

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
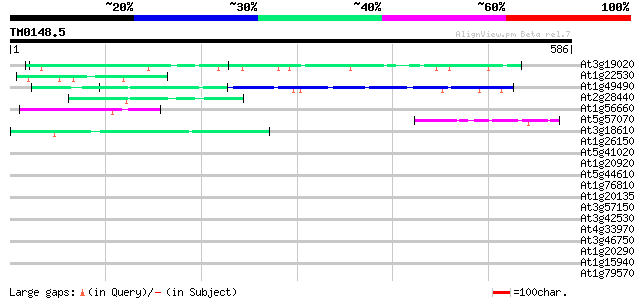
Query= TM0148.5
(586 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g19020 hypothetical protein 49 6e-06
At1g22530 unknown protein 47 3e-05
At1g49490 hypothetical protein 45 9e-05
At2g28440 En/Spm-like transposon protein 44 2e-04
At1g56660 hypothetical protein 44 3e-04
At5g57070 putative protein 43 4e-04
At3g18610 unknown protein 43 4e-04
At1g26150 Pto kinase interactor, putative 42 0.001
At5g41020 putative protein 41 0.002
At1g20920 putative RNA helicase 41 0.002
At5g44610 unknown protein 40 0.003
At1g76810 translation initiation factor IF-2 like protein 40 0.003
At1g20135 anter-specific proline-rich protein APG precursor 40 0.003
At3g57150 putative pseudouridine synthase (NAP57) 40 0.004
At3g42530 putative protein 40 0.004
At4g33970 extensin-like protein 40 0.005
At3g46750 hypothetical protein 40 0.005
At1g20290 hypothetical protein 40 0.005
At1g15940 T24D18.4 40 0.005
At1g79570 unknown protein 39 0.006
>At3g19020 hypothetical protein
Length = 951
Score = 49.3 bits (116), Expect = 6e-06
Identities = 57/231 (24%), Positives = 86/231 (36%), Gaps = 27/231 (11%)
Query: 17 REEKPEPLP-SPEQDQ--------EQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVV 67
+ E+P+P P SP+Q+ EQ K + K++ K+E + E + P+P
Sbjct: 469 KTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEES 528
Query: 68 ATAIPITLETPK-EQGAEPIEATVVHPVPDQRPAKTHRKKK--KKETVLEPQPEPEPEPK 124
P ETPK E+ +P P P++ P K++ K E +PQP + +P
Sbjct: 529 PKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQEQPP 588
Query: 125 ETSNTDHSEPTQLCSVKKK-------KKRKGAESNEAVAELATVCTEPPAVQGGVLPEPK 177
+T P V KKR+ + + E T + P V P
Sbjct: 589 KTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVH-----SPP 643
Query: 178 PKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPL 228
P P + P V PP + P P S P P PP+
Sbjct: 644 PPPPVHSPPPPVFSPPP---PMHSPPPPVYSPPPPVHSPPPPPVHSPPPPV 691
Score = 48.9 bits (115), Expect = 8e-06
Identities = 113/558 (20%), Positives = 190/558 (33%), Gaps = 99/558 (17%)
Query: 21 PEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPKE 80
P+P P+P K KK+I +E P+P E+PK
Sbjct: 404 PKPTPTP----------KAPEPKKEINPPNLEEPSK-----PKPE----------ESPKP 438
Query: 81 QGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSV 140
Q P T H + + K K++ +P+P+PE +E+ + +P Q
Sbjct: 439 QQPSPKPETPSHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPK 498
Query: 141 KKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIP 200
+ K++ ++ E + EPP + P+P +E T P P+P
Sbjct: 499 PESPKQESSKQEPPKPE-ESPKPEPPKPEESPKPQPPKQE---TPKPEESPKPQPPKQET 554
Query: 201 IDPEKNRKEAEPNESN---AEHPRACPEPPLGLAIPIDPEKNKG----ENLTPTDENELD 253
PE++ K P + E P+ P+PP P G E+ P D +
Sbjct: 555 PKPEESPKPQPPKQETPKPEESPK--PQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDAS 612
Query: 254 R-RNRREELVALVTSDEKIVNPRDHLV-------PLKSIPGFVLE----QQTQPGPVIPV 301
+ RR + + T + K +P+ V P+ S P V + P PV
Sbjct: 613 PIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSP 672
Query: 302 CPNSASLIGQSQTIPVVDTEQKINKRSRMSKMKRKRMNSMPDKHNTEPPETKK---RVLD 358
P + PV ++ ++S P ++ PP V
Sbjct: 673 PPP----VHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQS 728
Query: 359 REEPNKCNAKLPETLMLASNCHIDQAIPSDSNPAVPTNPEHKTPKKWKKRAKKKNVLLSE 418
P + P + + P +P P P H P
Sbjct: 729 PPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSP--PPPPVHSPPPP------------VH 774
Query: 419 GGKSNEHNGQPTETPVQKSVSPINAP-------SIDPTLPTDQPVPI-----------DL 460
H+ P PV P+++P S P + + P P+ +
Sbjct: 775 SPPPPVHSPPP---PVHSPPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANA 831
Query: 461 ATPKHSE-GKMSNAQRNNVLLSEGVKSNEQPTETPVQKS---VNPIKAPSIDPTLPTDQP 516
TP SE G++S + SE +++ +PV KS +P P ++ +P+ +P
Sbjct: 832 PTPSSSESGEISTPVQAPTPDSEDIEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEP 891
Query: 517 VPIDLATPKHSEGKMSKA 534
++ PK SE S +
Sbjct: 892 ---EVEAPKQSEATPSSS 906
Score = 38.1 bits (87), Expect = 0.014
Identities = 54/241 (22%), Positives = 80/241 (32%), Gaps = 44/241 (18%)
Query: 15 RRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPIT 74
++ KPE P P+ ++++ ++ K + K+E + E P+P P
Sbjct: 535 KQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEES-----PKPQPPKQEQPPK 589
Query: 75 LETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTD---- 130
E PK G+ P+E+ V + D P K R PQP P P +ET T
Sbjct: 590 TEAPK-MGSPPLESPVPNDPYDASPIKKRR----------PQP-PSPSTEETKTTSPQSP 637
Query: 131 -----------HSEPTQLCSVKKKKKRKGAESNEAVAELAT------------VCTEPPA 167
HS P + S + + V + PP
Sbjct: 638 PVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPP 697
Query: 168 VQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPP 227
V P P P + P V PP + P P + P S P P PP
Sbjct: 698 VHSPPPPVHSPPPPVHSPPPPVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPP 757
Query: 228 L 228
+
Sbjct: 758 V 758
Score = 37.7 bits (86), Expect = 0.018
Identities = 36/146 (24%), Positives = 53/146 (35%), Gaps = 16/146 (10%)
Query: 115 PQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEP--------- 165
P P+P P PK P L K K + + + + T EP
Sbjct: 402 PSPKPTPTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPE 461
Query: 166 -PAVQGGVL--PEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRA 222
P + P+PKP+ P + P+P P P++ + EP + E P+
Sbjct: 462 SPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPE-ESPK- 519
Query: 223 CPEPPLGLAIPIDPEKNKGENLTPTD 248
PEPP P P+ K E P +
Sbjct: 520 -PEPPKPEESP-KPQPPKQETPKPEE 543
Score = 29.6 bits (65), Expect = 4.9
Identities = 23/89 (25%), Positives = 39/89 (42%), Gaps = 4/89 (4%)
Query: 432 TPVQKSVSPI-NAPSIDPTLPTDQPVPIDLATPKHSEGKM-SNAQRNNVLLSEGVKS--N 487
TP+ + SP+ NAP+ + + P+ TP + + S++ + V S S +
Sbjct: 819 TPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSEDIEAPSDSNHSPVFKSSPAPSPDS 878
Query: 488 EQPTETPVQKSVNPIKAPSIDPTLPTDQP 516
E E PV S ++AP P+ P
Sbjct: 879 EPEVEAPVPSSEPEVEAPKQSEATPSSSP 907
>At1g22530 unknown protein
Length = 683
Score = 47.0 bits (110), Expect = 3e-05
Identities = 47/185 (25%), Positives = 73/185 (39%), Gaps = 35/185 (18%)
Query: 8 ELLHSKKRRRE---EKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEG----IESHEHDQVL 60
EL+ +RE P P P E+ E+ +++ KK+++K E E+ E ++
Sbjct: 103 ELVREALNKREFTAPPPPPAPVKEEKVEEKKTEETEEKKEEVKTEEKSLEAETKEEEKSA 162
Query: 61 LPEPV------VVATAIPITLETPKEQGAEPIEATVVHPVP-DQRPAKTHRKKKKKETVL 113
P V ++A PI ET KE+ T V P P + +PA + KKE +L
Sbjct: 163 APATVETKKEEILAAPAPIVAETKKEE-------TPVAPAPVETKPAAPVVAETKKEEIL 215
Query: 114 EPQP--------------EPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELA 159
P E P T+ T E + + K++ K A E A
Sbjct: 216 PAAPVTTETKVEEKVVPVETTPAAPVTTETKEEEKAAPVTTETKEEEKAAPGETKKEEKA 275
Query: 160 TVCTE 164
T T+
Sbjct: 276 TASTQ 280
Score = 32.0 bits (71), Expect = 0.98
Identities = 56/258 (21%), Positives = 90/258 (34%), Gaps = 38/258 (14%)
Query: 19 EKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETP 78
EK P+PE +E+ S+ + E + + + ++V + ++ + E+
Sbjct: 34 EKAVAAPAPEATEEKVVSEVA------VPETEVTAVKEEEVATGKEILQS-------ESF 80
Query: 79 KEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQ-- 136
KE+G E + R+ K P P P P +E +E T+
Sbjct: 81 KEEGYLASELQEAEKNALAELKELVREALNKREFTAPPPPPAPVKEEKVEEKKTEETEEK 140
Query: 137 LCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEP-PL 195
VK ++K AE+ E A E + P P E E P V P P
Sbjct: 141 KEEVKTEEKSLEAETKEEEKSAAPATVETKKEEILAAPAPIVAETKKEETP-VAPAPVET 199
Query: 196 GLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLTP---TDENEL 252
A P+ E ++E P A P+ E E + P T +
Sbjct: 200 KPAAPVVAETKKEEILP------------------AAPVTTETKVEEKVVPVETTPAAPV 241
Query: 253 DRRNRREELVALVTSDEK 270
+ EE A VT++ K
Sbjct: 242 TTETKEEEKAAPVTTETK 259
>At1g49490 hypothetical protein
Length = 847
Score = 45.4 bits (106), Expect = 9e-05
Identities = 48/207 (23%), Positives = 77/207 (37%), Gaps = 17/207 (8%)
Query: 23 PLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPKEQG 82
P P+P + E SK + K E+ + + P+P + PK +
Sbjct: 394 PSPNPPRTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPSPKP-----------QPPKHES 442
Query: 83 AEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKK 142
+P E H +P Q+ + + K +++ QP+PE PK EPT+ S
Sbjct: 443 PKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEESPK-PEQPQIPEPTKPVSPPN 501
Query: 143 KKKRKGAES-NEAVAELATVCTEPPAVQGGVLPEPKPKEPC-STEHPRVCPEPPLGLAIP 200
+ + + +A PP V+ +P P+P P S P P PP+
Sbjct: 502 EAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSP-- 559
Query: 201 IDPEKNRKEAEPNESNAEHPRACPEPP 227
P P+ + P A P PP
Sbjct: 560 -PPPVYSSPPPPHVYSPPPPVASPPPP 585
Score = 44.7 bits (104), Expect = 1e-04
Identities = 96/476 (20%), Positives = 142/476 (29%), Gaps = 82/476 (17%)
Query: 95 PDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEA 154
P P +T K K + P+P +P+ + P + K + E+
Sbjct: 394 PSPNPPRTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPSPKPQPPKHESPKPEEPENKH- 452
Query: 155 VAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNE 214
EL P PKP++P E P+ PE P PE + + PNE
Sbjct: 453 --ELPKQKESPKPQPSKPEDSPKPEQPKPEESPK--PEQPQ------IPEPTKPVSPPNE 502
Query: 215 SNAEHPRACPEPPLGLAIPIDPEKNKGENLTPTDENELDRRNRREELVALVTSDEKIVNP 274
+ P P D K P + E R + + + I +P
Sbjct: 503 AQGPTPDD----------PYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSP 552
Query: 275 RDHLVPLKSIPGFVLEQQTQP---GPVIPVC----PNSASLIGQSQTIPVVDTEQKINKR 327
P+ S P V P P PV P+ + PV +
Sbjct: 553 PP---PVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSP 609
Query: 328 SRMSKMKRKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCHIDQAIPS 387
S + + P H+ PP P N P + + PS
Sbjct: 610 PPPSPV----YSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPP---MGAPTPTQAPTPS 662
Query: 388 DSNPAVPTNPEHKTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSPINAPSID 447
VPT + + + +S+ + +PT+ P S AP++
Sbjct: 663 SETTQVPTPSSESDQSQILSPVQAPTPV-----QSSTPSSEPTQVPTPSSSESYQAPNLS 717
Query: 448 PTL---------------------------PTDQPVPIDLATPKHSEGKMSNAQRNNVLL 480
P P+ P PI P H+ S ++
Sbjct: 718 PVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPI--LEPVHAPTPNSKPVQSPTPS 775
Query: 481 SEGVKSNEQ------PTETPVQKSVNPIKAPSIDPTLP----TDQPVPIDLATPKH 526
SE V S EQ P TPV S P +PS D ++P D D P H
Sbjct: 776 SEPVSSPEQSEEVEAPEPTPVNPSSVPSSSPSTDTSIPPPENNDDDDDGDFVLPPH 831
Score = 36.2 bits (82), Expect = 0.052
Identities = 58/252 (23%), Positives = 86/252 (34%), Gaps = 43/252 (17%)
Query: 16 RREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITL 75
+++E P+P PS +D S K + K + ES + +Q +PEP +
Sbjct: 456 KQKESPKPQPSKPED-----SPKPEQPKPE------ESPKPEQPQIPEPTK-------PV 497
Query: 76 ETPKE-QGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEP 134
P E QG P + PV ++R K ++T + P P P P S +S P
Sbjct: 498 SPPNEAQGPTPDDPYDASPVKNRRSPPP---PKVEDTRVPPPQPPMPSPSPPSPI-YSPP 553
Query: 135 TQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPP 194
+ S + V + PP V P P P P P PP
Sbjct: 554 PPVHSPPPPVYSSPPPPH--------VYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPP 605
Query: 195 LGLAIP----IDPEKNRKEAEPNESNAEHPRACP-------EPPLGLAIPID-PEKNKGE 242
+ P P P + P P +PP+G P P +
Sbjct: 606 VFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSET 665
Query: 243 NLTPTDENELDR 254
PT +E D+
Sbjct: 666 TQVPTPSSESDQ 677
Score = 34.7 bits (78), Expect = 0.15
Identities = 38/192 (19%), Positives = 63/192 (32%), Gaps = 14/192 (7%)
Query: 59 VLLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPE 118
V P P V + P + +P P P P P TH T P
Sbjct: 599 VFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTH------NTNQPPMGA 652
Query: 119 PEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKP 178
P P T +++ ++ S + + + +T +EP V P P
Sbjct: 653 PTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQV-------PTP 705
Query: 179 KEPCSTEHPRVCP-EPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPE 237
S + P + P + P + P + + P+ + + P P P L P
Sbjct: 706 SSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPN 765
Query: 238 KNKGENLTPTDE 249
++ TP+ E
Sbjct: 766 SKPVQSPTPSSE 777
Score = 32.0 bits (71), Expect = 0.98
Identities = 26/116 (22%), Positives = 39/116 (33%), Gaps = 7/116 (6%)
Query: 23 PLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETP---K 79
P+ +P Q + S + + E ++ V P PV T T + P
Sbjct: 683 PVQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSS 742
Query: 80 EQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPT 135
E P +A P P P K P EP P+++ + EPT
Sbjct: 743 ESNQSPSQA----PTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEPT 794
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 43.9 bits (102), Expect = 2e-04
Identities = 42/200 (21%), Positives = 70/200 (35%), Gaps = 33/200 (16%)
Query: 62 PEPVVVATAIPITLETPKEQGAEPIEATVVHPV--PDQRPAKTHRKKKKKETVLEPQPEP 119
P P V+ + ++P + P E + + P P++ ++++ L P P
Sbjct: 28 PSPAAVSPGREPSTDSPLSPSSSPEEDSPLSPSSSPEEDSPLPPSSSPEEDSPLAPSSSP 87
Query: 120 E---------------PEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTE 164
E P+P +S+ + P S + + S+ LA +
Sbjct: 88 EVDSPLAPSSSPEVDSPQPP-SSSPEADSPLPPSSSPEANSPQSPASSPKPESLADSPSP 146
Query: 165 PPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACP 224
PP P P+P+ P S +P P P P + + +P P P
Sbjct: 147 PP-------PPPQPESPSSPSYPEPAPVPA--------PSDDDSDDDPEPETEYFPSPAP 191
Query: 225 EPPLGLAIPIDPEKNKGENL 244
P LG+A I GE L
Sbjct: 192 SPELGMAQDIKASDAAGEEL 211
>At1g56660 hypothetical protein
Length = 522
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/150 (24%), Positives = 67/150 (44%), Gaps = 8/150 (5%)
Query: 11 HSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATA 70
H + + ++ + + ++++++SC+++K++K K K+E ES E + L
Sbjct: 235 HDETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEK 294
Query: 71 IPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKK---KKKETVLEPQPEPEPEPKETS 127
E K + + E + D + K + K KKKETV++ E KET
Sbjct: 295 PEKEDEGKKTKEHDATEQEMDDEAADHKEGKKKKNKDKAKKKETVIDEVCE-----KETK 349
Query: 128 NTDHSEPTQLCSVKKKKKRKGAESNEAVAE 157
+ D E KKK++K + + V E
Sbjct: 350 DKDDDEGETKQKKNKKKEKKSEKGEKDVKE 379
Score = 41.6 bits (96), Expect = 0.001
Identities = 35/148 (23%), Positives = 61/148 (40%), Gaps = 17/148 (11%)
Query: 14 KRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPI 73
K+ + EKPE ++ +E ++++ + +EG + D+ E V+
Sbjct: 288 KKGKGEKPEKEDEGKKTKEHDATEQEMDDEAADHKEGKKKKNKDKAKKKETVIDEVCEKE 347
Query: 74 TLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQ--------PEPEPEPKE 125
T + ++G + ++ K ++ KKKE LE + EPE E KE
Sbjct: 348 TKDKDDDEGETKQKKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDIKLEEPEAEKKE 407
Query: 126 TSNTDHSEPTQLCSVKKKKKRKGAESNE 153
+T+ KKK K +G ES E
Sbjct: 408 EDDTEE---------KKKSKVEGGESEE 426
Score = 38.9 bits (89), Expect = 0.008
Identities = 88/429 (20%), Positives = 158/429 (36%), Gaps = 74/429 (17%)
Query: 12 SKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEG------------IESHEHDQV 59
SKK + ++K + + S ++ + KK + K EEG +E HE +
Sbjct: 57 SKKDKEKKKGKNVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDLEVKESDVKVEEHEKEHK 116
Query: 60 LLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEP 119
E LE KE + + P+++ K ++KK ++ E +
Sbjct: 117 KGKEKK------HEELEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELE 170
Query: 120 EPEPKETSNTDHSEP-TQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKP 178
E + K+ + E T+ K KK++K E +++ + + +G + E +
Sbjct: 171 EEDGKKNKKKEKDESGTEEKKKKPKKEKKQKEESKSNEDKKVKGKKEKGEKGDLEKEDEE 230
Query: 179 KEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACP--------EPPLGL 230
K+ E + E D +KN+K+ E +ES AE + P E
Sbjct: 231 KKKEHDETDQEMKEK--------DSKKNKKK-EKDESCAEEKKKKPDKEKKEKDESTEKE 281
Query: 231 AIPIDPEKNKGENLTPTDE----NELDRRNRREELVALVTSDEKIVNPRDHLVPLKSIPG 286
+ +K KGE DE E D + + A + K +D +++
Sbjct: 282 DKKLKGKKGKGEKPEKEDEGKKTKEHDATEQEMDDEAADHKEGKKKKNKDKAKKKETVID 341
Query: 287 FVLEQQTQPGPVIPVCPNSASLIGQSQTIPVVDTEQKINKRSRMSKMKRKRMNSMPDKHN 346
V E++T + + +T+QK NK+ + K ++ + DK
Sbjct: 342 EVCEKET-----------------KDKDDDEGETKQKKNKK-KEKKSEKGEKDVKEDKKK 383
Query: 347 TEPPETK--KRVLDREEPNKCNAKLPETLMLASNCHIDQAIPSDSNPAVPTNPEHKTPKK 404
P ET+ R + EEP + +T + + E + KK
Sbjct: 384 ENPLETEVMSRDIKLEEPEAEKKEEDDT--------------EEKKKSKVEGGESEEGKK 429
Query: 405 WKKRAKKKN 413
KK+ KKKN
Sbjct: 430 KKKKDKKKN 438
Score = 38.5 bits (88), Expect = 0.010
Identities = 40/152 (26%), Positives = 67/152 (43%), Gaps = 31/152 (20%)
Query: 8 ELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVV 67
++ K+ EE + E+D+ + KKK+ KK+K ++E +S+E +V
Sbjct: 161 DVSQEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQKEESKSNEDKKV-------- 212
Query: 68 ATAIPITLETPKEQG------AEPIEATVVHPVPDQ----RPAKTHRKKKKKETVL-EPQ 116
+ KE+G E E H DQ + +K ++KK+K E+ E +
Sbjct: 213 --------KGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESCAEEKK 264
Query: 117 PEPEPEPKETSNTDHSEPTQLCSVKKKKKRKG 148
+P+ E KE + E +L K KK KG
Sbjct: 265 KKPDKEKKEKDESTEKEDKKL----KGKKGKG 292
Score = 37.0 bits (84), Expect = 0.031
Identities = 104/567 (18%), Positives = 192/567 (33%), Gaps = 146/567 (25%)
Query: 28 EQDQEQSCSKKKRRKKKK-------IKEEGIESHEHDQVLLPEPVVVATAIPITLETPKE 80
++D+E S KK ++KKK +KE+ + + D ++ + E
Sbjct: 48 KKDEESSGKSKKDKEKKKGKNVDSEVKEDKDDDKKKDGKMVSKK--------------HE 93
Query: 81 QGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSV 140
+G +E V + K H+K K+K+ + E E E K+ N
Sbjct: 94 EGHGDLEVKE-SDVKVEEHEKEHKKGKEKK---HEELEEEKEGKKKKN------------ 137
Query: 141 KKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIP 200
KK+K G E A+ + K E S E + E
Sbjct: 138 KKEKDESGPEEKNKKAD-----------------KEKKHEDVSQEKEELEEE-------- 172
Query: 201 IDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLTPTDENELDRRNRREE 260
D +KN+K+ E +ES E + P+ EK + E ++ ++ + + E
Sbjct: 173 -DGKKNKKK-EKDESGTEEKKKKPKK----------EKKQKEESKSNEDKKVKGKKEKGE 220
Query: 261 LVALVTSDEKIVNPRDHLVPLKSIPGFVLEQQTQPGPVIPVCPNSASLIGQSQTIPVVDT 320
L DE+ D +Q+ + +
Sbjct: 221 KGDLEKEDEEKKKEHDET-----------DQEMK------------------------EK 245
Query: 321 EQKINKRSRMSKMKRKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCH 380
+ K NK+ + + PDK E E+ ++ + + K + PE
Sbjct: 246 DSKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTK 305
Query: 381 IDQAIPSDSNPAVPTNPEHKTPKKWKKRAKKKNVLLSE--GGKSNEHNGQPTETPVQKSV 438
A + + + E K KK K +AKKK ++ E ++ + + ET +K+
Sbjct: 306 EHDATEQEMDDEAADHKEGK-KKKNKDKAKKKETVIDEVCEKETKDKDDDEGETKQKKN- 363
Query: 439 SPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNVLLSEGVKSNEQPTETPVQKS 498
K E K +++ + K E P ET V
Sbjct: 364 -------------------------KKKEKKSEKGEKD----VKEDKKKENPLETEVMSR 394
Query: 499 VNPIKAPSIDPTL--PTDQPVPIDLATPKHSEGKMSKAQRRKKNKR--ALKSAWSESVHH 554
++ P + T++ + + EGK K + +KKNK+ + +E
Sbjct: 395 DIKLEEPEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDTKEPKMTEDEEE 454
Query: 555 SSDQNAKTPVQRTESLKLVEAKSVKAK 581
D + ++ +++ + + K VK K
Sbjct: 455 KKDDSKDVKIEGSKAKEEKKDKDVKKK 481
Score = 37.0 bits (84), Expect = 0.031
Identities = 40/167 (23%), Positives = 67/167 (39%), Gaps = 9/167 (5%)
Query: 18 EEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLET 77
+E E + D E +KK +KK+K E+G + + D+ + I LE
Sbjct: 341 DEVCEKETKDKDDDEGETKQKKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDIKLEE 400
Query: 78 PK-----EQGAEPIEATVVHPVPDQ---RPAKTHRKKKKKETVLEPQ-PEPEPEPKETSN 128
P+ E E + + V + + K +KK KK+ EP+ E E E K+ S
Sbjct: 401 PEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDTKEPKMTEDEEEKKDDSK 460
Query: 129 TDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPE 175
E ++ KK K K + + +L T + G ++ E
Sbjct: 461 DVKIEGSKAKEEKKDKDVKKKKGGNDIGKLKTKLAKIDEKIGALMEE 507
>At5g57070 putative protein
Length = 607
Score = 43.1 bits (100), Expect = 4e-04
Identities = 42/157 (26%), Positives = 64/157 (40%), Gaps = 18/157 (11%)
Query: 424 EHNGQPTETPVQKSV-SPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNVLLSE 482
E +P E + V P + P P P P P + P+ + + RN L
Sbjct: 224 EEESEPKEIQIDTFVVKPSSPPQQPPATPPPPPPPPPVEVPQKPR-RTHRSVRNRDL--- 279
Query: 483 GVKSNEQPTETPVQKSVNPIKAPSIDPTLPTDQPVPIDLATPKHSEGKMSKAQRRKKN-- 540
+ N + +ET +++ P PS P P P P+ ATP +G + QRRK N
Sbjct: 280 --QENAKRSETKFKRTFQP--PPSPPPPPPPPPPQPLIAATPPRKQGTL---QRRKSNAA 332
Query: 541 ---KRALKSAWSESVHHSSDQNAKTPVQRTESLKLVE 574
K S +++ Q +K +R ES +VE
Sbjct: 333 KEIKMVFASLYNQGKKKKKLQKSKRK-ERIESSPMVE 368
Score = 35.4 bits (80), Expect = 0.089
Identities = 36/160 (22%), Positives = 59/160 (36%), Gaps = 33/160 (20%)
Query: 386 PSDSNPAVPTNPEHKTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSPINAPS 445
P PA P P P + ++ ++ + S + + N + +ET +++ P PS
Sbjct: 244 PPQQPPATPPPPPPPPPVEVPQKPRRTH--RSVRNRDLQENAKRSETKFKRTFQP--PPS 299
Query: 446 IDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNV---------LLSEGVKSN--------- 487
P P P P+ ATP +G + + N L ++G K
Sbjct: 300 PPPPPPPPPPQPLIAATPPRKQGTLQRRKSNAAKEIKMVFASLYNQGKKKKKLQKSKRKE 359
Query: 488 --------EQPTETPVQKSVNPIKAPSIDPTLPTDQPVPI 519
E TE P +S+ P +P P P P P+
Sbjct: 360 RIESSPMVEDVTEPPQYQSLIPPPSP---PPPPPPPPPPL 396
Score = 31.6 bits (70), Expect = 1.3
Identities = 42/192 (21%), Positives = 66/192 (33%), Gaps = 39/192 (20%)
Query: 37 KKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPD 96
KKK++ +K ++E IES + + E P+ Q P + P P
Sbjct: 347 KKKKKLQKSKRKERIES--------------SPMVEDVTEPPQYQSLIPPPSPPPPPPPP 392
Query: 97 QRPAKTHRK----------KKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKR 146
P ++ + K K+ P P P P P+ T + P ++ S + +
Sbjct: 393 PPPLRSSQSVFYGLFKKGVKSNKKIHSVPAPPPPPPPRYTQFDPQTPPRRVKSGRPPRPT 452
Query: 147 KGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKN 206
K NE + P +Q + P P P P PPL + D K
Sbjct: 453 KPKNFNEENNGQGS-----PLIQ--ITPPPPPPPPFRV--------PPLKYVVSGDFAKI 497
Query: 207 RKEAEPNESNAE 218
R S+ E
Sbjct: 498 RSNQSSRCSSPE 509
>At3g18610 unknown protein
Length = 636
Score = 43.1 bits (100), Expect = 4e-04
Identities = 50/274 (18%), Positives = 99/274 (35%), Gaps = 15/274 (5%)
Query: 2 DSHSQSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKK---IKEEGIESHEHDQ 58
DS S E+ +KKR K + S D + + ++ KK+ +++ +ES D
Sbjct: 96 DSSSDEEIAPAKKRPEPIKKAKVESSSSDDDSTSDEETAPVKKQPAVLEKAKVESSSSDD 155
Query: 59 -VLLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQP 117
E V P LE K + + D + KK+T + +
Sbjct: 156 DSSSDEETVPVKKQPAVLEKAKIESSSS---------DDDSSSDEETVPMKKQTAVLEKA 206
Query: 118 EPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPK 177
+ E + ++ EPT K+ ++ + + E V +P V E
Sbjct: 207 KAESSSSDDGSSSDEEPTPAKKEPIVVKKDSSDESSSDEETPVVKKKPTTVVKDAKAESS 266
Query: 178 PKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPE 237
E S+ P P + + + K++ +E +++ + E P + +
Sbjct: 267 SSEEESSSDDE--PTPAKKPTVVKNAKPAAKDSSSSEEDSDEEESDDEKPPTKKAKVSSK 324
Query: 238 KNKGENLTPTDENELDRRNRREELVALVTSDEKI 271
+K E+ + +E D+ ++E V D +
Sbjct: 325 TSKQESSSDESSDESDKEESKDEKVTPKKKDSDV 358
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 41.6 bits (96), Expect = 0.001
Identities = 43/189 (22%), Positives = 59/189 (30%), Gaps = 3/189 (1%)
Query: 62 PEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEP 121
P P PIT +P P E+ P PD K + P+ P P
Sbjct: 127 PPPTEAPPTTPITSPSPPTNPPPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSP 186
Query: 122 EPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEP 181
E P+ S + +E E P G P P P P
Sbjct: 187 PASEIPPPPRHLPSPPASERPSTPPSDSEHPSPPPPGHPKRREQPPPPGSKRPTPSPPSP 246
Query: 182 CSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKG 241
++ P V P PP + P K +P SN+ P P ++ P P K+
Sbjct: 247 SDSKRP-VHPSPPSPPEETLPPPK--PSPDPLPSNSSSPPTLLPPSSVVSPPSPPRKSVS 303
Query: 242 ENLTPTDEN 250
P+ N
Sbjct: 304 GPDNPSPNN 312
Score = 33.9 bits (76), Expect = 0.26
Identities = 40/149 (26%), Positives = 51/149 (33%), Gaps = 27/149 (18%)
Query: 85 PIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKK 144
P E + P+P + P + P P P P T T S PT
Sbjct: 95 PPEPSPPPPLPTEAPPPANPVSSPPPESSPPPPPPTEAPPTTPITSPSPPT--------N 146
Query: 145 KRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLG------LA 198
ES ++ +PP+ LP PK P S PR P PP
Sbjct: 147 PPPPPESPPSLP-----APDPPS---NPLPPPKLVPP-SHSPPRHLPSPPASEIPPPPRH 197
Query: 199 IPIDPEKNRKEAEPNESNAEHPRACPEPP 227
+P P R P++S EHP P PP
Sbjct: 198 LPSPPASERPSTPPSDS--EHP--SPPPP 222
Score = 33.9 bits (76), Expect = 0.26
Identities = 27/95 (28%), Positives = 36/95 (37%), Gaps = 3/95 (3%)
Query: 433 PVQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNVLLSEGVKSNEQPTE 492
P VSP PS P LPT+ P P + + E + + S PT
Sbjct: 87 PTTIPVSPPPEPSPPPPLPTEAPPPANPVSSPPPESSPPPPPPTEAPPTTPITSPSPPTN 146
Query: 493 TPVQKSVNPIKAPSIDPTLPTDQPVPIDLATPKHS 527
P +P P+ DP P++ P L P HS
Sbjct: 147 PPPPPE-SPPSLPAPDP--PSNPLPPPKLVPPSHS 178
Score = 30.4 bits (67), Expect = 2.9
Identities = 69/308 (22%), Positives = 91/308 (29%), Gaps = 45/308 (14%)
Query: 110 ETVLEPQPEP-EPEPKETSNTD-HSEPTQLCSVKKKKKRKGAESNEAVAEL-ATVCTEPP 166
+ P EP P ET+NT S P + S T+ PP
Sbjct: 36 DNATSPTREPTNGNPPETTNTPAQSSPPPETPLSSPPPEPSPPSPSLTGPPPTTIPVSPP 95
Query: 167 AVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEP 226
PEP P P TE P PP PE + P E+ P P P
Sbjct: 96 -------PEPSPPPPLPTEAP-----PPANPVSSPPPESSPPPPPPTEAPPTTPITSPSP 143
Query: 227 PL----------GLAIPIDPEKNKGENLTPTDENELDRRNRREELVALVTSDEKIVNPRD 276
P L P DP N P +L + +I P
Sbjct: 144 PTNPPPPPESPPSLPAP-DPPSN------PLPPPKLVPPSHSPPRHLPSPPASEIPPPPR 196
Query: 277 HL--VPLKSIPGFVLEQQTQPGPVIPVCPNSASLIGQSQTIPVVDTEQKINKRSRMSKMK 334
HL P P P P P P + + P +++ S K
Sbjct: 197 HLPSPPASERPSTPPSDSEHPSPPPPGHPK------RREQPPPPGSKRPTPSPPSPSDSK 250
Query: 335 RKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCHIDQAIPSDSNPAVP 394
R S P + P ET +P N+ P TL+ S+ + P + + P
Sbjct: 251 RPVHPSPP----SPPEETLPPPKPSPDPLPSNSSSPPTLLPPSSV-VSPPSPPRKSVSGP 305
Query: 395 TNPEHKTP 402
NP P
Sbjct: 306 DNPSPNNP 313
>At5g41020 putative protein
Length = 588
Score = 40.8 bits (94), Expect = 0.002
Identities = 43/172 (25%), Positives = 72/172 (41%), Gaps = 41/172 (23%)
Query: 8 ELLHSKKRRREEKPEPLPSPEQDQEQSCSKKK----------------RRKKKKIKEEGI 51
E+ KK+++ +KP + S D + SKK+ ++ K+K KE +
Sbjct: 68 EVFIVKKKKKSKKPIRIDSEAVDAVKKKSKKRSKETKADSEAEDDGVEKKSKEKSKETKV 127
Query: 52 ESHEHDQVLLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKET 111
+S HD V + ++ KE G + IE T V D++ K+K+ +T
Sbjct: 128 DSEAHDGVKRKKK-----------KSKKESGGDVIENTESSKVSDKKKG----KRKRDDT 172
Query: 112 VLEPQPEPEPEPKETSN------TDHSEPTQLCSV----KKKKKRKGAESNE 153
L + + E K +N E L S KK+KK+K +E +E
Sbjct: 173 DLGAEENIDKEVKRKNNKKKPSVDSDVEDINLDSTNDGKKKRKKKKQSEDSE 224
Score = 37.7 bits (86), Expect = 0.018
Identities = 40/158 (25%), Positives = 66/158 (41%), Gaps = 15/158 (9%)
Query: 9 LLHSKKRRREEKPEPLPSPEQDQE----QSCSKKKRRKKKKIKEEGIE--SHEHDQVLLP 62
+ KK ++++ + S E +E S K K +K+KK K E + + E ++
Sbjct: 1 MAEEKKNKKKKSDAKVDSEETGEEFISEHSSMKDKEKKRKKNKRENKDGFTGEDMEITGR 60
Query: 63 EPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEP--- 119
E + + I + K + I++ V V K KK+ KET + + E
Sbjct: 61 ESEKLGDEVFIVKKKKKSKKPIRIDSEAVDAV------KKKSKKRSKETKADSEAEDDGV 114
Query: 120 EPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAE 157
E + KE S + VK+KKK+ ES V E
Sbjct: 115 EKKSKEKSKETKVDSEAHDGVKRKKKKSKKESGGDVIE 152
Score = 30.4 bits (67), Expect = 2.9
Identities = 15/46 (32%), Positives = 26/46 (55%)
Query: 13 KKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQ 58
KKR+++++ E + E + KKRRKKKK K++ S ++
Sbjct: 212 KKRKKKKQSEDSETEENGLNSTKDAKKRRKKKKKKKQSEVSEAEEK 257
Score = 30.4 bits (67), Expect = 2.9
Identities = 36/155 (23%), Positives = 65/155 (41%), Gaps = 16/155 (10%)
Query: 1 MDSHSQSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIK---EEGIESHEHD 57
+DS + + KK+ ++E + + + S KK +RK+ EE I+
Sbjct: 127 VDSEAHDGVKRKKKKSKKESGGDVIENTESSKVSDKKKGKRKRDDTDLGAEENIDKEVKR 186
Query: 58 QVLLPEPVVVATAIPITLETP--------KEQGAEPIEATVVHPVPDQRPAKTHRKKKKK 109
+ +P V + I L++ K++ +E E T + + + AK RKKKKK
Sbjct: 187 KNNKKKPSVDSDVEDINLDSTNDGKKKRKKKKQSEDSE-TEENGLNSTKDAKKRRKKKKK 245
Query: 110 ETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKK 144
+ Q E +++ +D T S K+ K
Sbjct: 246 ----KKQSEVSEAEEKSDKSDEDLTTPSTSSKRVK 276
>At1g20920 putative RNA helicase
Length = 1166
Score = 40.8 bits (94), Expect = 0.002
Identities = 53/281 (18%), Positives = 105/281 (36%), Gaps = 31/281 (11%)
Query: 13 KKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKE-EGIESHEHDQVLLPEPVVVATAI 71
+ R REE+ + E+D+E+ +++ R+K+++KE E E + ++ +
Sbjct: 136 RDREREERKDKEREREKDRERREREREEREKERVKERERREREDGERDRREREKERGSRR 195
Query: 72 PITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDH 131
E +E G E + V + +R RK+K++E + E PK S D+
Sbjct: 196 NRERERSREVGNEESDDDVKRDLKRRRKEGGERKEKEREKSVGRSSRHEDSPKRKSVEDN 255
Query: 132 SEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCP 191
E + KK + + E + + + L K + ++
Sbjct: 256 GEKKE-----KKTREEELEDEQKKLDEEVEKRRRRVQEWQELKRKKEEAESESKGDADGN 310
Query: 192 EPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLTPTDENE 251
EP G A ++ E + +E P E + + E + E P ++ +
Sbjct: 311 EPKAGKAWTLEGESDDEEGHPEEKS------------------ETEMDVDEETKPENDGD 352
Query: 252 LDRRNRREELVALVTS-------DEKIVNPRDHLVPLKSIP 285
+ E A V+ DE+ ++P D + +P
Sbjct: 353 AKMVDLENETAATVSESGGDGAVDEEEIDPLDAFMNTMVLP 393
>At5g44610 unknown protein
Length = 168
Score = 40.4 bits (93), Expect = 0.003
Identities = 37/152 (24%), Positives = 62/152 (40%), Gaps = 24/152 (15%)
Query: 38 KKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIP------------------ITLETPK 79
KK +K K+E +E + +V + E VVV T P I + K
Sbjct: 13 KKLFEKSPAKKEVVEEEKPREVEVVEEVVVKTEEPAKEGETKPEEIIATGEKEIEIVEEK 72
Query: 80 EQGAEPIEATVVHPVPDQRPAKTHRKK----KKKETVLEPQPEPEPEPKETSNTDHSEPT 135
++ A+P+E V+ +++PA KK ++K+ +E + +P E K+ + E
Sbjct: 73 KEEAKPVEVPVLAAAEEKKPAVEEEKKTAPVEEKKPAVEEEKKPAVEEKKPVEEEKKEVV 132
Query: 136 QLCSVKKKKKRKGAESNEAVAELATVCTEPPA 167
V + K E+ V E E PA
Sbjct: 133 AAVPVAETPSTKAPET--PVVETPAKAPETPA 162
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 40.4 bits (93), Expect = 0.003
Identities = 45/185 (24%), Positives = 71/185 (38%), Gaps = 30/185 (16%)
Query: 79 KEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLC 138
+E+ + + A + +RPA + ++K QPEP + + E T
Sbjct: 299 EEEDLDKLLAALGETPAAERPASSTPVEEKAA-----QPEPVAPVENAGEKEGEEETAAA 353
Query: 139 SVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLA 198
KKKKK K E A A AT E + E K +E +
Sbjct: 354 KKKKKKKEKEKEKKAAAAAAATSSVE--------VKEEKQEE---------------SVT 390
Query: 199 IPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLTPTDENELDRRNRR 258
P+ P+K K+A+ + + P+ E LA + E+ K + E + R R+
Sbjct: 391 EPLQPKK--KDAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEEEKLRKEEEERRRQ 448
Query: 259 EELVA 263
EEL A
Sbjct: 449 EELEA 453
Score = 34.7 bits (78), Expect = 0.15
Identities = 62/279 (22%), Positives = 105/279 (37%), Gaps = 48/279 (17%)
Query: 20 KPEPLPSPEQ------DQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPI 73
+PEP+ E ++E + +KKK++KK+K KE+ + +++ +
Sbjct: 331 QPEPVAPVENAGEKEGEEETAAAKKKKKKKEKEKEKKAAA----------AAAATSSVEV 380
Query: 74 TLETPKEQGAEPI------------EATVVHPVPDQRPAKTHR-----KKKKKETVLEPQ 116
E +E EP+ E + V + + A R +KKK+E +
Sbjct: 381 KEEKQEESVTEPLQPKKKDAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEEEKLRK 440
Query: 117 PEPEPEPKETSNTDHSEPTQLCSVKKKKK--RKGAESNEAVAELATVCTEPPAVQ----- 169
E E +E E + K+K+K RK E A+ T + A +
Sbjct: 441 EEEERRRQEELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAKQKTEAQKREAFKNQLLA 500
Query: 170 -GGVLP-EPKPKEPCSTEHPRVCPEPPLGLAIPIDPE-KNRKEAEPNESNAEHPRACPEP 226
GG LP + S++ P + ID + E EP E+ A+ E
Sbjct: 501 AGGGLPVADNDGDATSSKRPIYANKKKSSRQKGIDTSVQGEDEVEPKENQADEQDTLGEV 560
Query: 227 PLGLAIPIDPEK--NKGENLTPTD---ENELDRRNRREE 260
L +D + N EN P D EN ++ + +E
Sbjct: 561 GLTDTGKVDLIELVNTDENSGPADVAQENGVEEDDEEDE 599
Score = 32.7 bits (73), Expect = 0.58
Identities = 96/501 (19%), Positives = 161/501 (31%), Gaps = 89/501 (17%)
Query: 101 KTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKR----KGAESNEAVA 156
K KKK + + + +TS+T + E + KKKKK + + E +
Sbjct: 245 KKSSKKKGGSVLASVGDDSVADETKTSDTKNVEVVETGKSKKKKKNNKSGRTVQEEEDLD 304
Query: 157 ELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESN 216
+L E PA + +P E PE P+ P +N E E E
Sbjct: 305 KLLAALGETPAAE-------RPASSTPVEEKAAQPE-------PVAPVENAGEKEGEEET 350
Query: 217 AEHPRACPEPPLGLAIPIDPEKNKGENLTPTDENELDRRNRREELVALVTSDEKIVNPRD 276
A + + + EK T E+ + E + + +K +
Sbjct: 351 AAAKKKKKKKEK------EKEKKAAAAAAATSSVEVKEEKQEESVTEPLQPKKKDAKGK- 403
Query: 277 HLVPLKSIPGFVLEQQTQPGPVIPVCPNSASLIGQSQTIPVVDTEQKINKRSRMSKMKRK 336
K IP V E Q + + Q + E++ K K +
Sbjct: 404 --AAEKKIPKHVREMQ--------------EALARRQ-----EAEERKKKEEEEKLRKEE 442
Query: 337 RMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLAS--------NCHIDQAIPSD 388
++ + E K++ ++E+ KL L+ A +Q + +
Sbjct: 443 EERRRQEELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAKQKTEAQKREAFKNQLLAAG 502
Query: 389 SNPAVPTNPEHKTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSP-INAPSID 447
V N T K A KK KS+ G T + V P N
Sbjct: 503 GGLPVADNDGDATSSKRPIYANKK--------KSSRQKGIDTSVQGEDEVEPKENQADEQ 554
Query: 448 PTL------PTDQPVPIDLATPKHSEGKMSNAQRNNV---------------LLSEGVKS 486
TL T + I+L + G AQ N V + +K
Sbjct: 555 DTLGEVGLTDTGKVDLIELVNTDENSGPADVAQENGVEEDDEEDEWDAKSWGTVDLNLKG 614
Query: 487 NEQPTETPVQKSVNPIKAPSIDPTLPTDQP-VPIDLATPKHSEGKMSKAQRRKK--NKRA 543
+ E Q V +I + +P + ATP+ + +K R K +K+
Sbjct: 615 DFDDEEEEAQPVVKKELKDAISKAHDSGKPLIAAVKATPEVEDATRTKRATRAKDASKKG 674
Query: 544 LKSAWSESVHHSSDQNAKTPV 564
A SES+ ++N ++P+
Sbjct: 675 KGLAPSESI--EGEENLRSPI 693
Score = 31.2 bits (69), Expect = 1.7
Identities = 32/130 (24%), Positives = 54/130 (40%), Gaps = 14/130 (10%)
Query: 6 QSELLHSKKRRREE---KPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLP 62
+ E KK+ EE K E +++ E + KR++K+K KE+ + ++L
Sbjct: 423 RQEAEERKKKEEEEKLRKEEEERRRQEELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTA 482
Query: 63 EPVVVATAIPITLETPKEQ-----GAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEP-- 115
+ A E K Q G P+ +RP ++KK ++ ++
Sbjct: 483 KQKTEAQ----KREAFKNQLLAAGGGLPVADNDGDATSSKRPIYANKKKSSRQKGIDTSV 538
Query: 116 QPEPEPEPKE 125
Q E E EPKE
Sbjct: 539 QGEDEVEPKE 548
>At1g20135 anter-specific proline-rich protein APG precursor
Length = 534
Score = 40.4 bits (93), Expect = 0.003
Identities = 36/126 (28%), Positives = 44/126 (34%), Gaps = 24/126 (19%)
Query: 113 LEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGV 172
L P+P P+P P D S K G S + C PP
Sbjct: 47 LWPRPYPQPWPMNPPTPDPSP--------KPVAPPGPSSKPVAPPGPSPCPSPPPK---- 94
Query: 173 LPEPKPK-----EPCSTEHPRVCPEP-PLGLAIPIDPEKNRKEAEPNESN-----AEHPR 221
P+PKP PC + P+ P+P P P P+ K A P E A P
Sbjct: 95 -PQPKPPPAPSPSPCPSPPPKPQPKPVPPPACPPTPPKPQPKPAPPPEPKPAPPPAPKPV 153
Query: 222 ACPEPP 227
CP PP
Sbjct: 154 PCPSPP 159
Score = 39.3 bits (90), Expect = 0.006
Identities = 38/141 (26%), Positives = 49/141 (33%), Gaps = 12/141 (8%)
Query: 93 PVPDQRPAKTHRKKKKKETVLEPQPE--PEPEPKETSNTDHSEPTQLCSVKKKKKRKGAE 150
P PD P + V P P P P PK + C K +
Sbjct: 61 PTPDPSPKPVAPPGPSSKPVAPPGPSPCPSPPPKPQPKPPPAPSPSPCPSPPPKPQP--- 117
Query: 151 SNEAVAELATVCTEP-PAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLA---IPIDPEKN 206
+ V A T P P + PEPKP P + + P CP PP A P+ P
Sbjct: 118 --KPVPPPACPPTPPKPQPKPAPPPEPKPAPPPAPK-PVPCPSPPKPPAPTPKPVPPHGP 174
Query: 207 RKEAEPNESNAEHPRACPEPP 227
+ P + A P+ P PP
Sbjct: 175 PPKPAPAPTPAPSPKPAPSPP 195
Score = 34.3 bits (77), Expect = 0.20
Identities = 37/163 (22%), Positives = 52/163 (31%), Gaps = 26/163 (15%)
Query: 84 EPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKK 143
+P V P P +P +PQP+P P P P+ S K
Sbjct: 64 DPSPKPVAPPGPSSKPVAPPGPSPCPSPPPKPQPKPPPAPS---------PSPCPSPPPK 114
Query: 144 KKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEP-PLGLAIPID 202
+ K PP P+P+PK P P+ P P P + P
Sbjct: 115 PQPKPV---------------PPPACPPTPPKPQPK-PAPPPEPKPAPPPAPKPVPCPSP 158
Query: 203 PEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLT 245
P+ +P + P+ P P + P K EN T
Sbjct: 159 PKPPAPTPKPVPPHGPPPKPAPAPTPAPSPKPAPSPPKPENKT 201
>At3g57150 putative pseudouridine synthase (NAP57)
Length = 565
Score = 40.0 bits (92), Expect = 0.004
Identities = 32/118 (27%), Positives = 59/118 (49%), Gaps = 23/118 (19%)
Query: 5 SQSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEP 64
++ ++ SKK+++++K E E+++E KK+++KKK KEE IE +V P+
Sbjct: 469 AEEKVKSSKKKKKKDKEE-----EKEEEAGSEKKEKKKKKDKKEEVIE-----EVASPKS 518
Query: 65 VVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPE 122
E K++ ++ EA V D+ A+ KKKKK+ + + E +
Sbjct: 519 -----------EKKKKKKSKDTEAAV--DAEDESAAEKSEKKKKKKDKKKKNKDSEDD 563
Score = 32.0 bits (71), Expect = 0.98
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 13/89 (14%)
Query: 72 PITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDH 131
P+T + K + E EA + K+ +KKKKK+ E + E E KE
Sbjct: 453 PVTTKKSKTKEVEGEEA--------EEKVKSSKKKKKKDKEEEKEEEAGSEKKEKKKKKD 504
Query: 132 SEPTQLCSV-----KKKKKRKGAESNEAV 155
+ + V +KKKK+K ++ AV
Sbjct: 505 KKEEVIEEVASPKSEKKKKKKSKDTEAAV 533
Score = 31.6 bits (70), Expect = 1.3
Identities = 28/100 (28%), Positives = 40/100 (40%), Gaps = 3/100 (3%)
Query: 68 ATAIPITLETPKEQGAEPIEATVVHPVPDQRPAK-THRKKKKKETVLEPQPEPEPEPKET 126
A A P ++ E G H PA T +K K KE E E K+
Sbjct: 420 AAAAPEEIKADAENGEAGEARKRKHDDSSDSPAPVTTKKSKTKEVEGEEAEEKVKSSKKK 479
Query: 127 SNTDHSEPTQ--LCSVKKKKKRKGAESNEAVAELATVCTE 164
D E + S KK+KK+K + E + E+A+ +E
Sbjct: 480 KKKDKEEEKEEEAGSEKKEKKKKKDKKEEVIEEVASPKSE 519
>At3g42530 putative protein
Length = 889
Score = 40.0 bits (92), Expect = 0.004
Identities = 43/197 (21%), Positives = 76/197 (37%), Gaps = 21/197 (10%)
Query: 331 SKMKRKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCHIDQAIPS-DS 389
S+ KRK+ +P + T ++ L + + PE + H+D+A S DS
Sbjct: 437 SRSKRKKTMEVPIQSQTPETSSQSHTLSKHVFGQ-----PEVAQQVPDDHLDKAQDSSDS 491
Query: 390 NPAVPTNPEHKTPKKWKKRAKKKNVLLSEGGKSN--------EHNGQPTETPVQKSVSPI 441
P + T + + ++ + + N + N + Q V P
Sbjct: 492 TPLINTEITDDPMDVFVTPLQSEHSNNDDANEGNPVYDTDVKDQNANEEDVDSQMQVDPS 551
Query: 442 NAPSIDPTLPTDQPVPIDLAT---PKHSEGKMSNAQRNNVLLSEGVKSNEQPTETPVQKS 498
+ PS++ LP +Q D A+ P G + + N +E +SN + Q
Sbjct: 552 SNPSVEKVLPLNQDHISDDASERVPAIPSGLDLSKEHN----TEEQESNANEEDVDSQMK 607
Query: 499 VNPIKAPSIDPTLPTDQ 515
V+P PS++ LP Q
Sbjct: 608 VDPRSDPSVEKLLPLHQ 624
>At4g33970 extensin-like protein
Length = 699
Score = 39.7 bits (91), Expect = 0.005
Identities = 35/184 (19%), Positives = 61/184 (33%), Gaps = 9/184 (4%)
Query: 350 PETKKRVLDREEPNKCNAKLPE---------TLMLASNCHIDQAIPSDSNPAVPTNPEHK 400
P ++ + ++ C A PE + +C D+ S P+ P+
Sbjct: 369 PGGSRKEIALDDTRNCLASRPEQRSAQECAVVINRPVDCSKDKCAGGSSTPSKPSPVHKP 428
Query: 401 TPKKWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSPINAPSIDPTLPTDQPVPIDL 460
TP K V + K + P + P +P++ PS P D+P P+
Sbjct: 429 TPVPTTPVHKPTPVPTTPVQKPSPVPTTPVQKPSPVPTTPVHEPSPVLATPVDKPSPVPS 488
Query: 461 ATPKHSEGKMSNAQRNNVLLSEGVKSNEQPTETPVQKSVNPIKAPSIDPTLPTDQPVPID 520
+ + + Q ++ V P PV P+ +P P P P+
Sbjct: 489 RPVQKPQPPKESPQPDDPYDQSPVTKRRSPPPAPVNSPPPPVYSPPPPPPPVHSPPPPVH 548
Query: 521 LATP 524
P
Sbjct: 549 SPPP 552
Score = 36.2 bits (82), Expect = 0.052
Identities = 48/210 (22%), Positives = 71/210 (32%), Gaps = 20/210 (9%)
Query: 27 PEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPKEQGAEPI 86
PEQ Q C+ R K++ P PV T +P T P+
Sbjct: 389 PEQRSAQECAVVINRPVDCSKDKCAGGSSTPSK--PSPVHKPTPVPTT----------PV 436
Query: 87 -EATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKK 145
+ T V P Q+P+ +K + + P EP P + D P V+K +
Sbjct: 437 HKPTPVPTTPVQKPSPVPTTPVQKPSPVPTTPVHEPSPVLATPVDKPSPVPSRPVQKPQP 496
Query: 146 RKGAESNEAVAELATVC---TEPPAVQGGVLPE----PKPKEPCSTEHPRVCPEPPLGLA 198
K + + + + V + PPA P P P P + P V PP +
Sbjct: 497 PKESPQPDDPYDQSPVTKRRSPPPAPVNSPPPPVYSPPPPPPPVHSPPPPVHSPPPPPVY 556
Query: 199 IPIDPEKNRKEAEPNESNAEHPRACPEPPL 228
P P P + P P PP+
Sbjct: 557 SPPPPPPPVHSPPPPVFSPPPPVYSPPPPV 586
>At3g46750 hypothetical protein
Length = 415
Score = 39.7 bits (91), Expect = 0.005
Identities = 40/166 (24%), Positives = 72/166 (43%), Gaps = 16/166 (9%)
Query: 12 SKKRRREEKPEPLPSP---EQDQE------QSCSKKKRRKKKKIKEEGIESHEHDQVLLP 62
S K+R +E+ + SP +D E S S + + K +K++EEG E E D L
Sbjct: 78 STKKRHDEEGDATMSPFCRSEDHEVREVGYASLSPRDKSKDRKVREEGGEEEEEDPEYLG 137
Query: 63 EPVVVATAIPITL-ETPKEQGAE-PI--EATVVHPVPDQRPAKTHRKK---KKKETVLEP 115
P+ + P L ET ++ E P+ E V+ +P ++ A+ +K K E +
Sbjct: 138 APMYESKKAPEELKETARQHPRENPVITETNVLSVLPAKQDAEQEQKDCNGSKTEHPVIS 197
Query: 116 QPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATV 161
+ + K+ D T + +K +++ E ++ TV
Sbjct: 198 EKNVLSDVKQEKPADSDTTTIVTESSEKTRKECTSQQEPISPSKTV 243
>At1g20290 hypothetical protein
Length = 497
Score = 39.7 bits (91), Expect = 0.005
Identities = 29/142 (20%), Positives = 63/142 (43%), Gaps = 9/142 (6%)
Query: 6 QSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPV 65
Q+E ++ EE+ E E+++E+ +++ ++++ +EE E E ++ E
Sbjct: 362 QAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 421
Query: 66 VVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKE 125
E +E+ E E ++ + +++++E E + E E E +E
Sbjct: 422 E---------EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 472
Query: 126 TSNTDHSEPTQLCSVKKKKKRK 147
+ E + VKKKKK++
Sbjct: 473 EEEEEEEEEKKYMIVKKKKKKR 494
Score = 38.9 bits (89), Expect = 0.008
Identities = 28/147 (19%), Positives = 63/147 (42%), Gaps = 12/147 (8%)
Query: 1 MDSHSQSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVL 60
+ + + E ++ EE+ E E+++E+ +++ ++++ +EE E E ++
Sbjct: 361 VQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 420
Query: 61 LPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPE 120
E E +E+ E E ++ + +++++E E + E E
Sbjct: 421 EEE------------EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 468
Query: 121 PEPKETSNTDHSEPTQLCSVKKKKKRK 147
E +E + E + KKKKKR+
Sbjct: 469 EEEEEEEEEEEEEKKYMIVKKKKKKRR 495
>At1g15940 T24D18.4
Length = 1012
Score = 39.7 bits (91), Expect = 0.005
Identities = 109/633 (17%), Positives = 214/633 (33%), Gaps = 64/633 (10%)
Query: 1 MDSHSQSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVL 60
+ + +E R+R KP+ L +PE+ S K+ ++K++ + + +V
Sbjct: 352 LSESTDAETASGSTRKRGWKPKSLMNPEEGYSFKTSSSKKVQEKELGDSSLGKVAAKKVP 411
Query: 61 LPEPV---VVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQP 117
LP V + I ++ G+ T + D + + KK+TV + P
Sbjct: 412 LPSKVGQTNQSVVISLSSSGRARTGSRKRSRTKMEET-DHDVSSVATQPAKKQTVKKTNP 470
Query: 118 EPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPA---------- 167
E K S KK+K G A LA P+
Sbjct: 471 AKEDLTKSNVKKHEDGIKTGKSSKKEKADNGLAKTSAKKPLAETMMVKPSGKKLVHSDAK 530
Query: 168 ---VQGGVLPEPKPKEPCSTEHPRVCPEPPL---GLAIPIDPEKNRKEAEPNESNAEHPR 221
+G + P P+ S + P A P+ R E ESN
Sbjct: 531 KKNSEGASMDTPIPQSSKSKKKDSRATTPATKKSEQAPKSHPKMKRIAGEEVESNTNE-- 588
Query: 222 ACPEPPLGLAI----PIDPEKNKG-----------ENLTPTDENELDRRNRREELVALVT 266
E +G + P+D + +G +T +D + + ++E +
Sbjct: 589 -LGEELVGKRVNVWWPLDKKFYEGVIKSYCRVKKMHQVTYSDGDVEELNLKKERFKIIED 647
Query: 267 SDEKIVNPRDHLVPLKSIPGFVLEQQTQPGPVIP--VCPNSASLIGQS-QTIPVVDTEQK 323
+ D L+ + F+ ++++ ++ V P+S+ + S QT+ D+
Sbjct: 648 KSSASEDKEDDLLESTPLSAFIQREKSKKRKIVSKNVEPSSSPEVRSSMQTMKKKDSVTD 707
Query: 324 INKRSRMSKMKRKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCHIDQ 383
K+++ +K K +++ P+ + ++ K++ EP+K + + + H
Sbjct: 708 SIKQTKRTKGALKAVSNEPESTTGKNLKSLKKL--NGEPDKTRGRTGKKQKVTQAMHRKI 765
Query: 384 AIPSDSNPAVPTNPEHKTPKKWKKR-------------AKKKNVLLSEGGKSNEHNGQPT 430
D + T E + K K+ + NV G+ E +PT
Sbjct: 766 EKDCDEQEDLETKDEEDSLKLGKESDAEPDRMEDHQELPENHNVETKTDGEEQEAAKEPT 825
Query: 431 ETPVQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRNNVLLSEGVKSNEQP 490
P P D P A PK + A+ N L ++ E
Sbjct: 826 AESKTNGEEPNAEPETDGKEHKSLKEP--NAEPKSDGEEQEAAKEPNAELKTDGENQEAA 883
Query: 491 TETPVQKSVNPIKAPSIDPTLPTDQPVPIDLATPKHSEGKMSKAQRRKKNKRALKSAWSE 550
E ++ + + D +Q + +EG+ K+ + K E
Sbjct: 884 KELTAERKTDEEEHKVAD---EVEQKSQKETNVEPEAEGEEQKSVEEPNAEPKTKVEEKE 940
Query: 551 SVHHSSDQNAKTPVQRTESLKLVEAKSVKAKSF 583
S + +Q A T + E + + + + +++
Sbjct: 941 S---AKEQTADTKLIEKEDMSKTKGEEIDKETY 970
Score = 39.3 bits (90), Expect = 0.006
Identities = 79/398 (19%), Positives = 149/398 (36%), Gaps = 63/398 (15%)
Query: 8 ELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQV-----LLP 62
+++ K E+K + L E + ++++ KK+KI + +E +V +
Sbjct: 643 KIIEDKSSASEDKEDDLL--ESTPLSAFIQREKSKKRKIVSKNVEPSSSPEVRSSMQTMK 700
Query: 63 EPVVVATAIPITLETP---KEQGAEPIEAT--------VVHPVPDQRPAKTHRKKKKKET 111
+ V +I T T K EP T ++ PD+ +T +K+K +
Sbjct: 701 KKDSVTDSIKQTKRTKGALKAVSNEPESTTGKNLKSLKKLNGEPDKTRGRTGKKQKVTQA 760
Query: 112 VLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGG 171
+ + E + + +E T E + + + E ++ + E V T+ +
Sbjct: 761 -MHRKIEKDCDEQEDLETKDEEDSLKLGKESDAEPDRMEDHQELPENHNVETKTDGEEQE 819
Query: 172 VLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPN---ESNAEHPRACPEPPL 228
EP + + E P PE D ++++ EPN +S+ E A EP
Sbjct: 820 AAKEPTAESKTNGEEPNAEPE--------TDGKEHKSLKEPNAEPKSDGEEQEAAKEPNA 871
Query: 229 GLAIPIDPEKNKGENLTPTDENELDRRNRREE-----LVALVTSDEKIVNPRDHLVPLKS 283
L K GEN E +R+ EE V + E V P KS
Sbjct: 872 EL-------KTDGENQEAAKELTAERKTDEEEHKVADEVEQKSQKETNVEPEAEGEEQKS 924
Query: 284 IPGFVLEQQTQPGPVIPVCPNSASLIGQSQTIPVVDTEQKINKRSRMSKMK-----RKRM 338
+ E +T+ + + ++ + K+ ++ MSK K ++
Sbjct: 925 VEEPNAEPKTK--------------VEEKESAKEQTADTKLIEKEDMSKTKGEEIDKETY 970
Query: 339 NSMPD--KHNTEPPETKKRVLDREEPNKCNAKLPETLM 374
+S+P+ K E E +RV+ E A++ T++
Sbjct: 971 SSIPETGKVGNEAEEDDQRVIKELEEESDKAEVSTTVL 1008
>At1g79570 unknown protein
Length = 1248
Score = 39.3 bits (90), Expect = 0.006
Identities = 27/112 (24%), Positives = 50/112 (44%), Gaps = 10/112 (8%)
Query: 92 HPVPDQRPAKTHR--KKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGA 149
+P P P T +++K + ++ + E EPE ++T DH P Q+ V+
Sbjct: 492 YPYPGSTPKSTQALAEEQKVSSDMKIREEVEPENRKTPGNDHQNPPQIDDVE-------V 544
Query: 150 ESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPI 201
++ V E+A V T PP+ +LP + +T P + + +P+
Sbjct: 545 RNHNQVREMA-VATTPPSQDAHLLPPSRDPRQNTTAKPATYRDAVITGQVPL 595
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.306 0.126 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,405,815
Number of Sequences: 26719
Number of extensions: 804141
Number of successful extensions: 4637
Number of sequences better than 10.0: 269
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 218
Number of HSP's that attempted gapping in prelim test: 3329
Number of HSP's gapped (non-prelim): 895
length of query: 586
length of database: 11,318,596
effective HSP length: 105
effective length of query: 481
effective length of database: 8,513,101
effective search space: 4094801581
effective search space used: 4094801581
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0148.5


