

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0148.17
(215 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
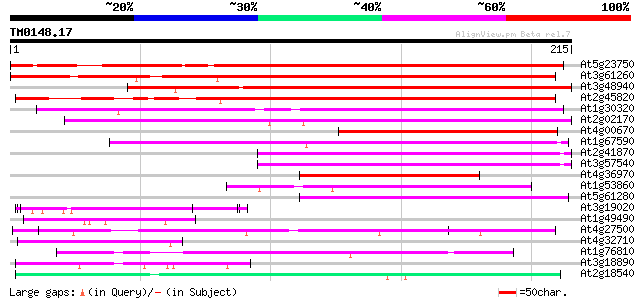
Score E
Sequences producing significant alignments: (bits) Value
At5g23750 Unknown protein (MRO11.3) 226 5e-60
At3g61260 putative DNA-binding protein 216 9e-57
At3g48940 remorin -like protein 200 5e-52
At2g45820 remorin 184 3e-47
At1g30320 unknown protein 80 8e-16
At2g02170 unknown protein 79 2e-15
At4g00670 unknown protein 77 7e-15
At1g67590 unknown protein 77 7e-15
At2g41870 unknown protein 75 3e-14
At3g57540 unknown protein 72 2e-13
At4g36970 unknown protein 53 1e-07
At1g53860 unknown protein 53 1e-07
At5g61280 putative protein 47 6e-06
At3g19020 hypothetical protein 47 8e-06
At1g49490 hypothetical protein 46 2e-05
At4g27500 unknown protein 45 2e-05
At4g32710 putative protein kinase 45 3e-05
At1g76810 translation initiation factor IF-2 like protein 45 4e-05
At3g18890 unknown protein 44 8e-05
At2g18540 putative vicilin storage protein (globulin-like) 43 1e-04
>At5g23750 Unknown protein (MRO11.3)
Length = 202
Score = 226 bits (577), Expect = 5e-60
Identities = 125/212 (58%), Positives = 150/212 (69%), Gaps = 13/212 (6%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M EE+PKKV +E+ S P P P E KP AP+E ++P P+
Sbjct: 1 MAEEEPKKV-TETVSEPTPTPEVPVE---------KPAAAADVAPQEKPVAPPPVLPSPA 50
Query: 61 PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI 120
P E K +DSKAIV V + EE+ EGS+NRDAVL RV TEKR+SLIKAWEE+EK
Sbjct: 51 PAEEKQ-EDSKAIVPV--VPKEVEEEKKEGSVNRDAVLARVETEKRMSLIKAWEEAEKCK 107
Query: 121 ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR 180
+NKA KKLS I +WEN+K AA E EL+K+EE LEKKKA YVE++KNKIA +H+EAEEKR
Sbjct: 108 VENKAEKKLSSIGSWENNKKAAVEAELKKMEEQLEKKKAEYVEQMKNKIAQIHKEAEEKR 167
Query: 181 AFIEAKKGEDLLKAEELAAKYRATGTAPKKPF 212
A IEAK+GE++LKAEELAAKYRATGTAPKK F
Sbjct: 168 AMIEAKRGEEILKAEELAAKYRATGTAPKKLF 199
>At3g61260 putative DNA-binding protein
Length = 212
Score = 216 bits (549), Expect = 9e-57
Identities = 119/213 (55%), Positives = 146/213 (67%), Gaps = 11/213 (5%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKE---DVAEEKSIIP 57
M EEQ +ESESP+ PAPA TPAP P P P + DVAEEK
Sbjct: 1 MAEEQKIALESESPAKVTTPAPA---DTPAPAPAEIPAPAPAPTPADVTKDVAEEKI--- 54
Query: 58 QPSPPESKPVDDSKAIVKVEK-TQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEES 116
+PP + DDSKA+ VEK +E A KP S++RD L ++ EKRLS ++AWEES
Sbjct: 55 -QNPPPEQIFDDSKALTVVEKPVEEPAPAKPASASLDRDVKLADLSKEKRLSFVRAWEES 113
Query: 117 EKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREA 176
EKS A+NKA KK++D+ AWENSK AA E +L+KIEE LEKKKA Y E++KNK+A +H+EA
Sbjct: 114 EKSKAENKAEKKIADVHAWENSKKAAVEAQLKKIEEQLEKKKAEYAERMKNKVAAIHKEA 173
Query: 177 EEKRAFIEAKKGEDLLKAEELAAKYRATGTAPK 209
EE+RA IEAK+GED+LKAEE AAKYRATG PK
Sbjct: 174 EERRAMIEAKRGEDVLKAEETAAKYRATGIVPK 206
>At3g48940 remorin -like protein
Length = 175
Score = 200 bits (508), Expect = 5e-52
Identities = 103/171 (60%), Positives = 132/171 (76%), Gaps = 2/171 (1%)
Query: 46 KEDVAEEKSIIPQPSPP-ESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATE 104
++ V +++ +PSPP + + DDSKAIV V +E E+K + GS++RDAVL R+ +
Sbjct: 6 QKKVIMPEAVASEPSPPSKEEKSDDSKAIVLVVAAKEPTEDKKV-GSVHRDAVLVRLEQD 64
Query: 105 KRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEK 164
KR+SLIKAWEE+EKS +NKA KK+S + AWENSK A+ E EL+KIEE L KKKA Y E+
Sbjct: 65 KRISLIKAWEEAEKSKVENKAQKKISSVGAWENSKKASVEAELKKIEEQLNKKKAHYTEQ 124
Query: 165 LKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
+KNKIA +H+EAEEKRA EAK+GED+LKAEE+AAKYRATGTAP K F FF
Sbjct: 125 MKNKIAQIHKEAEEKRAMTEAKRGEDVLKAEEMAAKYRATGTAPTKLFGFF 175
>At2g45820 remorin
Length = 190
Score = 184 bits (467), Expect = 3e-47
Identities = 109/208 (52%), Positives = 134/208 (64%), Gaps = 28/208 (13%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
E++ KV+ ESP+ AP E P PV +VA+EK I P P
Sbjct: 4 EQKTSKVDVESPA------------VLAPAKEPTPAPV-------EVADEK--IHNPPPV 42
Query: 63 ESKPVDDSKAIVKVEKT-QEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIA 121
ESK A+ VEK +E +K GS +RD +L + EK+ S IKAWEESEKS A
Sbjct: 43 ESK------ALAVVEKPIEEHTPKKASSGSADRDVILADLEKEKKTSFIKAWEESEKSKA 96
Query: 122 DNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRA 181
+N+A KK+SD+ AWENSK AA E +LRKIEE LEKKKA Y EK+KNK+A +H+ AEEKRA
Sbjct: 97 ENRAQKKISDVHAWENSKKAAVEAQLRKIEEKLEKKKAQYGEKMKNKVAAIHKLAEEKRA 156
Query: 182 FIEAKKGEDLLKAEELAAKYRATGTAPK 209
+EAKKGE+LLKAEE+ AKYRATG PK
Sbjct: 157 MVEAKKGEELLKAEEMGAKYRATGVVPK 184
>At1g30320 unknown protein
Length = 509
Score = 80.1 bits (196), Expect = 8e-16
Identities = 60/203 (29%), Positives = 94/203 (45%), Gaps = 7/203 (3%)
Query: 11 SESPSNPPPPAPAPTESTPAPEAEAKPEPV-VHDAPKEDVAEEKSIIPQPSPPESKPVDD 69
S +P P +PT S P+ +PE + + +++EE+ + V
Sbjct: 305 SVTPVGATTPLRSPTSSLPSTPRGGQPEESSMSKNTRRELSEEEEKAKTRREIVALGVQL 364
Query: 70 SKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKL 129
K + ++E E K G ++ EKR + AWEE+EKS + + ++
Sbjct: 365 GKMNIAAWASKEEEENKKNNGDAEE---AQKIEFEKRAT---AWEEAEKSKHNARYKREE 418
Query: 130 SDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGE 189
I AWE+ + A E E+R+IE +E+ KA K+ KIA+ + +EEKRA EA+K
Sbjct: 419 IRIQAWESQEKAKLEAEMRRIEAKVEQMKAEAEAKIMKKIALAKQRSEEKRALAEARKTR 478
Query: 190 DLLKAEELAAKYRATGTAPKKPF 212
D KA A R TG P +
Sbjct: 479 DAEKAVAEAQYIRETGRIPASSY 501
>At2g02170 unknown protein
Length = 486
Score = 79.0 bits (193), Expect = 2e-15
Identities = 59/226 (26%), Positives = 93/226 (41%), Gaps = 32/226 (14%)
Query: 22 PAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQE 81
P+ S + + P+ P + ++ P SP S+P + + +
Sbjct: 257 PSTARSVSMRDMGTEMTPIASQEPSRNGTPIRATTPIRSPISSEPSSPGRQASASPMSNK 316
Query: 82 AAEEKPLEGSINRDAVL------------------------TRVATEKRLSLIK------ 111
EK L+ R+ ++ T + T+ L K
Sbjct: 317 ELSEKELQMKTRREIMVLGTQLGKFNIAAWASKEDEDKDASTSLKTKASLQTSKSVSEAR 376
Query: 112 --AWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKI 169
AWEE+EK+ + ++ I AWEN + A E E++K E +E+ K ++L K+
Sbjct: 377 ATAWEEAEKAKHMARFRREEMKIQAWENHQKAKSEAEMKKTEVKVERIKGRAQDRLMKKL 436
Query: 170 AMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
A + R+AEEKRA EAKK K E+ A + R TG P FS F
Sbjct: 437 ATIERKAEEKRAAAEAKKDHQAAKTEKQAEQIRRTGKVPSLLFSCF 482
>At4g00670 unknown protein
Length = 116
Score = 77.0 bits (188), Expect = 7e-15
Identities = 36/84 (42%), Positives = 56/84 (65%)
Query: 127 KKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAK 186
KKL DIS WE K E EL +I+ ++ KK EKL+N+ A VH +A++K+A ++ +
Sbjct: 27 KKLLDISGWEKKKTTKIESELARIQRKMDSKKMEKSEKLRNEKAAVHAKAQKKKADVQTR 86
Query: 187 KGEDLLKAEELAAKYRATGTAPKK 210
+ +++L AEE AA+++A G PKK
Sbjct: 87 RAQEILDAEEAAARFQAAGKIPKK 110
>At1g67590 unknown protein
Length = 347
Score = 77.0 bits (188), Expect = 7e-15
Identities = 57/180 (31%), Positives = 85/180 (46%), Gaps = 5/180 (2%)
Query: 39 PVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVL 98
P+ P ++ P P + PV S+ V E E S N + V
Sbjct: 158 PIGSQEPSRTATPVRATTPVGRSPVTSPVRASQRGEAVGVVMETVTEVRRVESNNSEKVN 217
Query: 99 TRVATEKRLSLIKA----WEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENL 154
V ++K +S ++A W+E+E++ + ++ I AWEN + E+E++K+E
Sbjct: 218 GFVESKKAMSAMEARAMAWDEAERAKFMARYKREEVKIQAWENHEKRKAEMEMKKMEVKA 277
Query: 155 EKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSF 214
E+ KA EKL NK+A R AEE+RA EAK E +K E A R +G P FSF
Sbjct: 278 ERMKARAEEKLANKLAATKRIAEERRANAEAKLNEKAVKTSEKADYIRRSGHLPSS-FSF 336
>At2g41870 unknown protein
Length = 274
Score = 74.7 bits (182), Expect = 3e-14
Identities = 42/120 (35%), Positives = 71/120 (59%), Gaps = 1/120 (0%)
Query: 96 AVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLE 155
+ + RV E+ + I AW+ ++ + +N+ ++ + I+ W N ++ ++KIE LE
Sbjct: 153 STVQRVKREEVEAKITAWQTAKLAKINNRFKREDAVINGWFNEQVNKANSWMKKIERKLE 212
Query: 156 KKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
++KA +EK +N +A R+AEE+RA EAK+G ++ K E+A RA G P K SFF
Sbjct: 213 ERKAKAMEKTQNNVAKAQRKAEERRATAEAKRGTEVAKVVEVANLMRALGRPPAKR-SFF 271
>At3g57540 unknown protein
Length = 296
Score = 72.4 bits (176), Expect = 2e-13
Identities = 41/120 (34%), Positives = 70/120 (58%), Gaps = 1/120 (0%)
Query: 96 AVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLE 155
A + RV E+ + I AW+ ++ + +N+ ++ + I+ W N ++ ++KIE LE
Sbjct: 175 ASVQRVKREEVEAKITAWQTAKVAKINNRFKRQDAVINGWLNEQVHRANSWMKKIERKLE 234
Query: 156 KKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
++A +EK +NK+A R+AEE+RA E K+G ++ + E+A RA G P K SFF
Sbjct: 235 DRRAKAMEKTQNKVAKAQRKAEERRATAEGKRGTEVARVLEVANLMRAVGRPPAKR-SFF 293
>At4g36970 unknown protein
Length = 427
Score = 53.1 bits (126), Expect = 1e-07
Identities = 25/69 (36%), Positives = 44/69 (63%)
Query: 112 AWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAM 171
+W+ SE ++ +K ++ + I+AWEN + A E +RK+E LEKKK+ ++K+ NK+
Sbjct: 305 SWDISEPAMTLSKLQREEAKIAAWENLQKAKAEAAIRKLEVKLEKKKSASMDKILNKLQT 364
Query: 172 VHREAEEKR 180
+A+E R
Sbjct: 365 AKIKAQEMR 373
>At1g53860 unknown protein
Length = 442
Score = 53.1 bits (126), Expect = 1e-07
Identities = 36/121 (29%), Positives = 62/121 (50%), Gaps = 7/121 (5%)
Query: 84 EEKPLEGSINR---DAVLTRVATEKRLSLIKAWEESEKSIAD-NKAHKKLSDISAWENSK 139
EE+ + S+ ++ L R +E + L W++ + I + ++ + I AW N +
Sbjct: 300 EEEEISKSLRHFDMESELRRSVSESKAPL---WDDEDDKIKFCQRYQREEAKIQAWVNLE 356
Query: 140 IAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAA 199
A E + RK+E ++K ++ EKL ++ MVHR AE+ RA + E + KA E A
Sbjct: 357 NAKAEAQSRKLEVKIQKMRSNLEEKLMKRMDMVHRRAEDWRATARQQHVEQMQKAAETAR 416
Query: 200 K 200
K
Sbjct: 417 K 417
>At5g61280 putative protein
Length = 263
Score = 47.4 bits (111), Expect = 6e-06
Identities = 27/103 (26%), Positives = 47/103 (45%)
Query: 112 AWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAM 171
+WE+S+ + K +DI WEN + A + + K + LEK+K + + K+K+A
Sbjct: 158 SWEKSQIKKIRLRYEKMKADIVGWENERKLAATLLMEKRKSELEKRKGINNQHYKSKLAR 217
Query: 172 VHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSF 214
+ A+ + +E K+ + K TG P F F
Sbjct: 218 IQLIADGAKKQLEEKRRSKEAQVHGKVKKMSRTGKVPNNYFCF 260
>At3g19020 hypothetical protein
Length = 951
Score = 47.0 bits (110), Expect = 8e-06
Identities = 32/93 (34%), Positives = 42/93 (44%), Gaps = 8/93 (8%)
Query: 3 EEQPKKVESESPSNPPP---PAP-APTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQ 58
+E PK ES P P P P P P + TP PE KP+P + PK E+S PQ
Sbjct: 509 QEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPK----PEESPKPQ 564
Query: 59 PSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGS 91
P E+ ++S ++ Q E P GS
Sbjct: 565 PPKQETPKPEESPKPQPPKQEQPPKTEAPKMGS 597
Score = 45.8 bits (107), Expect = 2e-05
Identities = 24/66 (36%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Query: 5 QPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPES 64
QP K E+ P P P P P + TP PE KP+P + P + A + P SP +
Sbjct: 548 QPPKQETPKPEESPKPQP-PKQETPKPEESPKPQPPKQEQPPKTEAPKMGSPPLESPVPN 606
Query: 65 KPVDDS 70
P D S
Sbjct: 607 DPYDAS 612
Score = 42.7 bits (99), Expect = 1e-04
Identities = 25/84 (29%), Positives = 33/84 (38%), Gaps = 11/84 (13%)
Query: 5 QPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPES 64
QP K E+ P P P P P + TP PE KP+P + PK P+ SP
Sbjct: 532 QPPKQETPKPEESPKPQP-PKQETPKPEESPKPQPPKQETPK----------PEESPKPQ 580
Query: 65 KPVDDSKAIVKVEKTQEAAEEKPL 88
P + + K E P+
Sbjct: 581 PPKQEQPPKTEAPKMGSPPLESPV 604
Score = 42.4 bits (98), Expect = 2e-04
Identities = 20/85 (23%), Positives = 38/85 (44%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
+++ K E P P P P E +P P+ + P ++PK ++++ P+ SP
Sbjct: 503 KQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPK 562
Query: 63 ESKPVDDSKAIVKVEKTQEAAEEKP 87
P ++ + K Q +E+P
Sbjct: 563 PQPPKQETPKPEESPKPQPPKQEQP 587
Score = 42.4 bits (98), Expect = 2e-04
Identities = 30/89 (33%), Positives = 39/89 (43%), Gaps = 9/89 (10%)
Query: 4 EQPK-KVES----ESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQ 58
EQPK K ES S PP P +P P PE KP+P + PK E+S PQ
Sbjct: 493 EQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKP----EESPKPQ 548
Query: 59 PSPPESKPVDDSKAIVKVEKTQEAAEEKP 87
P E+ ++S ++ EE P
Sbjct: 549 PPKQETPKPEESPKPQPPKQETPKPEESP 577
Score = 40.0 bits (92), Expect = 0.001
Identities = 25/91 (27%), Positives = 42/91 (45%), Gaps = 7/91 (7%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTEST-----PAPEAEAKPE-PVVHDAPKEDVAEEKSII 56
+E PK+ E+ P P P +P + + P PE KPE P ++PK ++++
Sbjct: 482 QESPKQ-EAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPK 540
Query: 57 PQPSPPESKPVDDSKAIVKVEKTQEAAEEKP 87
P+ SP P ++ + K Q +E P
Sbjct: 541 PEESPKPQPPKQETPKPEESPKPQPPKQETP 571
Score = 39.3 bits (90), Expect = 0.002
Identities = 24/87 (27%), Positives = 40/87 (45%), Gaps = 3/87 (3%)
Query: 4 EQPKKVESESPSNPPPPAPAPTEST--PAPEAEAKPEPVVHDAPKEDVAEEKSIIP-QPS 60
E+P K + E P P+P P + P+ E KPE ++PK + + K P Q S
Sbjct: 425 EEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQES 484
Query: 61 PPESKPVDDSKAIVKVEKTQEAAEEKP 87
P + P + QE+++++P
Sbjct: 485 PKQEAPKPEQPKPKPESPKQESSKQEP 511
Score = 37.7 bits (86), Expect = 0.005
Identities = 24/85 (28%), Positives = 36/85 (42%), Gaps = 2/85 (2%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
+E PK ES P P P P ES P P+ + +P +APK +S +P P
Sbjct: 552 QETPKPEESPKPQPPKQETPKPEES-PKPQPPKQEQPPKTEAPKMGSPPLESPVPN-DPY 609
Query: 63 ESKPVDDSKAIVKVEKTQEAAEEKP 87
++ P+ + T+E P
Sbjct: 610 DASPIKKRRPQPPSPSTEETKTTSP 634
Score = 37.7 bits (86), Expect = 0.005
Identities = 25/91 (27%), Positives = 37/91 (40%), Gaps = 9/91 (9%)
Query: 5 QPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQ------ 58
+PK ES P P P P+ P+ E KPE ++PK + + K P+
Sbjct: 429 KPKPEESPKPQQPSPKPETPSHEPSNPK-EPKPESPKQESPKTEQPKPKPESPKQESPKQ 487
Query: 59 --PSPPESKPVDDSKAIVKVEKTQEAAEEKP 87
P P + KP +S ++ EE P
Sbjct: 488 EAPKPEQPKPKPESPKQESSKQEPPKPEESP 518
Score = 37.4 bits (85), Expect = 0.006
Identities = 22/67 (32%), Positives = 33/67 (48%), Gaps = 7/67 (10%)
Query: 9 VESESPSNPPPPAPAPTESTPAPEAEAKPE---PVVHDAPKEDVAEEKSIIPQPSPPESK 65
+E+ S SN P +S+PAP +++PE PV P+ + ++ P SPP S
Sbjct: 856 IEAPSDSNHSPVF----KSSPAPSPDSEPEVEAPVPSSEPEVEAPKQSEATPSSSPPSSN 911
Query: 66 PVDDSKA 72
P D A
Sbjct: 912 PSPDVTA 918
Score = 37.4 bits (85), Expect = 0.006
Identities = 28/95 (29%), Positives = 48/95 (50%), Gaps = 15/95 (15%)
Query: 3 EEQPK--------KVESESPSNPPPPAP-APTESTPAPEAEAKPEPVVHDAPKEDVAEEK 53
EE PK + S PSNP P P +P + +P E + KP+P ++PK++ +++
Sbjct: 433 EESPKPQQPSPKPETPSHEPSNPKEPKPESPKQESPKTE-QPKPKP---ESPKQESPKQE 488
Query: 54 SIIP-QPSPPESKPVDDSKAIVKVEKTQEAAEEKP 87
+ P QP P P +S + K +E+ + +P
Sbjct: 489 APKPEQPKPKPESPKQESSK-QEPPKPEESPKPEP 522
Score = 37.4 bits (85), Expect = 0.006
Identities = 27/91 (29%), Positives = 38/91 (41%), Gaps = 7/91 (7%)
Query: 4 EQPK------KVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPK-EDVAEEKSII 56
EQPK K ES P P P P +P E+ + P ++PK E E+S
Sbjct: 471 EQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPK 530
Query: 57 PQPSPPESKPVDDSKAIVKVEKTQEAAEEKP 87
PQP E+ ++S ++ EE P
Sbjct: 531 PQPPKQETPKPEESPKPQPPKQETPKPEESP 561
Score = 36.2 bits (82), Expect = 0.013
Identities = 22/86 (25%), Positives = 42/86 (48%), Gaps = 5/86 (5%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP- 61
+++ K E P P +P + P PE + KP+P ++PK++ ++++ P+ SP
Sbjct: 464 KQESPKTEQPKPKPESPKQESPKQEAPKPE-QPKPKP---ESPKQESSKQEPPKPEESPK 519
Query: 62 PESKPVDDSKAIVKVEKTQEAAEEKP 87
PE ++S ++ EE P
Sbjct: 520 PEPPKPEESPKPQPPKQETPKPEESP 545
Score = 35.8 bits (81), Expect = 0.017
Identities = 29/98 (29%), Positives = 37/98 (37%), Gaps = 12/98 (12%)
Query: 6 PKKVESESPSNPP----PPAPAPTESTPAPEAEAKPEPVVHDA--PKEDVAEE------K 53
PK E + NPP P P P ES + KPE H+ PKE E K
Sbjct: 410 PKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPESPKQESPK 469
Query: 54 SIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGS 91
+ P+P P K + K E+ + E E S
Sbjct: 470 TEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESS 507
Score = 35.8 bits (81), Expect = 0.017
Identities = 23/69 (33%), Positives = 29/69 (41%), Gaps = 9/69 (13%)
Query: 6 PKKVESESPS---NPPPPAPA-----PTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIP 57
P V+S P +PPPPAP P +P P + P P VH +P V +
Sbjct: 723 PPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVH-SPPPPVHSPPPPVH 781
Query: 58 QPSPPESKP 66
P PP P
Sbjct: 782 SPPPPVHSP 790
Score = 35.8 bits (81), Expect = 0.017
Identities = 23/70 (32%), Positives = 32/70 (44%), Gaps = 10/70 (14%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPA-PEAEAKPEPVVHDAPKEDVAEEKSIIPQP-- 59
+EQP K E+ +PP +P P + A P + +P+P P E K+ PQ
Sbjct: 584 QEQPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQP-----PSPSTEETKTTSPQSPP 638
Query: 60 --SPPESKPV 67
SPP PV
Sbjct: 639 VHSPPPPPPV 648
Score = 35.0 bits (79), Expect = 0.030
Identities = 19/63 (30%), Positives = 27/63 (42%), Gaps = 5/63 (7%)
Query: 7 KKVESESPSNPP---PPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPE 63
++ ++ SP +PP PP P P S P P P P +H P + + P PP
Sbjct: 627 EETKTTSPQSPPVHSPPPPPPVHSPPPP--VFSPPPPMHSPPPPVYSPPPPVHSPPPPPV 684
Query: 64 SKP 66
P
Sbjct: 685 HSP 687
Score = 35.0 bits (79), Expect = 0.030
Identities = 22/65 (33%), Positives = 28/65 (42%), Gaps = 5/65 (7%)
Query: 6 PKKVESESPS--NPPPP--APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
P V S P +PPPP +P P +P P + P P VH +P V + P P
Sbjct: 645 PPPVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVH-SPPPPVHSPPPPVHSPPP 703
Query: 62 PESKP 66
P P
Sbjct: 704 PVHSP 708
Score = 33.9 bits (76), Expect = 0.066
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 5/71 (7%)
Query: 18 PPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP-PESKPVDDSKAIVKV 76
P P P PT P P+ E P + + P + EE QPSP PE+ + S K
Sbjct: 402 PSPKPTPTPKAPEPKKEINPPNL--EEPSKPKPEESPKPQQPSPKPETPSHEPSNP--KE 457
Query: 77 EKTQEAAEEKP 87
K + +E P
Sbjct: 458 PKPESPKQESP 468
Score = 33.5 bits (75), Expect = 0.087
Identities = 21/68 (30%), Positives = 26/68 (37%), Gaps = 7/68 (10%)
Query: 6 PKKVESESPSNPPP-PAPAPTESTPAPEAEAKPEPV------VHDAPKEDVAEEKSIIPQ 58
P+ SP PPP +P P +P P + P PV VH P V +
Sbjct: 634 PQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHS 693
Query: 59 PSPPESKP 66
P PP P
Sbjct: 694 PPPPVHSP 701
Score = 33.1 bits (74), Expect = 0.11
Identities = 18/54 (33%), Positives = 21/54 (38%)
Query: 13 SPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKP 66
SP PP +P P +P P + P PV P S I P PP P
Sbjct: 760 SPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPSPIYSPPPPVFSP 813
Score = 32.7 bits (73), Expect = 0.15
Identities = 18/54 (33%), Positives = 22/54 (40%), Gaps = 1/54 (1%)
Query: 13 SPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKP 66
SP PP +P P +P P P P VH +P V + P PP P
Sbjct: 745 SPPPPPVHSPPPPVHSPPPPPVHSPPPPVH-SPPPPVHSPPPPVHSPPPPVHSP 797
Score = 32.7 bits (73), Expect = 0.15
Identities = 20/59 (33%), Positives = 23/59 (38%), Gaps = 3/59 (5%)
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPES 64
P V S P PP P+P S P P P+PV P + P PS ES
Sbjct: 784 PPPVHSPPPPVHSPPPPSPIYSPPPPVFSPPPKPV---TPLPPATSPMANAPTPSSSES 839
Score = 32.3 bits (72), Expect = 0.19
Identities = 22/65 (33%), Positives = 27/65 (40%), Gaps = 5/65 (7%)
Query: 6 PKKVESESPS--NPPPP--APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
P V S P +PPPP +P P +P P P P VH +P V + P P
Sbjct: 652 PPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVH-SPPPPVHSPPPPVHSPPP 710
Query: 62 PESKP 66
P P
Sbjct: 711 PVHSP 715
Score = 32.3 bits (72), Expect = 0.19
Identities = 22/66 (33%), Positives = 27/66 (40%), Gaps = 7/66 (10%)
Query: 6 PKKVESESP---SNPPPP--APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
P V S P S PPPP +P P +P P + P PV +P V + P
Sbjct: 666 PPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPV--HSPPPPVHSPPPPVHSPP 723
Query: 61 PPESKP 66
PP P
Sbjct: 724 PPVQSP 729
Score = 32.0 bits (71), Expect = 0.25
Identities = 21/66 (31%), Positives = 27/66 (40%), Gaps = 6/66 (9%)
Query: 6 PKKVESESPS---NPPPP--APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
P V S P +PPPP +P P +P P + P PV H P + + P
Sbjct: 673 PPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPV-HSPPPPVHSPPPPVQSPPP 731
Query: 61 PPESKP 66
PP P
Sbjct: 732 PPVFSP 737
Score = 31.6 bits (70), Expect = 0.33
Identities = 21/84 (25%), Positives = 33/84 (39%), Gaps = 5/84 (5%)
Query: 6 PKKVESESPS---NPPPP--APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
P V S P +PPPP +P P +P P + P PV P + + P
Sbjct: 755 PPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPSPIYSPPPPVFSPP 814
Query: 61 PPESKPVDDSKAIVKVEKTQEAAE 84
P P+ + + + T ++E
Sbjct: 815 PKPVTPLPPATSPMANAPTPSSSE 838
Score = 31.6 bits (70), Expect = 0.33
Identities = 22/68 (32%), Positives = 27/68 (39%), Gaps = 7/68 (10%)
Query: 6 PKKVESESPS--NPPPPA---PAPTESTPAPEAE--AKPEPVVHDAPKEDVAEEKSIIPQ 58
P V S P +PPPP P P +P P A + P P VH P + +
Sbjct: 709 PPPVHSPPPPVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHS 768
Query: 59 PSPPESKP 66
P PP P
Sbjct: 769 PPPPVHSP 776
Score = 30.8 bits (68), Expect = 0.56
Identities = 20/61 (32%), Positives = 26/61 (41%), Gaps = 5/61 (8%)
Query: 6 PKKVESESPS--NPPPP--APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
P V S P +PPPP +P P +P P + P P VH P + + P P
Sbjct: 681 PPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPP-VHSPPPPVQSPPPPPVFSPPP 739
Query: 62 P 62
P
Sbjct: 740 P 740
Score = 30.0 bits (66), Expect = 0.96
Identities = 27/85 (31%), Positives = 33/85 (38%), Gaps = 16/85 (18%)
Query: 14 PSNPPPPAPAPTESTPAPEAEAKPE---PVVHDAPKEDVAEEKS---------IIPQPSP 61
P P PPA +P + P P + E PV P + E S P PSP
Sbjct: 817 PVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSEDIEAPSDSNHSPVFKSSPAPSP 876
Query: 62 ---PE-SKPVDDSKAIVKVEKTQEA 82
PE PV S+ V+ K EA
Sbjct: 877 DSEPEVEAPVPSSEPEVEAPKQSEA 901
Score = 27.3 bits (59), Expect = 6.2
Identities = 21/65 (32%), Positives = 26/65 (39%), Gaps = 9/65 (13%)
Query: 6 PKKVESESP---SNPPPP-APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
P V S P S PPP +P P +P P + P PV P + P P+P
Sbjct: 688 PPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQSPPPPPVFSP-----PPPAP 742
Query: 62 PESKP 66
S P
Sbjct: 743 IYSPP 747
Score = 26.9 bits (58), Expect = 8.1
Identities = 24/74 (32%), Positives = 30/74 (40%), Gaps = 14/74 (18%)
Query: 6 PKKVESESPSNP----PPPA--PAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP 59
P V S P +P PPP P P TP P A + + +AP +E I
Sbjct: 791 PPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSP----MANAPTPSSSESGEI---- 842
Query: 60 SPPESKPVDDSKAI 73
S P P DS+ I
Sbjct: 843 STPVQAPTPDSEDI 856
>At1g49490 hypothetical protein
Length = 847
Score = 45.8 bits (107), Expect = 2e-05
Identities = 28/76 (36%), Positives = 38/76 (49%), Gaps = 10/76 (13%)
Query: 6 PKKVESESPSNPPPPAPAPTES----TP-APEAEA-KPEPVVHDAPKEDVAEEKSIIPQ- 58
P + PS P P P P++S TP PE + KP+P H++PK + E K +P+
Sbjct: 398 PPRTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPSPKPQPPKHESPKPEEPENKHELPKQ 457
Query: 59 ---PSPPESKPVDDSK 71
P P SKP D K
Sbjct: 458 KESPKPQPSKPEDSPK 473
Score = 37.4 bits (85), Expect = 0.006
Identities = 20/65 (30%), Positives = 25/65 (37%), Gaps = 4/65 (6%)
Query: 6 PKKVESESPSNPPPPAPAPTESTPA----PEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
P KVE P PP P+P+ +P P + P PV P V + P P
Sbjct: 525 PPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPP 584
Query: 62 PESKP 66
P P
Sbjct: 585 PSPPP 589
Score = 35.8 bits (81), Expect = 0.017
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 21/74 (28%)
Query: 5 QPKKVESESPSNP------------PPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEE 52
QP K ES P P P P P+ E +P PE + KPE ++PK E
Sbjct: 436 QPPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPE-QPKPE----ESPK----PE 486
Query: 53 KSIIPQPSPPESKP 66
+ IP+P+ P S P
Sbjct: 487 QPQIPEPTKPVSPP 500
Score = 35.8 bits (81), Expect = 0.017
Identities = 28/89 (31%), Positives = 39/89 (43%), Gaps = 9/89 (10%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
+E PK S+ +P P P P ES P PE PEP +P + P P P
Sbjct: 458 KESPKPQPSKPEDSPKPEQPKPEES-PKPEQPQIPEPTKPVSPPNEAQG-----PTPDDP 511
Query: 63 -ESKPVDD--SKAIVKVEKTQEAAEEKPL 88
++ PV + S KVE T+ + P+
Sbjct: 512 YDASPVKNRRSPPPPKVEDTRVPPPQPPM 540
Score = 35.8 bits (81), Expect = 0.017
Identities = 22/64 (34%), Positives = 23/64 (35%), Gaps = 3/64 (4%)
Query: 6 PKKVESESP---SNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
P V S P S PPP P P S P P + P PV P V P PP
Sbjct: 569 PPHVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPP 628
Query: 63 ESKP 66
P
Sbjct: 629 VYSP 632
Score = 34.3 bits (77), Expect = 0.051
Identities = 22/69 (31%), Positives = 27/69 (38%), Gaps = 6/69 (8%)
Query: 4 EQPKKVESESPSNPPPPAPA-----PTEST-PAPEAEAKPEPVVHDAPKEDVAEEKSIIP 57
EQPK ES P P P P P E+ P P+ PV + E + +P
Sbjct: 475 EQPKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVP 534
Query: 58 QPSPPESKP 66
P PP P
Sbjct: 535 PPQPPMPSP 543
Score = 33.9 bits (76), Expect = 0.066
Identities = 22/63 (34%), Positives = 28/63 (43%), Gaps = 9/63 (14%)
Query: 6 PKKVESESP---SNPPPP---APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP 59
P V S P S+PPPP +P P ++P P + P P VH P V + P
Sbjct: 553 PPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPS---PPPPVHSPPPPPVFSPPPPVFSP 609
Query: 60 SPP 62
PP
Sbjct: 610 PPP 612
Score = 33.9 bits (76), Expect = 0.066
Identities = 20/65 (30%), Positives = 24/65 (36%), Gaps = 4/65 (6%)
Query: 6 PKKVESESPSNPPPPA----PAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
P V S P +PPPP P P S P P P V+ P + + P P
Sbjct: 576 PPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPP 635
Query: 62 PESKP 66
S P
Sbjct: 636 TFSPP 640
Score = 33.9 bits (76), Expect = 0.066
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 8/63 (12%)
Query: 4 EQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPE 63
+ K ++P P P P +P PE +PE H+ PK +++S PQPS PE
Sbjct: 418 DSSKPETPKTPEQPSPKPQPPKHESPKPE---EPEN-KHELPK----QKESPKPQPSKPE 469
Query: 64 SKP 66
P
Sbjct: 470 DSP 472
Score = 32.7 bits (73), Expect = 0.15
Identities = 22/76 (28%), Positives = 30/76 (38%), Gaps = 5/76 (6%)
Query: 11 SESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDS 70
S S+P P P +E P+ KPEPV+ E QPSP P +S
Sbjct: 388 SNGGSSPSPNPPRTSEPKPS-----KPEPVMPKPSDSSKPETPKTPEQPSPKPQPPKHES 442
Query: 71 KAIVKVEKTQEAAEEK 86
+ E E ++K
Sbjct: 443 PKPEEPENKHELPKQK 458
Score = 32.7 bits (73), Expect = 0.15
Identities = 29/111 (26%), Positives = 43/111 (38%), Gaps = 29/111 (26%)
Query: 4 EQPKKVESESPSNPPP--PAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS- 60
+ P V+S +PS+ P P P+ +ES AP P AP + E S +P PS
Sbjct: 685 QAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQAP--TTSSETSQVPTPSS 742
Query: 61 --------------------PPESKPVD----DSKAIVKVEKTQEAAEEKP 87
P SKPV S+ + E+++E +P
Sbjct: 743 ESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEP 793
Score = 32.0 bits (71), Expect = 0.25
Identities = 21/69 (30%), Positives = 28/69 (40%), Gaps = 10/69 (14%)
Query: 4 EQPKKVESESPSNPPPPAPAPT-----ESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQ 58
EQP+ E P +PP A PT +++P + P P V ED P
Sbjct: 486 EQPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSPPPPKV-----EDTRVPPPQPPM 540
Query: 59 PSPPESKPV 67
PSP P+
Sbjct: 541 PSPSPPSPI 549
Score = 30.0 bits (66), Expect = 0.96
Identities = 21/69 (30%), Positives = 24/69 (34%), Gaps = 18/69 (26%)
Query: 5 QPKKVESESPS---NPPPPAPAPT----ESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIP 57
QP PS +PPPP +P S P P + P PV P P
Sbjct: 537 QPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPP-----------P 585
Query: 58 QPSPPESKP 66
P PP P
Sbjct: 586 SPPPPVHSP 594
Score = 28.1 bits (61), Expect = 3.6
Identities = 17/61 (27%), Positives = 24/61 (38%), Gaps = 6/61 (9%)
Query: 6 PKKVESESPS--NPPPPAPA----PTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP 59
P V S P +PPPP+P P +P P + P P P + + P P
Sbjct: 596 PPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTP 655
Query: 60 S 60
+
Sbjct: 656 T 656
Score = 27.7 bits (60), Expect = 4.8
Identities = 23/83 (27%), Positives = 29/83 (34%), Gaps = 19/83 (22%)
Query: 6 PKKVESESPSNPPPP------APAPTES-----TPAPEAEAKPEPVVHDAPKEDVAEEKS 54
P ++SPS P P AP P TP+ E + PE E S
Sbjct: 740 PSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPS 799
Query: 55 IIPQPS--------PPESKPVDD 69
+P S PPE+ DD
Sbjct: 800 SVPSSSPSTDTSIPPPENNDDDD 822
>At4g27500 unknown protein
Length = 612
Score = 45.4 bits (106), Expect = 2e-05
Identities = 43/170 (25%), Positives = 75/170 (43%), Gaps = 8/170 (4%)
Query: 2 TEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
TE PK P P AP P ++T A E + V +D +E + +P
Sbjct: 403 TEVVPKAKAKPQPKEEPVSAPKP-DATVAQNTEKAKDAVKVKNVADDDDDEVYGLGKPQ- 460
Query: 62 PESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIA 121
E KPVD + A ++ K +E A+ K + + + + A + + K E+ EK
Sbjct: 461 KEEKPVDAATA-KEMRKQEEIAKAK--QAMERKKKLAEKAAAKAAIRAQKEAEKKEKKEQ 517
Query: 122 DNKAHKKLSDISAWENSKI---AAKEVELRKIEENLEKKKAVYVEKLKNK 168
+ KA KK + E ++ + +E+E EE +K+K + ++N+
Sbjct: 518 EKKAKKKTGGNTETETEEVPEASEEEIEAPVQEEKPQKEKVFKEKPIRNR 567
Score = 44.3 bits (103), Expect = 5e-05
Identities = 48/203 (23%), Positives = 90/203 (43%), Gaps = 18/203 (8%)
Query: 12 ESPSNPPPPAPA---PTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVD 68
E P P AP+ P+E+ P+A+AKP+P EE P+P ++ +
Sbjct: 385 EKPLIAPEAAPSKATPSETEVVPKAKAKPQP----------KEEPVSAPKPDATVAQNTE 434
Query: 69 DSKAIVKVEKTQEAAEEKPLE-GSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHK 127
+K VKV+ + +++ G ++ AT K + + EE K+ + K
Sbjct: 435 KAKDAVKVKNVADDDDDEVYGLGKPQKEEKPVDAATAKEM---RKQEEIAKAKQAMERKK 491
Query: 128 KLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEK-RAFIEAK 186
KL++ +A + + A KE E ++ +E +K K + + V +EE+ A ++ +
Sbjct: 492 KLAEKAAAKAAIRAQKEAEKKEKKEQEKKAKKKTGGNTETETEEVPEASEEEIEAPVQEE 551
Query: 187 KGEDLLKAEELAAKYRATGTAPK 209
K + +E + R G P+
Sbjct: 552 KPQKEKVFKEKPIRNRTRGRGPE 574
>At4g32710 putative protein kinase
Length = 731
Score = 45.1 bits (105), Expect = 3e-05
Identities = 27/65 (41%), Positives = 29/65 (44%), Gaps = 2/65 (3%)
Query: 4 EQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS--P 61
E P ESPS P P AP+ S P P AKP P P E V +I P P P
Sbjct: 85 ESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISPPPRSLP 144
Query: 62 PESKP 66
ES P
Sbjct: 145 SESTP 149
Score = 37.4 bits (85), Expect = 0.006
Identities = 21/69 (30%), Positives = 27/69 (38%), Gaps = 8/69 (11%)
Query: 6 PKKVESESPSNPPPPAPAPT--------ESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIP 57
P + S P PPP APT ES P+P E+ P P++ P + P
Sbjct: 41 PPPLSSPPPLPSPPPLSAPTASPPPLPVESPPSPPIESPPPPLLESPPPPPLESPSPPSP 100
Query: 58 QPSPPESKP 66
S P P
Sbjct: 101 HVSAPSGSP 109
Score = 32.0 bits (71), Expect = 0.25
Identities = 21/67 (31%), Positives = 29/67 (42%), Gaps = 2/67 (2%)
Query: 6 PKKVESESPSNPP-PPAPAP-TESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPE 63
P + ESP +PP P P ES P P E+ P H + + +PSPP
Sbjct: 63 PPPLPVESPPSPPIESPPPPLLESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPP 122
Query: 64 SKPVDDS 70
S P ++
Sbjct: 123 SSPPSET 129
Score = 30.8 bits (68), Expect = 0.56
Identities = 21/62 (33%), Positives = 26/62 (41%), Gaps = 9/62 (14%)
Query: 10 ESESPSNP--PPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP---SPPES 64
E+ P N PPP P+ESTP + P P +P + K P P SPP
Sbjct: 128 ETVPPGNTISPPPRSLPSESTPPVNTASPPPP----SPPRRRSGPKPSFPPPINSSPPNP 183
Query: 65 KP 66
P
Sbjct: 184 SP 185
Score = 28.5 bits (62), Expect = 2.8
Identities = 22/71 (30%), Positives = 26/71 (35%), Gaps = 15/71 (21%)
Query: 11 SESPSNPPPPAPAP-------------TESTPAPEAEAKPEPVVH--DAPKEDVAEEKSI 55
S SPS+ P PA +P T S PAP P P+ P +
Sbjct: 2 SLSPSSSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAPTA 61
Query: 56 IPQPSPPESKP 66
P P P ES P
Sbjct: 62 SPPPLPVESPP 72
Score = 27.7 bits (60), Expect = 4.8
Identities = 19/59 (32%), Positives = 22/59 (37%), Gaps = 6/59 (10%)
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPES 64
P S P +PP P S P P + P P +P E S P P PP S
Sbjct: 150 PVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNP----SPNTPSLPETS--PPPKPPLS 202
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 44.7 bits (104), Expect = 4e-05
Identities = 42/176 (23%), Positives = 77/176 (42%), Gaps = 18/176 (10%)
Query: 19 PPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEK 78
P A P STP E A+PEPV AP E+ E++ +++ A K +K
Sbjct: 314 PAAERPASSTPVEEKAAQPEPV---APVENAGEKEG------------EEETAAAKKKKK 358
Query: 79 TQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKL-SDISAWEN 137
+E +EK + + + ++ S+ + + +K A KK+ + +
Sbjct: 359 KKEKEKEKKAAAAAAATSSVEVKEEKQEESVTEPLQPKKKDAKGKAAEKKIPKHVREMQE 418
Query: 138 SKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLK 193
+ +E E RK +E EK + E+ + + + +AEE + + K+ E LL+
Sbjct: 419 ALARRQEAEERKKKEEEEKLRKEEEERRRQE--ELEAQAEEAKRKRKEKEKEKLLR 472
>At3g18890 unknown protein
Length = 641
Score = 43.5 bits (101), Expect = 8e-05
Identities = 35/103 (33%), Positives = 52/103 (49%), Gaps = 16/103 (15%)
Query: 3 EEQPKKVESESPSNPPPPAPAPT-ESTPAPEAEAKPEPVVHDAPKEDVA--EEKSIIPQP 59
E+ +K+ S+ P PPP A T E P P EP APKED A +EK++ P+P
Sbjct: 320 EKLLEKIPSKRPYVPPPKASVATKEVKPVPTKPVTQEPT---APKEDEAPPKEKNVKPRP 376
Query: 60 -SP--------PESKPVDDSKAIVKVEKTQEA-AEEKPLEGSI 92
SP P + P+ +S V K++E A + P+E ++
Sbjct: 377 LSPYASYEDLKPPTSPIPNSTTSVSPAKSKEVDATQVPVEANV 419
Score = 26.9 bits (58), Expect = 8.1
Identities = 20/73 (27%), Positives = 30/73 (40%), Gaps = 4/73 (5%)
Query: 18 PPPAPAPTESTPAPEAEAKPE---PVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIV 74
PP +P P + P A + P ++ + V + IP PS PE+ PV S
Sbjct: 544 PPTSPTPASTGPKEAASVEDNSELPGGNNDVLKTVDGNLNTIP-PSTPEAVPVVSSAIDT 602
Query: 75 KVEKTQEAAEEKP 87
+ A+ KP
Sbjct: 603 SLASGDNTAQPKP 615
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 43.1 bits (100), Expect = 1e-04
Identities = 48/219 (21%), Positives = 89/219 (39%), Gaps = 13/219 (5%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
EE+ K+ E E+ E+ A + E + E A K + ++ +
Sbjct: 481 EEERKREEEEAKRREEERKKREEEAEQARKREEEREKEEEMAKKREEERQRK---EREEV 537
Query: 63 ESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIAD 122
E K ++ + + E+ ++ EE+ E + + R E+ K EE E+ +
Sbjct: 538 ERKRREEQERKRREEEARKREEERKREEEMAKRREQERQRKEREEVERKIREEQERKREE 597
Query: 123 NKAHKKLSDISAWENSKIAAK---------EVELRKI-EENLEKKKAVYVEKLKNKIAMV 172
A ++ + E ++ K E E+ KI EE ++K+ VE+ + + +
Sbjct: 598 EMAKRREQERQKKEREEMERKKREEEARKREEEMAKIREEERQRKEREDVERKRREEEAM 657
Query: 173 HREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKP 211
RE E KR AK+ E+ + +E + R PK P
Sbjct: 658 RREEERKREEEAAKRAEEERRKKEEEEEKRRWPPQPKPP 696
Score = 33.9 bits (76), Expect = 0.066
Identities = 38/121 (31%), Positives = 58/121 (47%), Gaps = 14/121 (11%)
Query: 92 INRD--AVLTRVATEKRLSLIKAWEESE----KSIADNKAHKKLSDISAWENSKIAAKEV 145
++RD AV ++ E L+KA +ES S A+ + K + +I E + +E+
Sbjct: 378 LDRDVVAVSFNLSNETIKGLLKAQKESVIFECASCAEGELSKLMREIE--ERKRREEEEI 435
Query: 146 ELRKIEENLEKKKAVYVEKLKNKIAMVHREAEE----KRAFIEAKKGEDLLKAEELAAKY 201
E R+ EE +K+ K + + RE EE KR EA+K E+ K EE AK
Sbjct: 436 ERRRKEEEEARKREE--AKRREEEEAKRREEEETERKKREEEEARKREEERKREEEEAKR 493
Query: 202 R 202
R
Sbjct: 494 R 494
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.302 0.122 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,329,555
Number of Sequences: 26719
Number of extensions: 266065
Number of successful extensions: 5486
Number of sequences better than 10.0: 569
Number of HSP's better than 10.0 without gapping: 218
Number of HSP's successfully gapped in prelim test: 359
Number of HSP's that attempted gapping in prelim test: 1998
Number of HSP's gapped (non-prelim): 2126
length of query: 215
length of database: 11,318,596
effective HSP length: 95
effective length of query: 120
effective length of database: 8,780,291
effective search space: 1053634920
effective search space used: 1053634920
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0148.17


