

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147b.4
(313 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
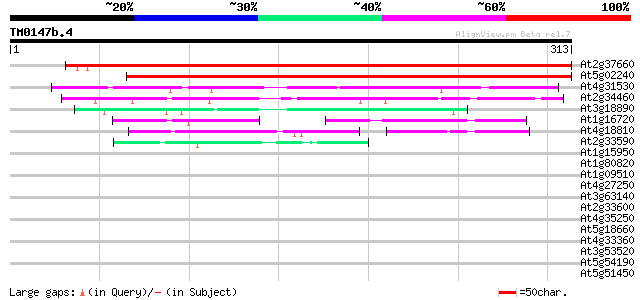
Score E
Sequences producing significant alignments: (bits) Value
At2g37660 unknown protein 432 e-122
At5g02240 unknown protein 388 e-108
At4g31530 unknown protein 106 2e-23
At2g34460 unknown protein 86 3e-17
At3g18890 unknown protein 67 2e-11
At1g16720 unknown protein (At1g16720) 55 5e-08
At4g18810 unknown protein 50 2e-06
At2g33590 putative cinnamoyl-CoA reductase 43 3e-04
At1g15950 unknown protein 40 0.002
At1g80820 cinnamoyl CoA reductase like protein 39 0.003
At1g09510 putative cinnamyl alcohol dehydrogenase 35 0.040
At4g27250 unknown protein 35 0.053
At3g63140 mRNA binding protein precursor - like 34 0.090
At2g33600 putative cinnamoyl-CoA reductase 34 0.090
At4g35250 unknown protein 33 0.26
At5g18660 unknown protein 32 0.58
At4g33360 putative dihydrokaempferol 4-reductase 32 0.58
At3g53520 UDP-glucuronic acid decarboxylase (UXS1) 31 0.76
At5g54190 NADPH:protochlorophyllide oxidoreductase A (gb|AAC4904... 31 0.99
At5g51450 unknown protein 30 1.7
>At2g37660 unknown protein
Length = 325
Score = 432 bits (1112), Expect = e-122
Identities = 216/289 (74%), Positives = 253/289 (86%), Gaps = 7/289 (2%)
Query: 32 TSLQF----SHSSSL---SLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKL 84
+SLQF S S+S+ S T K R +V A A + TVLVTGAGGRTG+IVYKKL
Sbjct: 37 SSLQFRSLVSDSTSICGPSKFTGKNRRVSVTVSAAATTEPLTVLVTGAGGRTGQIVYKKL 96
Query: 85 QERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMK 144
+ERS Q++ARGLVRT+ESK+ I D+V++GDIRDT SIAPA++GIDAL+ILTSAVP MK
Sbjct: 97 KERSEQFVARGLVRTKESKEKINGEDEVFIGDIRDTASIAPAVEGIDALVILTSAVPQMK 156
Query: 145 PGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLN 204
PGF+P+KG RPEF+F+DGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGT++N+PLN
Sbjct: 157 PGFDPSKGGRPEFFFDDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTNINHPLN 216
Query: 205 SLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIA 264
S+GN NILVWKRKAEQYLADSGIPYTIIRAGGLQDK+GG+REL++GKDDE+L+TETRTIA
Sbjct: 217 SIGNANILVWKRKAEQYLADSGIPYTIIRAGGLQDKDGGIRELLVGKDDELLETETRTIA 276
Query: 265 RPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITTRF 313
R DVAEVC+QAL EEA+FKA DLASKPEG GTPT+DFKALF+Q+TT+F
Sbjct: 277 RADVAEVCVQALQLEEAKFKALDLASKPEGTGTPTKDFKALFTQVTTKF 325
>At5g02240 unknown protein
Length = 253
Score = 388 bits (996), Expect = e-108
Identities = 189/248 (76%), Positives = 217/248 (87%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAP 125
TVLVTGA GRTG+IVYKKL+E S +++A+GLVR+ + K+ IG DV++GDI D SI P
Sbjct: 6 TVLVTGASGRTGQIVYKKLKEGSDKFVAKGLVRSAQGKEKIGGEADVFIGDITDADSINP 65
Query: 126 AIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGV 185
A QGIDAL+ILTSAVP MKPGF+PTKG RPEF FEDG YPEQVDWIGQKNQIDAAK AGV
Sbjct: 66 AFQGIDALVILTSAVPKMKPGFDPTKGGRPEFIFEDGQYPEQVDWIGQKNQIDAAKVAGV 125
Query: 186 KQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVR 245
K IV+VGSMGGT+ ++PLN LGNGNILVWKRKAEQYLADSG PYTIIRAGGL DKEGGVR
Sbjct: 126 KHIVVVGSMGGTNPDHPLNKLGNGNILVWKRKAEQYLADSGTPYTIIRAGGLLDKEGGVR 185
Query: 246 ELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKAL 305
EL++GKDDE+L+T+T+T+ R DVAEVCIQAL FEEA+ KAFDL SKPEG TPT+DFKAL
Sbjct: 186 ELLVGKDDELLQTDTKTVPRADVAEVCIQALLFEEAKNKAFDLGSKPEGTSTPTKDFKAL 245
Query: 306 FSQITTRF 313
FSQ+T+RF
Sbjct: 246 FSQVTSRF 253
>At4g31530 unknown protein
Length = 324
Score = 106 bits (264), Expect = 2e-23
Identities = 94/306 (30%), Positives = 148/306 (47%), Gaps = 45/306 (14%)
Query: 24 LPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKK 83
LPSS L+ + SS + TS +A SS VLV G G G++V
Sbjct: 35 LPSSFFLVRNEASLSSSITPVQAFTEEAVDTS--DLASSSSKLVLVVGGTGGVGQLVVAS 92
Query: 84 LQERS--SQYIARGLVRTEESKQTIGASDD----VYVGDIRDTGSIAPAI-QGIDALIIL 136
L +R+ S+ + R L +++ + G D+ V GD R+ + P++ +G+ +I
Sbjct: 93 LLKRNIRSRLLLRDL---DKATKLFGKQDEYSLQVVKGDTRNAEDLDPSMFEGVTHVICT 149
Query: 137 TSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGG 196
T P + + PE+VDW G KN I A ++ VK++VLV S+G
Sbjct: 150 TGTTAF------------PSKRWNEENTPEKVDWEGVKNLISALPSS-VKRVVLVSSVGV 196
Query: 197 TDLNN-PLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQD--------------KE 241
T N P + + +L +K+ E +L DSG+P+TIIR G L D
Sbjct: 197 TKSNELPWSIMNLFGVLKYKKMGEDFLRDSGLPFTIIRPGRLTDGPYTSYDLNTLLKATA 256
Query: 242 GGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLAS-KPEGAGTPTR 300
G R ++IG+ D ++ +R + VAE CIQAL+ E Q KA+++ S K +G G+ +
Sbjct: 257 GERRAVVIGQGDNLVGEVSRLV----VAEACIQALDIEFTQGKAYEINSVKGDGPGSDPQ 312
Query: 301 DFKALF 306
++ LF
Sbjct: 313 QWRELF 318
>At2g34460 unknown protein
Length = 280
Score = 85.5 bits (210), Expect = 3e-17
Identities = 85/301 (28%), Positives = 141/301 (46%), Gaps = 45/301 (14%)
Query: 30 LLTSLQFSHSSSLSLAT------HKRVRTRTSVVAMADSSRSTV-----LVTGAGGRTGK 78
+ TSL HSS++ + +K R+ TS+ + + V V GA G+TGK
Sbjct: 1 MATSLLLRHSSAVFFSQSSFFTKNKSFRSFTSIKMEKGEAENAVKTKKVFVAGATGQTGK 60
Query: 79 IVYKKLQERSSQYIARGLVRTEESKQTIGASD---DVYVGDIRDTGSIAPAIQGIDALII 135
+ ++L R + + VR E +T D + D+ + + G D+ +
Sbjct: 61 RIVEQLLSRG--FAVKAGVRDVEKAKTSFKDDPSLQIVRADVTEGPDKLAEVIGDDSQAV 118
Query: 136 LTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSM- 194
+ + G RP F D P +VD G N +DA + GV++ VLV S+
Sbjct: 119 ICAT------------GFRPGF---DIFTPWKVDNFGTVNLVDACRKQGVEKFVLVSSIL 163
Query: 195 -GGTDLNNPLNSLGN-----GNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELI 248
G + LN G LV K +AE+Y+ SGI YTI+R GGL++ + ++
Sbjct: 164 VNGAAMGQILNPAYLFLNLFGLTLVAKLQAEKYIKKSGINYTIVRPGGLKN-DPPTGNVV 222
Query: 249 IGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQ 308
+ +D + + +I+R VAEV ++AL EE+ FK ++ ++ E P R +K LF+
Sbjct: 223 MEPEDTLYE---GSISRDLVAEVAVEALLQEESSFKVVEIVARAE---APKRSYKDLFAS 276
Query: 309 I 309
+
Sbjct: 277 V 277
>At3g18890 unknown protein
Length = 641
Score = 66.6 bits (161), Expect = 2e-11
Identities = 66/245 (26%), Positives = 97/245 (38%), Gaps = 42/245 (17%)
Query: 37 SHSSSLSLATHKRVR-----TRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQE----- 86
S S LSL +R T S + V V GA G+ G ++L +
Sbjct: 49 SRSFDLSLRASGPIRASSVVTEASPTNLNSKEEDLVFVAGATGKVGSRTVRELLKLGFRV 108
Query: 87 ----RSSQYIAR--------GLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALI 134
RS+Q L T+E Q + + V D+ SI PA+ +I
Sbjct: 109 RAGVRSAQRAGSLVQSVKEMKLQNTDEGTQPVEKLEIVEC-DLEKKDSIQPALGNASVII 167
Query: 135 ILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSM 194
A E D P ++D++ KN +DAA +A V +LV S+
Sbjct: 168 CCIGA---------------SEKEISDITGPYRIDYLATKNLVDAATSAKVNNFILVTSL 212
Query: 195 GGTDLNNPLNSLGN-GNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRE---LIIG 250
G P L +L WKRKAE+ L +SG+ Y I+R GG++ +E L +
Sbjct: 213 GTNKFGFPAAILNLFWGVLCWKRKAEEALIESGLNYAIVRPGGMERPTDAYKETHNLTLA 272
Query: 251 KDDEI 255
DD +
Sbjct: 273 LDDTL 277
>At1g16720 unknown protein (At1g16720)
Length = 598
Score = 55.1 bits (131), Expect = 5e-08
Identities = 39/113 (34%), Positives = 59/113 (51%), Gaps = 11/113 (9%)
Query: 177 IDAAKAAGVKQIVLVGSMG-GTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAG 235
I A A +LV G G + N +L KR E L SG+ YTIIR G
Sbjct: 458 IKALPAGQETDFILVSCTGSGVEANR------REQVLKAKRAGEDSLRRSGLGYTIIRPG 511
Query: 236 GLQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDL 288
L+++ GG R LI + + I ++ I+ DVA++C++AL+ A+ K+FD+
Sbjct: 512 PLKEEPGGQRALIFDQGNRI----SQGISCADVADICVKALHDSTARNKSFDV 560
Score = 43.9 bits (102), Expect = 1e-04
Identities = 28/84 (33%), Positives = 43/84 (50%), Gaps = 4/84 (4%)
Query: 58 AMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVR--TEESKQTIGASDDVYVG 115
A+ + TVLV GA R G+IV +KL R Y + LVR EE + S D+ VG
Sbjct: 156 AVPGAQNVTVLVVGATSRIGRIVVRKLMLRG--YTVKALVRKQDEEVMSMLPRSVDIVVG 213
Query: 116 DIRDTGSIAPAIQGIDALIILTSA 139
D+ + ++ A++ +I +A
Sbjct: 214 DVGEPSTLKSAVESCSKIIYCATA 237
>At4g18810 unknown protein
Length = 596
Score = 49.7 bits (117), Expect = 2e-06
Identities = 30/80 (37%), Positives = 44/80 (54%), Gaps = 5/80 (6%)
Query: 211 ILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAE 270
IL +K K E + DSGIP+ I+R L ++ G +LI + D I T ++R +VA
Sbjct: 477 ILTYKLKGEDLIRDSGIPFAIVRPCALTEEPAGA-DLIFEQGDNI----TGKVSRDEVAR 531
Query: 271 VCIQALNFEEAQFKAFDLAS 290
+CI AL A K F++ S
Sbjct: 532 ICIAALESPYALNKTFEVKS 551
Score = 46.6 bits (109), Expect = 2e-05
Identities = 42/140 (30%), Positives = 70/140 (50%), Gaps = 16/140 (11%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEE-SKQTIGASDDVYVGDIRDTGSIAP 125
+LV GA G G+ + L++R + LVR EE +++ +G D+ V DI ++ P
Sbjct: 125 ILVAGATGGVGRRIVDILRKRGLP--VKALVRNEEKARKMLGPEIDLIVADITKENTLVP 182
Query: 126 A-IQGIDALIILTSAVPLMKPGFNPTKGERPEF-----YFED---GAYPEQVDWIGQKNQ 176
+G+ +I S + K G P ER ++ +FE G PE V++IG KN
Sbjct: 183 EKFKGVRKVINAVSVIVGPKEGDTP---ERQKYNQGVRFFEPEIKGDSPELVEYIGMKNL 239
Query: 177 IDAAK-AAGVKQIVLVGSMG 195
I+A + G++ L+ +G
Sbjct: 240 INAVRDGVGLENGKLIFGVG 259
>At2g33590 putative cinnamoyl-CoA reductase
Length = 321
Score = 42.7 bits (99), Expect = 3e-04
Identities = 44/150 (29%), Positives = 60/150 (39%), Gaps = 23/150 (15%)
Query: 59 MADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESK--------QTIGASD 110
MAD + V VTGAGG G V L S Y G VR +++ + G
Sbjct: 1 MADVHKGKVCVTGAGGFLGSWVVDLL--LSKDYFVHGTVRDPDNEKYAHLKKLEKAGDKL 58
Query: 111 DVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDW 170
++ D+ D GS+ AI G + + VP P PE A VD
Sbjct: 59 KLFKADLLDYGSLQSAIAGCSGVFHVACPVP-------PASVPNPEVELIAPA----VD- 106
Query: 171 IGQKNQIDAAKAAGVKQIVLVGSMGGTDLN 200
G N + A A VK++V V S+ +N
Sbjct: 107 -GTLNVLKACIEANVKRVVYVSSVAAAFMN 135
>At1g15950 unknown protein
Length = 344
Score = 40.0 bits (92), Expect = 0.002
Identities = 41/147 (27%), Positives = 62/147 (41%), Gaps = 28/147 (19%)
Query: 57 VAMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEES------KQTIGASD 110
V +A + TV VTGAGG + K L ER Y +G VR + ++ G +
Sbjct: 3 VDVASPAGKTVCVTGAGGYIASWIVKILLERG--YTVKGTVRNPDDPKNTHLRELEGGKE 60
Query: 111 DVYV--GDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQV 168
+ + D++D ++ AI G D + S V + PE E
Sbjct: 61 RLILCKADLQDYEALKAAIDGCDGVFHTASPVT-----------DDPEQMVEPAVN---- 105
Query: 169 DWIGQKNQIDAAKAAGVKQIVLVGSMG 195
G K I+AA A VK++V+ S+G
Sbjct: 106 ---GAKFVINAAAEAKVKRVVITSSIG 129
>At1g80820 cinnamoyl CoA reductase like protein
Length = 332
Score = 39.3 bits (90), Expect = 0.003
Identities = 41/142 (28%), Positives = 58/142 (39%), Gaps = 28/142 (19%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEES------KQTIGASD--DVYVGDIR 118
V VTGAGG + K L ER Y RG VR ++ GA + ++ D+
Sbjct: 8 VCVTGAGGYIASWIVKLLLERG--YTVRGTVRNPTDPKNNHLRELQGAKERLTLHSADLL 65
Query: 119 DTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQID 178
D ++ I G D + +A P+ + PE E G K ID
Sbjct: 66 DYEALCATIDGCDG--VFHTASPMT---------DDPETMLEPAVN-------GAKFVID 107
Query: 179 AAKAAGVKQIVLVGSMGGTDLN 200
AA A VK++V S+G +N
Sbjct: 108 AAAKAKVKRVVFTSSIGAVYMN 129
>At1g09510 putative cinnamyl alcohol dehydrogenase
Length = 322
Score = 35.4 bits (80), Expect = 0.040
Identities = 64/264 (24%), Positives = 99/264 (37%), Gaps = 60/264 (22%)
Query: 59 MADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVR-------TEESKQTIGASDD 111
MAD + V VTGA G + K L R Y R VR TE GA +
Sbjct: 1 MADGGKM-VCVTGASGYVASWIVKLLLLRG--YTVRATVRDPSDEKKTEHLLALDGAKEK 57
Query: 112 V--YVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVD 169
+ + D+ + GS AI+G DA+ S V L P+ D A ++
Sbjct: 58 LKLFKADLLEEGSFEQAIEGCDAVFHTASPVSLTVTD--------PQIELIDPAVKGTLN 109
Query: 170 WIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNIL---------------VW 214
+ AK + VK++++ SM P +LG +++ +W
Sbjct: 110 VLK-----TCAKVSSVKRVIVTSSMAAVLFREP--TLGPNDLVDESCFSDPNFCTEKKLW 162
Query: 215 --------KRKAEQYLADSGIPYTIIRAG---------GLQDKEGGVRELIIGKDDEILK 257
+ +A ++ + G+ +I G L + ELI GKD+ I K
Sbjct: 163 YALSKTLAEDEAWRFAKEKGLDLVVINPGLVLGPLLKPSLTFSVNVIVELITGKDNFINK 222
Query: 258 TETRTIARPDVAEVCIQALNFEEA 281
+ R + DVA I+A A
Sbjct: 223 -DFRLVDVRDVALAHIKAFETPSA 245
>At4g27250 unknown protein
Length = 354
Score = 35.0 bits (79), Expect = 0.053
Identities = 25/84 (29%), Positives = 42/84 (49%), Gaps = 4/84 (4%)
Query: 61 DSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIA--RGLVRTE--ESKQTIGASDDVYVGD 116
+S +T VTGA G G + K L +R A R L ++E +SK ++ D
Sbjct: 7 ESKTATYCVTGASGYIGSWLVKSLLQRGYTVHATLRDLAKSEYFQSKWKENERLRLFRAD 66
Query: 117 IRDTGSIAPAIQGIDALIILTSAV 140
+RD GS A++G D + + +++
Sbjct: 67 LRDDGSFDDAVKGCDGVFHVAASM 90
>At3g63140 mRNA binding protein precursor - like
Length = 406
Score = 34.3 bits (77), Expect = 0.090
Identities = 45/188 (23%), Positives = 72/188 (37%), Gaps = 14/188 (7%)
Query: 10 VSATLSPNQCHKYCLPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMADSSRSTVLV 69
+S L P H++ LPSS +SL S SSS SL T RTS + TV
Sbjct: 19 ISNLLIPPSLHRFSLPSSSSSFSSLSSSSSSSSSLLTFS---LRTS--RRLSPQKFTVKA 73
Query: 70 TGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQG 129
+ G + ++ + S + G +E A + VGD P
Sbjct: 74 SSVGEKKNVLI---VNTNSGGHAVIGFYFAKELLSAGHAVTILTVGDESSEKMKKPPFNR 130
Query: 130 IDALIILTSAVPLMKPG--FNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQ 187
++ P N GE + ++ + +D + + +D AK++GVKQ
Sbjct: 131 FSEIVSGGGKTVWGNPANVANVVGGETFDVVLDNNG--KDLDTV--RPVVDWAKSSGVKQ 186
Query: 188 IVLVGSMG 195
+ + S G
Sbjct: 187 FLFISSAG 194
>At2g33600 putative cinnamoyl-CoA reductase
Length = 321
Score = 34.3 bits (77), Expect = 0.090
Identities = 41/150 (27%), Positives = 57/150 (37%), Gaps = 23/150 (15%)
Query: 59 MADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQ--------TIGASD 110
MA + V VTGAGG G V L R Y G VR +++ G
Sbjct: 1 MAVVQKGKVCVTGAGGFLGSWVVNHLLSR--DYFVHGTVRDPGNEKYAHLKKLDKAGDKL 58
Query: 111 DVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDW 170
++ D+ + GS+ AI G + + VP NP E VD
Sbjct: 59 KLFKADLLNYGSLQSAIAGCSGVFHVACPVPSASVP-NP----------EVDLIAPAVD- 106
Query: 171 IGQKNQIDAAKAAGVKQIVLVGSMGGTDLN 200
G N + A A VK++V V S+ +N
Sbjct: 107 -GTLNVLKACVEAKVKRVVYVSSVSAVAMN 135
>At4g35250 unknown protein
Length = 395
Score = 32.7 bits (73), Expect = 0.26
Identities = 53/233 (22%), Positives = 93/233 (39%), Gaps = 39/233 (16%)
Query: 65 STVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTI----GASDDVYVGDIRDT 120
+++LV GA G G+ + ++ + Y R LVR + GA+ V D+
Sbjct: 80 TSILVVGATGTLGRQIVRRALDEG--YDVRCLVRPRPAPADFLRDWGAT--VVNADLSKP 135
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAA 180
+I + GI +I + P E P + VDW G+ I A
Sbjct: 136 ETIPATLVGIHTVIDCATGRP-----------EEPI---------KTVDWEGKVALIQCA 175
Query: 181 KAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDK 240
KA G+++ V S+ D + + ++ K E++L +SG+ + IR G
Sbjct: 176 KAMGIQKYVFY-SIHNCDKHPEV------PLMEIKYCTEKFLQESGLNHITIRLCGFMQG 228
Query: 241 EGGVRELIIGKDDEILKTETRT----IARPDVAEVCIQALNFEEAQFKAFDLA 289
G + I ++ + T+ T + D+A + + AL E+ K A
Sbjct: 229 LIGQYAVPILEEKSVWGTDAPTRVAYMDTQDIARLTLIALRNEKINGKLLTFA 281
>At5g18660 unknown protein
Length = 417
Score = 31.6 bits (70), Expect = 0.58
Identities = 59/246 (23%), Positives = 98/246 (38%), Gaps = 52/246 (21%)
Query: 31 LTSLQFSHSSSLSLATHKRVRTRT--------SVVAMADSSRS------TVLVTGAGGRT 76
L S ++ SS SL +KR R + S +A + S R+ VLV G+ G
Sbjct: 37 LASTFHTNESSTSLK-YKRARLKPISSLDSGISEIATSPSFRNKSPKDINVLVVGSTGYI 95
Query: 77 GKIVYKKLQERSSQYIA--------RGLVRTEES-KQTIGASDDVYVGDIRDTGSIAPAI 127
G+ V K++ +R IA RG EE+ KQ GA+ V D+ + + +I
Sbjct: 96 GRFVVKEMIKRGFNVIAVAREKSGIRGKNDKEETLKQLQGAN--VCFSDVTELDVLEKSI 153
Query: 128 QGID-ALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVK 186
+ + + ++ S + G + ++D+ KN + A K G K
Sbjct: 154 ENLGFGVDVVVSCLASRNGGIKDSW---------------KIDYEATKNSLVAGKKFGAK 198
Query: 187 QIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAE----QYLADSGIPYTIIRAGGLQDKEG 242
VL+ ++ + PL + K +AE DS Y+I+R G
Sbjct: 199 HFVLLSAI---CVQKPLLEFQRAKL---KFEAELMDLAEQQDSSFTYSIVRPTAFFKSLG 252
Query: 243 GVRELI 248
G E++
Sbjct: 253 GQVEIV 258
>At4g33360 putative dihydrokaempferol 4-reductase
Length = 344
Score = 31.6 bits (70), Expect = 0.58
Identities = 35/142 (24%), Positives = 57/142 (39%), Gaps = 21/142 (14%)
Query: 61 DSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDT 120
++ +LVTG+ G G + L R + R LVR + ++ GD+ D
Sbjct: 9 ETENMKILVTGSTGYLGARLCHVLLRRG--HSVRALVRRTSDLSDLPPEVELAYGDVTDY 66
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAA 180
S+ A G D I+ A L++P + D + V+ G KN ++A
Sbjct: 67 RSLTDACSGCD---IVFHAAALVEP------------WLPDPSRFISVNVGGLKNVLEAV 111
Query: 181 KAAGVKQIVLVGS----MGGTD 198
K Q ++ S +G TD
Sbjct: 112 KETKTVQKIIYTSSFFALGSTD 133
>At3g53520 UDP-glucuronic acid decarboxylase (UXS1)
Length = 433
Score = 31.2 bits (69), Expect = 0.76
Identities = 29/86 (33%), Positives = 40/86 (45%), Gaps = 4/86 (4%)
Query: 40 SSLSLATHKRVRTRTSVVAMADS--SRSTVLVTGAGGRTGKIVYKKLQERSSQYI--ARG 95
S L A + TR+ A+ DS SRST G GGRTG++ ++R + G
Sbjct: 70 SRLGAAESTSLITRSVSYAVTDSPPSRSTFNSGGGGGRTGRVPVGIGRKRLRIVVTGGAG 129
Query: 96 LVRTEESKQTIGASDDVYVGDIRDTG 121
V + + IG D+V V D TG
Sbjct: 130 FVGSHLVDKLIGRGDEVIVIDNFFTG 155
>At5g54190 NADPH:protochlorophyllide oxidoreductase A
(gb|AAC49043.1)
Length = 405
Score = 30.8 bits (68), Expect = 0.99
Identities = 39/170 (22%), Positives = 68/170 (39%), Gaps = 21/170 (12%)
Query: 43 SLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYI---ARGLVRT 99
++AT T++S+ + V+VTGA G K L E ++ R ++
Sbjct: 71 AIATSTPSVTKSSLDRKKTLRKGNVVVTGASSGLGLATAKALAETGKWHVIMACRDFLKA 130
Query: 100 EESKQTIGASDDVYVGDIRDTGSIAPAIQGID-------ALIILTSAVPLMKPGFNPTKG 152
E + Q+ G D Y D S+ Q +D L +L + +P N
Sbjct: 131 ERAAQSAGMPKDSYTVMHLDLASLDSVRQFVDNFRRAEMPLDVLVCNAAVYQPTAN---- 186
Query: 153 ERPEFYFEDGAYPEQVDWIG----QKNQIDAAKAAGV--KQIVLVGSMGG 196
+P F E ++ +G + ID K + K++++VGS+ G
Sbjct: 187 -QPTFTAEGFELSVGINHLGHFLLSRLLIDDLKNSDYPSKRLIIVGSITG 235
>At5g51450 unknown protein
Length = 577
Score = 30.0 bits (66), Expect = 1.7
Identities = 18/70 (25%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Query: 62 SSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTI-GASDDVYVGDIRDT 120
S+ ST + GAGGRTG + + + R++ +A L E ++ + D++ D++ T
Sbjct: 506 STSSTYVPPGAGGRTGGL-HLRTVSRAANNMASILAMAETVREVLPHVPDEIIFQDLQRT 564
Query: 121 GSIAPAIQGI 130
S++ + +
Sbjct: 565 NSVSVTVNNL 574
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,612,396
Number of Sequences: 26719
Number of extensions: 272955
Number of successful extensions: 706
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 683
Number of HSP's gapped (non-prelim): 31
length of query: 313
length of database: 11,318,596
effective HSP length: 99
effective length of query: 214
effective length of database: 8,673,415
effective search space: 1856110810
effective search space used: 1856110810
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0147b.4


