5/99 (5%)
Query: 1582 DLLSETRDEAHIRETAMKQRVAAKFNSRVRVRDMQVGDLVLKW----RSGASGNKLTPNW 1637
D + E RD A I+ +Q +NS +++R +VG+LVL+ + KL NW
Sbjct: 1465 DFIDERRDHAMIQVQNYQQPAVRYYNSNIKIRRFEVGELVLRKVFSNTRELNAGKLGTNW 1524
Query: 1638 EGPYRIIKVLGNEAYHLEEL-DGRRLPRSFNGLSLRYYY 1675
EGPYRI +V+ + Y L ++ +G R +N + L+ Y+
Sbjct: 1525 EGPYRITEVVRDGIYKLVKVFNGVPELRPWNAMHLKKYH 1563
Score = 52.4 bits (124), Expect = 2e-06
Identities = 81/377 (21%), Positives = 147/377 (38%), Gaps = 28/377 (7%)
Query: 94 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASVKVEDEEPRACALAFKNGL- 152
FL Q S Q + DL+++ + ES + Y+ ++ A K+ + A +NGL
Sbjct: 176 FLKQDSVLIEQRTSEADLWSLTLLQNESHRSYVEKFKAIKSKIANLNEEVAIAALRNGLW 235
Query: 153 LPGGLNSKLTR*PARSMEEMRARASTYILDEEDDAFKRKRAKLEKGDTSPKRAKKDKNGE 212
+LT S+++ +A + EE+ A + K K +P K E
Sbjct: 236 FSSRFREELTVRQPISLDDALHKALHFAKGEEELAVLALKFKESKTQNAPLAIKTPFKKE 295
Query: 213 DKGDGKQQRQDKGKAALRPTKEQLYPRRDDYEQRRPWQSKSHRQREEADMVMNTELTDML 272
++ G+ +AA + E + Y +R ++ R + +
Sbjct: 296 NQTQGQHTLFAIEEAAEDESPELDLGKYCKYHNKRGHSTEECR-------AVKKLIAAGG 348
Query: 273 RGARGANLVDEPEAPKYQSRDANPKKWCEFHRSAGHGTDDCWTLQREIDKLIRAGYQGNR 332
+ +G+N E P Q + PK+ + R+ G + ID + G +
Sbjct: 349 KTKKGSNPKVETPPPDEQEEEQTPKQ-KKRERTLEGGDSPPPARRERIDLVFTELDLGGK 407
Query: 333 QGQWRNNGDHNKTHKREEERVDTKGKKKQESAAIATRGADDTFAQHSGPPVGTINTIAGG 392
G R+ ++ R + K SA +H P I+ I GG
Sbjct: 408 TG--RSVTTPPSAPHKKNMRFPLRLSKFCRSA-----------TEHLSPK--RIDFIMGG 452
Query: 393 FGGGGDTHAARKRHVRAVNSVHEVAFGFVHPDRTISMADFEGI---KPHKDDPIVVQLRM 449
D+ + K H R +S + + PD I++ + E KPH DD IV+++ +
Sbjct: 453 SQLCNDSINSIKTHQRKADSYTKGKSLMMGPDHKITLWESETTDLDKPH-DDAIVIRIDV 511
Query: 450 NSFNVRRVLLDQGSSAD 466
++ + R+++D GSS D
Sbjct: 512 GNYKLSRIMIDTGSSVD 528
>At2g13330 F14O4.9
Length = 889
Score = 454 bits (1169), Expect = e-127
Identities = 241/528 (45%), Positives = 328/528 (61%), Gaps = 32/528 (6%)
Query: 733 NGKWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADED 792
NGKW +C+D+ DLNKACPKDS+PLP ID LV+ +E LS MDA+ GY+QI M ++
Sbjct: 291 NGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQE 350
Query: 793 KTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDH 852
KTAF+T R Y Y+ MPFGLKNAG TYQRL++R+F Q+G+ +EVY+DDM+VKS DH
Sbjct: 351 KTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDH 410
Query: 853 HQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSN 912
L E F + K M+LNPEKCSF VQ +FL +++T RGIE NP + A +M +P
Sbjct: 411 VPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSPKM 470
Query: 913 VKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPI 972
+EVQRLTGRI AL+ F+ +S ++ PF++ L+K F+W +CE+AF +LK L+ PPI
Sbjct: 471 AREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTEPPI 530
Query: 973 LSKPMQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAV 1032
L+KP +G PL+LY + R +EK AL V
Sbjct: 531 LAKPEKGEPLYLY------------------------------TNRDKRCPAMEKLALTV 560
Query: 1033 LVTARRLRPYFQSFPVRVRTDLPLRQVLQKPDMSGRLVAWSVELSEYGLQYDKRGKVGAQ 1092
+++ R+LR YFQS P+ V T P+R +L SGRL W++EL EY ++Y R + AQ
Sbjct: 561 VMSVRKLRLYFQSHPIVVMTSQPIRTILHSLTQSGRLAKWAIELREYDIEYRTRTSLKAQ 620
Query: 1093 SLADFVVELTPDRFERVDT--QWTLFVDGSSNSSGSGTGVTLEGPGDLVLEQSLKFEFKA 1150
LADFV++L + ++ +W L VDGSSN GSG G+ L P V+EQSL+ F A
Sbjct: 621 VLADFVIKLPLADLDGTNSNKKWLLHVDGSSNRQGSGVGIQLTSPTGEVIEQSLQLGFNA 680
Query: 1151 TNNQAEYEALIAGLKLAREVKIGSLLIRTDSQLVENQVKGTFQVKDPNLIKYLE*VRYLM 1210
+NN++EYEALIAG+KLA+E I + +DSQLV +Q G ++ KD + YLE V+ L
Sbjct: 681 SNNESEYEALIAGIKLAQEKGIREIHAYSDSQLVTSQFHGEYEAKDERMEAYLELVKTLA 740
Query: 1211 TLFQEVVVEYVPRTENQRADALAKLASTRKPDNNRSVIQETLAFPSIE 1258
F+ + +PR EN AD LA LAST P R + E + SI+
Sbjct: 741 QQFESFKLTRIPRGENTSADTLAALASTSDPFVKRIIPVEGIEHTSID 788
Score = 87.0 bits (214), Expect = 8e-17
Identities = 76/244 (31%), Positives = 116/244 (47%), Gaps = 27/244 (11%)
Query: 436 KPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDKDLTPDAGTLVGFAG 495
KPH DD +V+++ + ++ + +++D GSS D+++ DAF + G D L L GFAG
Sbjct: 59 KPH-DDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTGFAG 117
Query: 496 EQVMVRGYIDLDTIFGEDECARVLK--VRYLVLQVVASYNVIIGRNTLNRLCAVISTAHL 553
+ G I L TI AR ++ +LV+ A +N I+GR L+ + AV ST H
Sbjct: 118 DTTFSIGTIQLPTI------ARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQ 171
Query: 554 AVKYPLSSGKVGKLKVDQKMARECYNNCLNLYGKKSALVGHRCYEIEASD----ENLDPR 609
+K+P G + + Q+ +R+CY K +V E E ++ +LDP
Sbjct: 172 CIKFPSYKG-IAVVYGSQRSSRKCYMGSYEDIKKADPVV--LMIEDELAEMKTVRSLDPS 228
Query: 610 GEG-RVNRPTPI----EDTKALKFGDRTLKIGTRLTEEQETRLTKLLGENLDLFAWSCKD 664
G R + T + D K R + IG L L L EN D FAWS +
Sbjct: 229 QRGTRKSLITQVCIDESDPK------RCVGIGHDLDLTVREDLITFLKENKDSFAWSSAN 282
Query: 665 MPGI 668
+ GI
Sbjct: 283 LQGI 286
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 385 bits (989), Expect = e-106
Identities = 181/322 (56%), Positives = 245/322 (75%)
Query: 690 RRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKWLMCVDYTDLNKAC 749
RR+LG ++ KAV EVDKLL IREV+YP WLAN V+VKK NGK +C+D+TDLNKAC
Sbjct: 469 RRKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKAC 528
Query: 750 PKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAFMTARVNYCYRTMP 809
PKDS+PLP ID LV+ +GNELLS MDA+SGY+QI M+P D++KT F+T R YCY+ MP
Sbjct: 529 PKDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMP 588
Query: 810 FGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMR 869
FGL+NAGATY RL++++F+ VG+ MEVY+DDM++KS++ DH + LEE F + ++ M+
Sbjct: 589 FGLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMK 648
Query: 870 LNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEVQRLTGRIAALSRF 929
LNP KC+FGV G+FLG+++T RGIE NP++ A M +P N KEVQRLTGRIAAL+RF
Sbjct: 649 LNPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRF 708
Query: 930 LPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKPMQGHPLHLYFAVS 989
+ +S D+S PF++ L+ N F W +CEEAF +LK L+SPP+L KP L+LY +VS
Sbjct: 709 ISRSTDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVS 768
Query: 990 DSALSSVILQEGDGEHRVIYFV 1011
+ A+S V+++E GE + IY++
Sbjct: 769 NHAVSGVLIREDRGEQKPIYYI 790
Score = 275 bits (704), Expect = 1e-73
Identities = 181/548 (33%), Positives = 278/548 (50%), Gaps = 94/548 (17%)
Query: 1172 IGSLLIRTDS-------QLVENQVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEYVPRT 1224
+ +LIR D ++ NQ G ++ KD + YL V+ L F++ + +PR
Sbjct: 772 VSGVLIREDRGEQKPIYYIIVNQFNGDYEAKDSRMEAYLHVVKDLAKNFRKFELIRIPRG 831
Query: 1225 ENQRADALAKLASTRKPDNNRSVIQETLAFPSIEGELMACV---------DRG--ATWMG 1273
+N ADALA LAST P+ N + E ++ SI+ E A V DRG A +
Sbjct: 832 QNTTADALAALASTSDPEVNSIIPVECISERSIKEEKEAFVVTRSRAAARDRGDTAVELP 891
Query: 1274 PILSILAGD-----PAE--------VEQCTKEQRREASHYTLID---------------G 1305
P+ + + PA +E+ + + EA +D
Sbjct: 892 PVKRRKSKEATVPKPAVQENIGIELIEEVISDAKIEAETEPWVDEDILAPNEHPEPEALN 951
Query: 1306 HLYRRGFSAPLLKCVPPEKYEAIMSEVHEGMCASHIGGRSLACKVLRAGFYWPTLRKDCM 1365
LYRRG S P L + E +MSEVHEG+C SH GR++A K+ + +YWPT+ DC+
Sbjct: 952 SLYRRGVSDPYLLSIFGPVVEIVMSEVHEGLCGSHSSGRAMAFKIKKMCYYWPTMITDCV 1011
Query: 1366 DFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMWGVDLVGPFPIARSQMKFILVAVDYF 1425
+ ++CK+CQ+ A L P + ++SAP+PF W +D++GP + ++++LV DYF
Sbjct: 1012 KYAQRCKRCQLHAPLIHQPSELFSSISAPYPFMRWSMDIIGPLHRSTRGVQYLLVLTDYF 1071
Query: 1426 TKWIEAEPLAKITSAKIVNFYWKRIVCRFGIPRAIVSDNGTQFSSNQTREFCREMGIQMR 1485
+KWIEAE + ++N+ GI++
Sbjct: 1072 SKWIEAEAI-------LLNW-----------------------------------GIKVS 1089
Query: 1486 FASVEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTTRETPFR 1545
++++ +PQ NGQ E AN+ IL L++RL KG W +EL VLW+Y TT + +T ETPF
Sbjct: 1090 YSTLRYPQGNGQAEAANKTILSNLKKRLNHLKGGWYDELQPVLWAYRTTPRRSTGETPFS 1149
Query: 1546 MTYGVDAMLPAEID-NFTWRTE-PDFEGENQANMAAELDLLSETRDEAHIRETAMKQRVA 1603
+ YG+ A++PAE++ RTE P E EN A + LD ++E RD+A I+ + A
Sbjct: 1150 LVYGMKAVVPAELNVPGLRRTEAPLNEKENSAMLDDSLDTINERRDQALIQIQNYQHAAA 1209
Query: 1604 AKFNSRVRVRDMQVGDLVLKW----RSGASGNKLTPNWEGPYRIIKVLGNEAYHLEELDG 1659
+NS+V+ R VGD VLK + KL NWEGPY + +V+ N Y L++L+
Sbjct: 1210 RYYNSKVKSRPFFVGDYVLKRVFDNKKEEGAGKLGINWEGPYIVTEVVRNGVYRLKDLED 1269
Query: 1660 RRLPRSFN 1667
R + R +N
Sbjct: 1270 RPVQRPWN 1277
Score = 79.3 bits (194), Expect = 2e-14
Identities = 58/214 (27%), Positives = 106/214 (49%), Gaps = 9/214 (4%)
Query: 11 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVII----AASDAVKCRMFPS 66
PF+ + S+ I D+ K L L+SY+G DPK +L F + A D C++F
Sbjct: 144 PFTPQITSLRIRDSRK-LNLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSE 202
Query: 67 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 126
A+ WFT L GSISNF + S FL Q+S ++ ++ DL+N+ Q E+L+ ++
Sbjct: 203 NLCGQALMWFTQLEPGSISNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFI 262
Query: 127 ARYSAASVKVEDEEPRACALAFKNGL-LPGGLNSKLTR*PARSMEEMRARASTYI-LDEE 184
++ K+ ++ A + GL L ++++ RA+ ++ +++E
Sbjct: 263 TKFKYVLSKLSRISQQSALSALRKGLWYDSRFKEDLILHKLDTIQDALFRANNWMEVEDE 322
Query: 185 DDAFKRKRAKLEKGDTSPKRAKKDKNGEDKGDGK 218
++F ++ + + T P KK + E++G K
Sbjct: 323 KESFAKRDKQAKPAVTFP--PKKFEPRENQGPRK 354
Score = 40.8 bits (94), Expect = 0.007
Identities = 17/44 (38%), Positives = 30/44 (67%)
Query: 440 DDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTDKDL 483
DD +VV L + +F V R+L+D GSS D+I+ +++G++ D+
Sbjct: 424 DDALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISRADV 467
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 354 bits (908), Expect = 3e-97
Identities = 168/289 (58%), Positives = 217/289 (74%), Gaps = 1/289 (0%)
Query: 664 DMPGIDPNFICHRLALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWL 723
DM GIDP CH L ++P+ K V Q RR+LG ++ KAV EVDKLL A I EVKYP WL
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 724 ANVVMVKKANGKWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQ 783
AN V+VKK N KW +C+D+TDLNKACPKDS+PLP ID +V+ +GNELLS MDA+SGY+Q
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 784 IRMHPADEDKTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMI 843
I MH D++KT+F+ R YCY+ MPFGLKN GA YQRL++++FA Q+G+ MEVY+DDM+
Sbjct: 596 IPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDML 655
Query: 844 VKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKA 903
VKS R DH L+ F + K+NM+LNP KC FGV G+FLG+++T RGIE NP + +A
Sbjct: 656 VKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRA 715
Query: 904 IQQMKNPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLQK-NAAFE 951
I +++P N KEVQRLTGRIA L+RF+ +S D+S PF++ L+ N FE
Sbjct: 716 ILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLRSANKTFE 764
Score = 92.4 bits (228), Expect = 2e-18
Identities = 71/301 (23%), Positives = 132/301 (43%), Gaps = 31/301 (10%)
Query: 298 KWCEFHRSAGHGTDDCWTLQREIDKLIRAGYQGNRQGQWRNNGDHNKT------------ 345
++C+FH+ +GH T C LQ L+ +G+ + Q R HN T
Sbjct: 185 QYCDFHKRSGHSTAACRHLQ---SILLNKYKKGDIEVQHRQYKSHNNTYAARGGRDGNNV 241
Query: 346 ------HKREEERVDTKGKKKQESAAIATRGADDTFAQHSGPPVGT--INTIAGGFGGGG 397
H ++ ++++ + + QH+ PV +N I GG
Sbjct: 242 FHRLGPHTGRQQEAPPANEEERHPDMEPPKKNRENDQQHNDAPVPRRRVNMIMGGLTACR 301
Query: 398 DTHAARKRHVR--AVNSVHEVAFGFVHPDRTISMADFEGIKPHKDDPIVVQLRMNSFNVR 455
D+ + K +++ A A + P T + D G+ +DP+V++L + V
Sbjct: 302 DSFRSIKEYIKSGAATLWSSPATKEMTP-LTFTSEDLFGVDLPHNDPLVIELHIGESEVT 360
Query: 456 RVLLDQGSSADIIYGDAFDKLGLTDKDLTPDAGTLVGFAGEQVMVRGYIDLDTIFGEDEC 515
R+L+D GSS ++++ D K+ + D+ + P L GF G +M G I L G
Sbjct: 361 RILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTNGTIKLPIYLG---- 416
Query: 516 ARVLKVRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKVGKLKVDQKMAR 575
+++V+ YN+I+G ++ + A+ S+ H +K P S G + ++ +Q +A
Sbjct: 417 GAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG-IETIRGNQNLAH 475
Query: 576 E 576
+
Sbjct: 476 D 476
>At4g03800
Length = 637
Score = 322 bits (826), Expect = 9e-88
Identities = 163/336 (48%), Positives = 230/336 (67%), Gaps = 21/336 (6%)
Query: 665 MPGIDPNFICHRLALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLA 724
MP IDP+ ICH L ++P KP+ Q RR+LG ++ KAV ++DKLL IREV+YP W+A
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 725 NVVMVKKANGKWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQI 784
V+VKK NGK +C+D+TDLNKACPKDS+PLP ID LV+ +GNELL+ MDA+ GY+QI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 785 RMHPADEDKTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIV 844
M+P D++KT+F+T R GATYQ L++++F + + MEV +DD +V
Sbjct: 441 MMNPEDQEKTSFITDR---------------GATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 845 KSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAI 904
KS++ DH + L E F + ++ M+LN KC+FGV G+FLG+++T RGIE NP++ A
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINAF 545
Query: 905 QQMKNPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLK 964
+ + N KEVQRLTGRIA+ +S D+S PF++ L+ N F W +CEEAF +LK
Sbjct: 546 LKTPSLRNFKEVQRLTGRIAS------RSTDKSLPFYQILKGNNGFLWDEKCEEAFRQLK 599
Query: 965 ELLSSPPILSKPMQGHPLHLYFAVSDSALSSVILQE 1000
L++PP+LSKP L+LY VS+ A+S V+++E
Sbjct: 600 AYLTTPPVLSKPEADEKLYLYVFVSNHAVSGVLVRE 635
Score = 63.5 bits (153), Expect = 1e-09
Identities = 68/262 (25%), Positives = 116/262 (43%), Gaps = 37/262 (14%)
Query: 304 RSAGHGTDDCWTLQREIDKLIRAGYQGNRQGQWRNNGDHNKTHKREEERVDTKGKKKQES 363
R H T DC L+R + +L +G N D K + + +T+ K+
Sbjct: 77 RVTEHLTKDCIVLKRYLAELWASGDLSKF-----NIDDFVKQYHEARDNSETQNSKRPRL 131
Query: 364 AAIATRGADDTFAQHSGPPVGTINTIAGGFGGGGDTHAARKRHVRAVNSVHEVAFGFVHP 423
T + G IN I GG ++ +A K+H V ++ +
Sbjct: 132 ----------TNEEEPRSSKGKINVILGGSKLCHNSVSAIKKHRPNVLLKSSLSEEVNYQ 181
Query: 424 DRTISMADFEGI---KPHKDDPIVVQLRMNSFNVRRVLLDQGSSADIIYGDAFDKLGLTD 480
++S + E +PH DD +++ L + +F + R+L+D GSS D+I+
Sbjct: 182 CSSVSFDEEETRHIERPH-DDALIITLDVANFKISRILVDTGSSVDLIF----------- 229
Query: 481 KDLTPDAGTLVGFAGEQVMVRGYIDLDTIFGEDECARVLKVRYLVLQVVASYNVIIGRNT 540
L P + LV F E M G I L + G + +++ V ++V A+YN+I+G
Sbjct: 230 --LGPPS-PLVAFTSESAMSLGTIKLPVLAG--KMSKI--VDFVVFDKPATYNIILGTPW 282
Query: 541 LNRLCAVISTAHLAVKYPLSSG 562
+ ++ AV ST H +K+P S+G
Sbjct: 283 IFQMKAVPSTYHQWLKFPTSNG 304
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 291 bits (744), Expect = 3e-78
Identities = 159/417 (38%), Positives = 233/417 (55%), Gaps = 21/417 (5%)
Query: 1266 DRGATWMGPILS-ILAGDPAEVEQCTKEQRREASHYTLIDGHLYRRGFSAPLLKCVPPEK 1324
D GA W PI IL G + ++ + + + + + LYRR FSAP C+ E+
Sbjct: 926 DWGADWREPIRDYILNGTLPAEKWAARKLKATCARFCIANDILYRRIFSAPDAVCIFGEQ 985
Query: 1325 YEAIMSEVHEGMCASHIGGRSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAP 1384
+M EVH+G C +H GGRSLA KV + G+YWPTL DC + +KC++CQ A L P
Sbjct: 986 TRTVMKEVHDGTCGNHTGGRSLAFKVRKYGYYWPTLVADCEAYARKCEQCQKHAPLILQP 1045
Query: 1385 PKELVTMSAPWPFAMWGVDLVGPFPIARSQMKFILVAVDYFTKWIEAEPLAKITSAKIVN 1444
+ L T+SAP+PF W +D+VGP ++ T+ +EA + IT ++ N
Sbjct: 1046 AELLTTVSAPYPFMKWLMDIVGPLHVS--------------TRGVEAAAYSNITHVQVWN 1091
Query: 1445 FYWKRIVCRFGIPRAIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRV 1504
F WK I+CR G+P IV+DNG+QF S Q FC E I++ ++ +PQ NGQ E N+
Sbjct: 1092 FIWKDIICRHGLPYEIVTDNGSQFISEQFEVFCEEWQIRLSHSTPRYPQGNGQAEAMNKT 1151
Query: 1505 ILRGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWR 1564
I+ L+++L KGAW EL VLW+ TT + T ETPF + YG++A++PAEI + R
Sbjct: 1152 IISNLKKKLNAYKGAWFGELQNVLWAVRTTPRRATDETPFSLIYGMEAVIPAEIKVPSAR 1211
Query: 1565 --TEPDFEGENQANMAAELDLLSETRDEAHIRETAMKQRVAAKFNSRVRVRDMQVGDLVL 1622
P E EN + +D + E R+ A R A +NS VR R +VG LVL
Sbjct: 1212 RIRNPQNETENNEMIIDVIDTIDERRNRALARMQNYHNAGARYYNSNVRNRSFEVGTLVL 1271
Query: 1623 KW----RSGASGNKLTPNWEGPYRIIKVLGNEAYHLEELDGRRLPRSFNGLSLRYYY 1675
+ ++ KL +WEGPY+I V+ N Y L ++G+ + R++N + L+ +Y
Sbjct: 1272 RRVQQNKAEKGAGKLGISWEGPYKITHVVRNGVYRLINMEGKTVRRAWNSMHLKRFY 1328
Score = 185 bits (470), Expect = 2e-46
Identities = 89/151 (58%), Positives = 112/151 (73%)
Query: 665 MPGIDPNFICHRLALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLA 724
M GI I H L ++P+ KPV Q RR+LG D+ +AV EV +LL IREVKYP WLA
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 725 NVVMVKKANGKWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQI 784
N V+VKK NGKW +CVD+TDLNKAC KD +PLP ID LV+ +G+E+LS MDA+SGY+QI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 785 RMHPADEDKTAFMTARVNYCYRTMPFGLKNA 815
M+P D++KT+F+T Y Y+ MPFGLKNA
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
Score = 123 bits (309), Expect = 8e-28
Identities = 68/150 (45%), Positives = 91/150 (60%), Gaps = 8/150 (5%)
Query: 938 FPFFKCLQK-NAAFEWTAECEEAFVRLKELLSSPPILSKPMQGHPLHLYFAVSDSALSSV 996
FPF K + F W CEEAF +LK LS P +L+K G L LY AVS+SA++ V
Sbjct: 616 FPFLHFTAKISKGFIWNESCEEAFKQLKRYLSEPSVLAKREFGEQLFLYIAVSESAVTGV 675
Query: 997 ILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTARRLRPYFQSFPVRVRTDLPL 1056
++ + R I++V + RY +EK ALAV+ AR+LRPYFQS P+ V T LPL
Sbjct: 676 QVRVERSDKRPIFYV-------KTRYPMMEKLALAVVTAARKLRPYFQSHPIVVLTSLPL 728
Query: 1057 RQVLQKPDMSGRLVAWSVELSEYGLQYDKR 1086
R +L P SGRL W++ELSE+ L++ R
Sbjct: 729 RTILHSPTQSGRLGKWAIELSEFDLEFRAR 758
Score = 62.0 bits (149), Expect = 3e-09
Identities = 39/126 (30%), Positives = 58/126 (45%), Gaps = 13/126 (10%)
Query: 11 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAA--------SDAVKCR 62
PF+ + + I + + L Y+G D K+HL F +IA DA C+
Sbjct: 157 PFTPRISKLRIRE-FRDFKLPVYNGKGDLKEHLTSFQ----VIAGRVPLEPHEEDAGLCK 211
Query: 63 MFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESL 122
+F A+ WFT L GSI NF+ S+ F+ Q+ N +T L+N Q E L
Sbjct: 212 LFSENLFGLALTWFTQLEEGSIDNFKQLSTAFIKQYEYFINSDITEAHLWNFSQSADEPL 271
Query: 123 KEYMAR 128
+ Y+ R
Sbjct: 272 RTYIYR 277
Score = 47.0 bits (110), Expect = 9e-05
Identities = 27/72 (37%), Positives = 41/72 (56%)
Query: 1186 NQVKGTFQVKDPNLIKYLE*VRYLMTLFQEVVVEYVPRTENQRADALAKLASTRKPDNNR 1245
N+ G ++ KD + YL VR + F++ + +PR EN A+ALA LAST + R
Sbjct: 790 NRYSGEYEAKDACMEAYLNLVREVSGRFEQFELTRIPRAENSAANALAALASTFEVTLPR 849
Query: 1246 SVIQETLAFPSI 1257
+ ET++ PSI
Sbjct: 850 VIPVETISQPSI 861
Score = 34.3 bits (77), Expect = 0.63
Identities = 19/47 (40%), Positives = 25/47 (52%)
Query: 518 VLKVRYLVLQVVASYNVIIGRNTLNRLCAVISTAHLAVKYPLSSGKV 564
V V + V A+YNVI+G L + V ST H VK+P GK+
Sbjct: 366 VKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKM 412
>At2g10780 pseudogene
Length = 1611
Score = 231 bits (589), Expect = 3e-60
Identities = 157/532 (29%), Positives = 260/532 (48%), Gaps = 24/532 (4%)
Query: 677 LALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKW 736
+ L P P+S+ R+ + +++++++LL FIR P W A V+ VKK +G +
Sbjct: 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSF 709
Query: 737 LMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAF 796
+C+DY LNK K+ YPLP ID L+D G + S +D SGYHQI + P D KTAF
Sbjct: 710 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 769
Query: 797 MTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 856
T ++ + MPFGL NA A + ++M+ VF + + ++++D++V S H + L
Sbjct: 770 RTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHL 829
Query: 857 EEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEV 916
+R+H + KCSF + FLG +I+ +G+ ++P+K ++I++ P N E+
Sbjct: 830 RAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 889
Query: 917 QRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKP 976
+ G RF+ + P + K+ AF W+ ECE++F+ LK +L++ P+L P
Sbjct: 890 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLP 949
Query: 977 MQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTA 1036
+G P +Y S L V++Q+G VI + S L+ E Y + AV+
Sbjct: 950 EEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKHEKNYPTHDLEMAAVVFFL 1005
Query: 1037 RRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAWSVELSEYGLQYDKR-GKVGAQSL 1094
+ R Y V++ TD L+ + +P+++ R W +++Y L GK A +
Sbjct: 1006 KIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVADYNLDIAYHPGK--ANQV 1063
Query: 1095 ADFV------VELTPDRFERVDTQWTLFVDGSSNS---SGSGTGVTLEGPGDLVLEQSLK 1145
AD + VE + + V+ TL V+ S G G + + L Q
Sbjct: 1064 ADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLGLGAADQADLLSRIRLAQERD 1123
Query: 1146 FEFK--ATNNQAEYEALIAGLKLAREVKIGSLLIRTDSQLVENQVKGTFQVK 1195
E K A NN+ EY+ G + G + + D L E ++ Q K
Sbjct: 1124 EEIKGWAQNNKTEYQTSNNGTIVVN----GRVCVPNDRALKEEILREAHQSK 1171
Score = 115 bits (288), Expect = 2e-25
Identities = 141/565 (24%), Positives = 243/565 (42%), Gaps = 51/565 (9%)
Query: 1113 WTLFVDGSSNSSGSGTGVTLEGPGDLVL---EQSLKFEFKATNNQAEYEALIAGLKLARE 1169
+T++ D S G G L G ++ Q K E + E A++ LK+ R
Sbjct: 955 YTVYTDASI----VGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRS 1010
Query: 1170 VKIGSLL-IRTDSQLVENQVKGTFQVKDPNLIKYLE*VRYLMTLFQE--VVVEYVPRTEN 1226
G+ + I TD + +K F + NL + R M L + + + Y P N
Sbjct: 1011 YLYGAKVQIYTDHK----SLKYIFTQPELNLRQ-----RRWMELVADYNLDIAYHPGKAN 1061
Query: 1227 QRADALAKLASTRKPDNNRSVIQE---TLAFPSIEGELMACVDRGATWMGPILS-ILAGD 1282
Q ADAL++ S + + ++ + TL ++ E+ + GA +LS I
Sbjct: 1062 QVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEP-LGLGAADQADLLSRIRLAQ 1120
Query: 1283 PAEVEQCTKEQRREASHYTLIDGHLYRRGFSAPLLKCVPPEKY--EAIMSEVHEGMCASH 1340
+ E Q + + T +G + G CVP ++ E I+ E H+ + H
Sbjct: 1121 ERDEEIKGWAQNNKTEYQTSNNGTIVVNG-----RVCVPNDRALKEEILREAHQSKFSIH 1175
Query: 1341 IGGRSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAP-WPFAM 1399
G + + L+ ++W ++KD +V KC CQ+ + P L + P W +
Sbjct: 1176 PGSNKMY-RDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPIPEWKWDH 1234
Query: 1400 WGVDLVGPFPIA-RSQMKFILVAVDYFTKWIEAEPLAKITSAKIV-NFYWKRIVCRFGIP 1457
+D V P +S+ + V VD TK ++ A+I+ Y IV GIP
Sbjct: 1235 ITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIVRLHGIP 1294
Query: 1458 RAIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAK 1517
+IVSD T+F+S + F + +G ++ ++ HPQ + Q E + + LR + +
Sbjct: 1295 VSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQTLEDMLRACVLDWG 1354
Query: 1518 GAWLEELPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWRTEPDFEGENQANM 1577
G W + L +V ++YN + Q++ +P+ YG P W GE +
Sbjct: 1355 GNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTP-----LCWTP----VGERRLFG 1405
Query: 1578 AAELDLLSETRDEAHIRETAMKQRVAAKFNSRVRVRDMQVGDLV----LKWRSG---ASG 1630
+D +E I+ + R + N R + + QVGDLV + ++ S
Sbjct: 1406 PTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSR 1465
Query: 1631 NKLTPNWEGPYRIIKVLGNEAYHLE 1655
KL+P + GPY++I+ +G AY L+
Sbjct: 1466 KKLSPRYVGPYKVIERVGAVAYKLD 1490
>At2g13230 pseudogene
Length = 353
Score = 228 bits (581), Expect = 2e-59
Identities = 128/358 (35%), Positives = 198/358 (54%), Gaps = 28/358 (7%)
Query: 1202 YLE*VRYLMTLFQEVVVEYVPRTENQRADALAKLASTRKPDNNRSVIQETLAFPSIE--- 1258
YL V+ F E + +PR EN ADALA LAST + R + E + PSIE
Sbjct: 4 YLALVQDHAKQFHEFKLTRIPRGENTSADALAALASTSDLNMRRVIPVEFIEKPSIELDK 63
Query: 1259 ----------------GELMACVDRGATWMGPILSILAGDPAEVEQC-TKEQRREASHYT 1301
+ + + W+ PI + ++ +++ ++ + EA+ +
Sbjct: 64 VEHAFPIQVVAGYEDASDNSPSLGCDSEWIEPIRNFISKRKVRLDKWEARKLKAEAARFV 123
Query: 1302 LIDGHLYRRGFSAPLLKCVPPEKYEAIMSEVHEGMCASHIGGRSLACKVLRAGFYWPTLR 1361
L++ L++ S PL+ CV E +M EVH G C ++ GGR+LA K+ R G++WPT+
Sbjct: 124 LVEEKLFKWRLSGPLMTCVEKEAARKVMKEVHGGSCGNNSGGRALAIKIKRLGYFWPTMI 183
Query: 1362 KDCMDFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMWGVDLVGPFPIARSQMKFILVA 1421
KDC +KCQ A P + L ++++P+P W +D++GP A Q K +LV
Sbjct: 184 KDC-------EKCQRNAPTIHLPAELLSSIASPYPLMRWSMDIIGPMH-ASKQKKLVLVL 235
Query: 1422 VDYFTKWIEAEPLAKITSAKIVNFYWKRIVCRFGIPRAIVSDNGTQFSSNQTREFCREMG 1481
DYF+K EAE A I A++ +F WK I+CR G+P IV+DNG+QF S + + FC + G
Sbjct: 236 TDYFSKLREAESYASIKEAQVESFMWKHILCRHGVPYEIVTDNGSQFISTRFQGFCDKWG 295
Query: 1482 IQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTT 1539
I++ ++ + Q NGQ + AN+ IL GL++RL KG+W++EL VLWS+ TT + T
Sbjct: 296 IRLSKSTPRYLQGNGQADAANKTILDGLKKRLTAKKGSWVDELEGVLWSHRTTPRRAT 353
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 223 bits (568), Expect = 7e-58
Identities = 132/407 (32%), Positives = 211/407 (51%), Gaps = 6/407 (1%)
Query: 677 LALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKW 736
+ L P P+S+ R+ + +++++ LL FIR P W A V+ VKK +G +
Sbjct: 483 IELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 541
Query: 737 LMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAF 796
+C+DY +LN+ K+ YPLP ID L+D G S +D SGYHQI + AD KTAF
Sbjct: 542 RLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 601
Query: 797 MTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 856
T ++ + MPFGL NA A + RLM+ VF + + +++DD++V S + L
Sbjct: 602 RTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHL 661
Query: 857 EEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEV 916
++R+ + KCSF + FLG ++++ G+ ++P+K +AI+ P+N E+
Sbjct: 662 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 721
Query: 917 QRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKP 976
+ G RF+ + P K K+ F W+ ECEE FV LKE+L+S P+L+ P
Sbjct: 722 RSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALP 781
Query: 977 MQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTA 1036
G P +Y S L V++Q G +VI + S L E Y + AV+
Sbjct: 782 EHGQPYMVYTDASRVGLGCVLMQHG----KVIAYASRQLMKHEGNYPTHDLEMAAVIFAL 837
Query: 1037 RRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAWSVELSEYGLQ 1082
+ R Y V+V TD L+ + +P+++ R W +++Y L+
Sbjct: 838 KIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLE 884
Score = 33.5 bits (75), Expect = 1.1
Identities = 38/171 (22%), Positives = 69/171 (40%), Gaps = 20/171 (11%)
Query: 1351 LRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAP-WPFAMWGVDLVGPFP 1409
L+ + W ++ D ++V +C CQ+ + L ++ P W + +DLV
Sbjct: 965 LKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGLR 1024
Query: 1410 IARSQMKFILVAVDYFTKWIEAEPLAKITSAKIV-NFYWKRIVCRFGIPRAI--VSDNGT 1466
++R++ I V VD TK + K A ++ + IV G+P + D
Sbjct: 1025 VSRTK-DAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMKEAQDRQR 1083
Query: 1467 QFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAK 1517
++ + RE E+G ++ Y +LRG R + E K
Sbjct: 1084 SYADKRRRELEFEVGDRV---------------YLKMAMLRGPNRSISETK 1119
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 220 bits (560), Expect = 6e-57
Identities = 130/407 (31%), Positives = 210/407 (50%), Gaps = 6/407 (1%)
Query: 677 LALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKW 736
+ L P P+S+ R+ + ++++++ LL FIR P W A V+ VKK +G +
Sbjct: 509 IELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 567
Query: 737 LMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAF 796
+C+DY LN K+ YPLP ID L+D G S +D SGYHQI + AD KTAF
Sbjct: 568 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 627
Query: 797 MTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 856
T ++ + MPF L NA A + RLM+ VF + + +++DD++V S +H L
Sbjct: 628 RTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHL 687
Query: 857 EEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEV 916
++R+ + KCSF + FLG ++++ G+ ++P+K +AI+ P+N E+
Sbjct: 688 RRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 747
Query: 917 QRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKP 976
+ RF+ + P K K+ F W+ ECEE FV LKE+L+S P+L+ P
Sbjct: 748 RSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 807
Query: 977 MQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTA 1036
G P +Y S L V++Q G +VI + S L+ E Y + V+
Sbjct: 808 EHGQPYMVYTDASRVGLGCVLMQRG----KVIAYASRQLRKHEGNYPTHDLEMAVVIFAL 863
Query: 1037 RRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAWSVELSEYGLQ 1082
+ R Y V+V TD L+ + +P+++ R + W +++Y L+
Sbjct: 864 KIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADYDLE 910
Score = 100 bits (249), Expect = 7e-21
Identities = 142/584 (24%), Positives = 233/584 (39%), Gaps = 101/584 (17%)
Query: 1121 SNSSGSGTGVTLEGPGDLVL---EQSLKFEFKATNNQAEYEALIAGLKLAREVKIGSLL- 1176
+++S G G L G ++ Q K E + E +I LK+ R G +
Sbjct: 817 TDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQ 876
Query: 1177 IRTDSQLVENQVKGTFQVKDPNL--IKYLE*VRYLMTLFQEVVVEYVPRTENQRADALAK 1234
+ TD + +K F + NL ++++E + ++ + Y P N ADAL++
Sbjct: 877 VFTDHK----SLKYIFNQPELNLRQMRWME-----LVADYDLEIAYHPGKANVVADALSR 927
Query: 1235 LASTRKPDNNRSVIQETLAFPSIEGELMACVDRGATWMGPILSILAGDPAEV-------- 1286
P Q A S G L CV L + A D A++
Sbjct: 928 KRVGAAPG------QSVEALVSEIGALRLCVVAREP-----LGLEAVDRADLLTRARLAQ 976
Query: 1287 ---EQCTKEQRREASHYTLI-DGHLYRRGFSAPLLKCVPPEKY--EAIMSEVHEGMCASH 1340
E + E S Y +G ++ G CVP ++ I+SE H M + H
Sbjct: 977 EKDEGLIAASKAEGSEYQFAANGTIFVYG-----RVCVPKDEELRREILSEAHASMFSIH 1031
Query: 1341 IGGRSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMW 1400
G + + L+ + W +++D ++V +C CQ LV P A+W
Sbjct: 1032 PGATKMY-RDLKRYYQWVGMKRDVANWVAECDVCQ------------LVKAEHQVPDAIW 1078
Query: 1401 GVDLVGPFPIARSQMKFILVAVDYFTKWIEAEPLAKITSAKIV-NFYWKRIVCRFGIPRA 1459
V +D TK + K A ++ Y IV G+P +
Sbjct: 1079 -------------------VIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVS 1119
Query: 1460 IVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKGA 1519
IVSD ++F+ R F +MG +++ ++ HPQ +GQ E + + LR + + G
Sbjct: 1120 IVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGH 1179
Query: 1520 WLEELPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWRTEPDFEGENQANMAA 1579
W + L +V ++YN + Q++ PF YG P +T E G
Sbjct: 1180 WADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPL---RWTQVEERSIYG-------- 1228
Query: 1580 ELDLLSETRDEAHIRETAMKQ---RVAAKFNSRVRVRDMQVGDLVLKWRSGASG------ 1630
D + ET + + + MK+ R + + R R + +VGD V + G
Sbjct: 1229 -ADYVQETTERIRVLKLNMKEAQARQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSIL 1287
Query: 1631 -NKLTPNWEGPYRIIKVLGNEAYHLEELD-GRRLPRSFNGLSLR 1672
KL+P + GP+RI++ +G AY LE D R + F+ L LR
Sbjct: 1288 ETKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVLMLR 1331
>At3g11970 hypothetical protein
Length = 1499
Score = 214 bits (544), Expect = 4e-55
Identities = 138/417 (33%), Positives = 222/417 (53%), Gaps = 10/417 (2%)
Query: 675 HRLALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANG 734
H++ L PV+Q R + + + V+ LL ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 735 KWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKT 794
W +CVDY +LN KDS+P+P I+ L+D G + S +D +GYHQ+RM P D KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 795 AFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQ 854
AF T ++ Y MPFGL NA AT+Q LM+ +F + + + V+ DD++V S +H Q
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 855 DLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVK 914
L++ F +R + + KC+F V ++LG I+++GIE +P K KA+++ P+ +K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 915 EVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILS 974
+++ G RF+ G + P L K AFEWTA ++AF LK L P+LS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 975 KPMQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLV 1034
P+ + + +V++QEG H + Y +S L+G ++ EK LAV+
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQEG---HPLAY-ISRQLKGKQLHLSIYEKELLAVIF 944
Query: 1035 TARRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAWSVELSE--YGLQYDKRGK 1088
R+ R Y ++TD L+ +L++ + W +L E Y +QY ++GK
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY-RQGK 1000
Score = 103 bits (256), Expect = 1e-21
Identities = 89/329 (27%), Positives = 144/329 (43%), Gaps = 34/329 (10%)
Query: 1343 GRSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMWG- 1401
GR + + ++ FYW + KD +++ C CQ A P L + P P +W
Sbjct: 1098 GRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPL--PIPDTIWSE 1155
Query: 1402 --VDLVGPFPIARSQMKFILVAVDYFTKWIEAEPLAKITSA-KIVNFYWKRIVCRFGIPR 1458
+D + P++ + I+V VD +K L+ SA + + Y + G P
Sbjct: 1156 VSMDFIEGLPVSGGKT-VIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPT 1214
Query: 1459 AIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKG 1518
+IVSD F+S REF G+ ++ S HPQ++GQ E NR + LR +
Sbjct: 1215 SIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQ 1274
Query: 1519 AWLEELPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWRTEPDFEGENQANMA 1578
W + L + + YNT S++R TPF + YG + P + P GE++ +
Sbjct: 1275 LWSKWLALAEYWYNTNYHSSSRMTPFEIVYG--QVPPVHL--------PYLPGESKVAVV 1324
Query: 1579 AELDLLSETRD-----EAHIRETAMKQRVAAKFNSRVRVRDMQVGDLVL----KWRSGA- 1628
A L E D + H+ + + A + R+ ++GD V +R +
Sbjct: 1325 AR--SLQEREDMLLFLKFHLMRAQHRMKQFA--DQHRTEREFEIGDYVYVKLQPYRQQSV 1380
Query: 1629 ---SGNKLTPNWEGPYRIIKVLGNEAYHL 1654
+ KL+P + GPY+II G AY L
Sbjct: 1381 VMRANQKLSPKYFGPYKIIDRCGEVAYKL 1409
>At1g36590 hypothetical protein
Length = 1499
Score = 214 bits (544), Expect = 4e-55
Identities = 138/417 (33%), Positives = 222/417 (53%), Gaps = 10/417 (2%)
Query: 675 HRLALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANG 734
H++ L PV+Q R + + + V+ LL ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 735 KWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKT 794
W +CVDY +LN KDS+P+P I+ L+D G + S +D +GYHQ+RM P D KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 795 AFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQ 854
AF T ++ Y MPFGL NA AT+Q LM+ +F + + + V+ DD++V S +H Q
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 855 DLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVK 914
L++ F +R + + KC+F V ++LG I+++GIE +P K KA+++ P+ +K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 915 EVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILS 974
+++ G RF+ G + P L K AFEWTA ++AF LK L P+LS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 975 KPMQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLV 1034
P+ + + +V++QEG H + Y +S L+G ++ EK LAV+
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQEG---HPLAY-ISRQLKGKQLHLSIYEKELLAVIF 944
Query: 1035 TARRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAWSVELSE--YGLQYDKRGK 1088
R+ R Y ++TD L+ +L++ + W +L E Y +QY ++GK
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQY-RQGK 1000
Score = 97.4 bits (241), Expect = 6e-20
Identities = 88/329 (26%), Positives = 142/329 (42%), Gaps = 34/329 (10%)
Query: 1343 GRSLACKVLRAGFYWPTLRKDCMDFVKKCKKCQVFADLSKAPPKELVTMSAPWPFAMWG- 1401
GR + + ++ FY + KD +++ C CQ A P L + P P +W
Sbjct: 1098 GRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPL--PIPDTIWSE 1155
Query: 1402 --VDLVGPFPIARSQMKFILVAVDYFTKWIEAEPLAKITSA-KIVNFYWKRIVCRFGIPR 1458
+D + P++ + I+V VD +K L+ SA + Y + G P
Sbjct: 1156 VSMDFIEGLPVSGGKT-VIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKLHGCPT 1214
Query: 1459 AIVSDNGTQFSSNQTREFCREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKG 1518
+IVSD F+S REF G+ ++ S HPQ++GQ E NR + LR +
Sbjct: 1215 SIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQ 1274
Query: 1519 AWLEELPVVLWSYNTTVQSTTRETPFRMTYGVDAMLPAEIDNFTWRTEPDFEGENQANMA 1578
W + L + + YNT S++R TPF + YG + P + P GE++ +
Sbjct: 1275 LWSKWLALAEYWYNTNYHSSSRMTPFEIVYG--QVPPVHL--------PYLPGESKVAVV 1324
Query: 1579 AELDLLSETRD-----EAHIRETAMKQRVAAKFNSRVRVRDMQVGDLVL----KWRSGA- 1628
A L E D + H+ + + A + R+ ++GD V +R +
Sbjct: 1325 AR--SLQEREDMLLFLKFHLMRAQHRMKQFA--DQHRTEREFEIGDYVYVKLQPYRQQSV 1380
Query: 1629 ---SGNKLTPNWEGPYRIIKVLGNEAYHL 1654
+ KL+P + GPY+II G AY L
Sbjct: 1381 VMRANQKLSPKYFGPYKIIDRCGEVAYKL 1409
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 213 bits (543), Expect = 6e-55
Identities = 119/366 (32%), Positives = 194/366 (52%), Gaps = 5/366 (1%)
Query: 677 LALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKW 736
+ L P P+S+ R+ + +++++++LLA FIR P W A V+ VKK +G +
Sbjct: 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSF 204
Query: 737 LMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAF 796
+C+DY LNK K+ YPLP ID L+D G + S +D SGYHQI + P D KTAF
Sbjct: 205 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 264
Query: 797 MTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 856
T ++ + MPFGL NA A + ++M+ VF + + +++DD++V S H + L
Sbjct: 265 RTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHL 324
Query: 857 EEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEV 916
+R+H + K SF + FLG +I+ +G+ ++P+K ++I++ P N E+
Sbjct: 325 RAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 384
Query: 917 QRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKP 976
+ G RF+ + P + K+ AF W+ ECE++F+ LK +L + P+L P
Sbjct: 385 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLP 444
Query: 977 MQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTA 1036
+G P +Y S L V++Q+G VI + S L+ E Y + AV+
Sbjct: 445 EEGEPYTVYTDASIVGLGCVLMQKGS----VIAYASRQLRKHEKNYPTHDLEMAAVVFAL 500
Query: 1037 RRLRPY 1042
+ R Y
Sbjct: 501 KIWRSY 506
Score = 112 bits (280), Expect = 2e-24
Identities = 103/414 (24%), Positives = 184/414 (43%), Gaps = 29/414 (7%)
Query: 1254 FPSIEGELMACVDRGATWMGPILSILAGDPAEVEQCTKEQRREASHYTLIDGHLYRRGFS 1313
+P+ + E+ A V W ++ + E++ T + E + T +G + G
Sbjct: 486 YPTHDLEMAAVVFALKIWRSYLIRLAQERDEEIKGWTLNNKTE--YQTSNNGTIVVNG-- 541
Query: 1314 APLLKCVPPEKY--EAIMSEVHEGMCASHIGGRSLACKVLRAGFYWPTLRKDCMDFVKKC 1371
CVP ++ E I+ E H+ + H G + + L+ ++W +RKD +V KC
Sbjct: 542 ---RVCVPNDRALKEEILREAHQSKFSIHPGSNKMY-RDLKRYYHWVGMRKDVARWVAKC 597
Query: 1372 KKCQVFADLSKAPPKELVTMS-APWPFAMWGVDLVGPFPIA-RSQMKFILVAVDYFTKWI 1429
CQ+ + P L + + W + +D V P +S+ + V VD TK
Sbjct: 598 PTCQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSA 657
Query: 1430 EAEPLAKITSAKIV-NFYWKRIVCRFGIPRAIVSDNGTQFSSNQTREFCREMGIQMRFAS 1488
++ A+I+ Y I+ GIP +IVSD T+F+S F + +G ++ ++
Sbjct: 658 HFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLST 717
Query: 1489 VEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTTRETPFRMTY 1548
HPQ +GQ E + + LR + + G W + L ++ ++YN + Q++ +P+ Y
Sbjct: 718 AYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALY 777
Query: 1549 GVDAMLPAEIDNFTWRTEPDFEGENQANMAAELDLLSETRDEAHIRETAMKQRVAAKFNS 1608
G P W GE + +D +E I+ + R + N
Sbjct: 778 GRACRTP-----LCWTP----VGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANK 828
Query: 1609 RVRVRDMQVGDLV----LKWRSG---ASGNKLTPNWEGPYRIIKVLGNEAYHLE 1655
R + + QVGDLV + ++ S KL+P + GPY++I+ +G AY L+
Sbjct: 829 RRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLD 882
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 210 bits (535), Expect = 5e-54
Identities = 127/407 (31%), Positives = 206/407 (50%), Gaps = 18/407 (4%)
Query: 677 LALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANGKW 736
+ L P P+S+ R+ + ++++++ LL A V+ VKK +G +
Sbjct: 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAP-------------VLFVKKKDGSF 516
Query: 737 LMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKTAF 796
+C+DY LN K+ YPLP ID L+D G S +D SGYH I + AD KTAF
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 797 MTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQDL 856
T ++ + MPFGL NA A + RLM+ VF + + +++DD++V S +H L
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 857 EEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVKEV 916
++R+ + KCSF + FLG ++++ G+ ++P+K +AI+ P+N E+
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEI 696
Query: 917 QRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILSKP 976
+ G RF+ + P K K+ F W+ ECEE FV LKE+L+S P+L+ P
Sbjct: 697 RSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 756
Query: 977 MQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLVTA 1036
G P +Y S L V++Q G +VI + S L+ E Y + AV+
Sbjct: 757 EHGEPYMVYTDASGVGLGCVLMQRG----KVIAYASRQLRKHEGNYPTHDLEMAAVIFAL 812
Query: 1037 RRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAWSVELSEYGLQ 1082
+ R Y V+V TD L+ + +P+++ R W +++Y L+
Sbjct: 813 KIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLE 859
Score = 97.4 bits (241), Expect = 6e-20
Identities = 87/312 (27%), Positives = 141/312 (44%), Gaps = 25/312 (8%)
Query: 1362 KDCMDFVKKCKKCQVFADLSKAPPKELVTMSAP-WPFAMWGVDLVGPFPIARSQMKFILV 1420
+D ++V +C CQ+ + P L ++ P W + +D V P++R++ I V
Sbjct: 941 EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTK-DAIWV 999
Query: 1421 AVDYFTKWIEAEPLAKITSAKIV-NFYWKRIVCRFGIPRAIVSDNGTQFSSNQTREFCRE 1479
VD TK + K A ++ Y IV G+P +IVSD ++F+S R F E
Sbjct: 1000 IVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAE 1059
Query: 1480 MGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTT 1539
MG +++ ++ HPQ GQ E + + LR + + G W + L +V ++YN + ++
Sbjct: 1060 MGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASI 1119
Query: 1540 RETPFRMTYGVDAMLPAEIDNFTWRTEPDFEGENQANMAAELDLLSETRDEAHIRETAMK 1599
PF Y P + GE A D + ET + + + MK
Sbjct: 1120 GMAPFEALYERPCRTPLCLTQV---------GERSIYGA---DYVQETTERIRVLKLNMK 1167
Query: 1600 Q---RVAAKFNSRVRVRDMQVGDLV-LKW------RSGASGNKLTPNWEGPYRIIKVLGN 1649
+ R + + R R + +VGD V LK S KL+P + GP+RI++ +G
Sbjct: 1168 EAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGP 1227
Query: 1650 EAYHLEELDGRR 1661
AY LE D R
Sbjct: 1228 VAYRLELPDVMR 1239
>At2g13060 pseudogene
Length = 930
Score = 181 bits (458), Expect = 4e-45
Identities = 113/388 (29%), Positives = 184/388 (47%), Gaps = 15/388 (3%)
Query: 1244 NRSVIQETLAFPSIEGELMACVDRGATWMGPILSILAGDPAEVEQCT----KEQRREASH 1299
+R + T + S E + V++ W I++ LA D E + T K RE
Sbjct: 111 DRHPSESTRKWSSTENCAVTAVEKDYPWYADIVNYLAAD-VEPDNFTDYNKKRFLREIRR 169
Query: 1300 YTLIDGHLYRRGFSAPLLKCVPPEKYEAIMSEVHEGMCASHIGGRSLACKVLRAGFYWPT 1359
Y + +LY+ + +C+ + +I+S H H KVL+A F+WPT
Sbjct: 170 YQWDEPYLYKHSYDGIYRRCIAATEVPSILSHCHSSSYGGHFATFKTVSKVLQADFWWPT 229
Query: 1360 LRKDCMDFVKKCKKCQVFADLSKA---PPKELVTMSAPWPFAMWGVDLVGPFPIARSQMK 1416
+ +D F+ +C CQ +SK PP + + F WG+D +GPFP + +
Sbjct: 230 MFRDTQKFISQCDPCQRRGKISKRNEMPPNFRLEVEV---FDRWGIDFMGPFPPSNKNL- 285
Query: 1417 FILVAVDYFTKWIEAEPLAKITSAKIVNFYWKRIVCRFGIPRAIVSDNGTQFSSNQTREF 1476
+ILV VDY +KW+EA K SA ++ + I RFG+PR ++SD F + +
Sbjct: 286 YILVDVDYVSKWVEAIASLKNDSAVVMKLFKSIIFPRFGVPRIVISDGDKHFINKILEKL 345
Query: 1477 CREMGIQMRFASVEHPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLWSYNTTVQ 1536
+ G+Q R A+ HPQ +GQVE +NR I L + + +AK W +L LW+Y T +
Sbjct: 346 LLQYGVQHRVATPYHPQTSGQVEVSNRQIKEILEKTVGKAKKEWSYKLYDALWAYKTAFK 405
Query: 1537 STTRETPFRMTYGVDAMLPAEIDNFTWRTEPDFEGENQANMAAELDLLSETRDEAHIRET 1596
+ TPF + YG LP E+++ + ++ +ELD E R A+
Sbjct: 406 TPLGTTPFHLLYGKACHLPVELEHKAAWAVKMMNFDIKSAGESELD---EIRIHAYDNSK 462
Query: 1597 AMKQRVAAKFNSRVRVRDMQVGDLVLKW 1624
K+R A + ++ R + D +L++
Sbjct: 463 LYKERTKAYHDKKILTRTFEPNDQILRF 490
>At1g35370 hypothetical protein
Length = 1447
Score = 180 bits (456), Expect = 7e-45
Identities = 127/417 (30%), Positives = 211/417 (50%), Gaps = 18/417 (4%)
Query: 675 HRLALNPSVKPVSQLRRRLGGDKGKAVQQEVDKLLAAEFIREVKYPTWLANVVMVKKANG 734
H++ L PV+Q R + + + V ++ + I+ P + + VV+VKK +G
Sbjct: 570 HKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSP-FASPVVLVKKKDG 628
Query: 735 KWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLMDAYSGYHQIRMHPADEDKT 794
W +CVDYT+LN KD + +P I+ L+D G+ + S +D +GYHQ+RM P D KT
Sbjct: 629 TWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKT 688
Query: 795 AFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNMEVYVDDMIVKSVRGLDHHQ 854
AF T ++ Y M FGL NA AT+Q LM+ VF + + + V+ DD+++ S +H +
Sbjct: 689 AFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKE 748
Query: 855 DLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIEINPDKCKAIQQMKNPSNVK 914
L F +R H + F + LG I++R IE +P K +A+++ P+ VK
Sbjct: 749 HLRLVFEVMRLHKL--------FAKGSKEHLGHFISAREIETDPAKIQAVKEWPTPTTVK 800
Query: 915 EVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAECEEAFVRLKELLSSPPILS 974
+V+ G RF+ G + P L K F W+ E + AF LK +L + P+L+
Sbjct: 801 QVRGFLGFAGYYRRFVRNFGVIAGP-LHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLA 859
Query: 975 KPMQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTLQGAEVRYKKIEKAALAVLV 1034
P+ + + +V++Q+G H + Y +S L+G ++ EK LA +
Sbjct: 860 LPVFDKQFMVETDACGQGIRAVLMQKG---HPLAY-ISRQLKGKQLHLSIYEKELLAFIF 915
Query: 1035 TARRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAWSVELSE--YGLQYDKRGK 1088
R+ R Y ++TD L+ +L++ + W +L E Y +QY ++GK
Sbjct: 916 AVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPKLLEFDYEIQY-RQGK 971
Score = 99.4 bits (246), Expect = 2e-20
Identities = 91/345 (26%), Positives = 158/345 (45%), Gaps = 26/345 (7%)
Query: 1318 KCVPPEKYEAIMSEVHEGMCASHIGGRS---LACKVLRAGFYWPTLRKDCMDFVKKCKKC 1374
K V P E I +++ + + S +GGRS + + +++ FYW + KD F++ C C
Sbjct: 1042 KIVVPNDVE-ITNKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTC 1100
Query: 1375 QVFADLSKAPPKELVTMSAPWPFAMW---GVDLVGPFPIARSQMKFILVAVDYFTKWIEA 1431
Q + A P L + P P +W +D + P + + I+V VD +K
Sbjct: 1101 QQCKSDNAAYPGLLQPL--PIPDKIWCDVSMDFIEGLPNSGGK-SVIMVVVDRLSKAAHF 1157
Query: 1432 EPLAKITSA-KIVNFYWKRIVCRFGIPRAIVSDNGTQFSSNQTREFCREMGIQMRFASVE 1490
LA SA + + + G P +IVSD F+S+ +EF + G+++R +S
Sbjct: 1158 VALAHPYSALTVAQAFLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAY 1217
Query: 1491 HPQANGQVEYANRVILRGLRRRLKEAKGAWLEELPVVLWSYNTTVQSTTRETPFRMTYGV 1550
HPQ++GQ E NR + LR W + LP+ + YNT S+++ TPF + YG
Sbjct: 1218 HPQSDGQTEVVNRCLENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYG- 1276
Query: 1551 DAMLPAEIDNFTWRTEPDFEG---ENQANMAAELDLLSETRDEAHIRETAMKQRVAAKFN 1607
P + +++ + + NM L R + +++ A + R F+
Sbjct: 1277 -QAPPIHLPYLPGKSKVAVVARSLQERENMLLFLK-FHLMRAQHRMKQFADQHRTERTFD 1334
Query: 1608 ----SRVRVRDMQVGDLVLKWRSGASGNKLTPNWEGPYRIIKVLG 1648
V+++ + +VL+ KL+P + GPY+II+ G
Sbjct: 1335 IGDFVYVKLQPYRQQSVVLR-----VNQKLSPKYFGPYKIIEKCG 1374
>At2g06890 putative retroelement integrase
Length = 1215
Score = 179 bits (453), Expect = 2e-44
Identities = 114/376 (30%), Positives = 188/376 (49%), Gaps = 12/376 (3%)
Query: 720 PTWLANVVMVKKA----NGKWLMCVDYTDLNKACPKDSYPLPSIDSLVDGASGNELLSLM 775
P + N V K+ +G W MC D +N K +P+P +D ++D G+ + S +
Sbjct: 449 PAYRTNPVETKELQRQKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKI 508
Query: 776 DAYSGYHQIRMHPADEDKTAFMTARVNYCYRTMPFGLKNAGATYQRLMDRVFAGQVGRNM 835
D SGYHQIRM+ DE KTAF T Y + MPFGL +A +T+ RLM+ V +G +
Sbjct: 509 DLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFV 568
Query: 836 EVYVDDMIVKSVRGLDHHQDLEEAFGEIRKHNMRLNPEKCSFGVQGGKFLGFMITSRGIE 895
VY DD++V S +H + L+ +RK + N +KC+F FLGF++++ G++
Sbjct: 569 IVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVK 628
Query: 896 INPDKCKAIQQMKNPSNVKEVQRLTGRIAALSRFLPKSGDRSFPFFKCLQKNAAFEWTAE 955
++ +K KAI+ +P V EV+ G RF P + ++K+ F+W
Sbjct: 629 VDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKA 688
Query: 956 CEEAFVRLKELLSSPPILSKPMQGHPLHLYFAVSDSALSSVILQEGDGEHRVIYFVSHTL 1015
EEAF LK+ L++ P+L + S + +V++Q + ++I F S L
Sbjct: 689 QEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQ----DQKLIAFFSEKL 744
Query: 1016 QGAEVRYKKIEKAALAVLVTARRLRPYFQSFPVRVRTD-LPLRQVLQKPDMSGRLVAW-- 1072
GA + Y +K A++ +R + Y + TD L+ + + ++ R W
Sbjct: 745 GGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLNKRHARWVE 804
Query: 1073 SVELSEYGLQYDKRGK 1088
+E Y ++Y K+GK
Sbjct: 805 FIETFAYVIKY-KKGK 819
Score = 39.7 bits (91), Expect = 0.015
Identities = 31/122 (25%), Positives = 57/122 (46%), Gaps = 11/122 (9%)
Query: 1542 TPFRMTYGVDAMLPAEIDNFTWRTEPDFEGENQANMAAELDLLSETRDEAHIRETAMKQR 1601
+PF++ YG + + P ++ +G+ +A +L+ + + A +
Sbjct: 988 SPFQIVYGFNPISPFDLIPLPLSERVSIDGKKKA------ELVQQIHENARRNIEEKTKL 1041
Query: 1602 VAAKFNSRVRVRDMQVGDLVL----KWRSGASG-NKLTPNWEGPYRIIKVLGNEAYHLEE 1656
A + N R + +VGD+V K R A +KL P +GP++IIK + + AY L+
Sbjct: 1042 YAKQANKGRREQIFEVGDMVWIHLRKERFPAQRKSKLMPRIDGPFKIIKRINDNAYQLDL 1101
Query: 1657 LD 1658
D
Sbjct: 1102 QD 1103
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,876,206
Number of Sequences: 26719
Number of extensions: 1670458
Number of successful extensions: 5102
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 141
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 4537
Number of HSP's gapped (non-prelim): 433
length of query: 1675
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1562
effective length of database: 8,299,349
effective search space: 12963583138
effective search space used: 12963583138
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0151.7


