

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147a.4
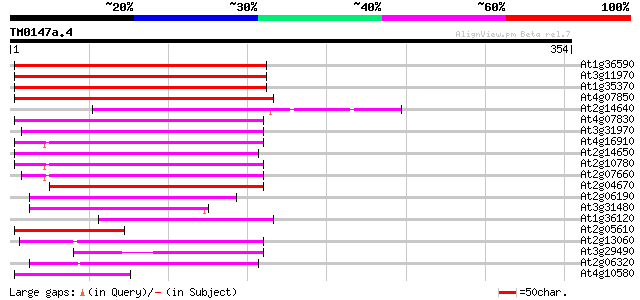
(354 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g36590 hypothetical protein 166 2e-41
At3g11970 hypothetical protein 165 3e-41
At1g35370 hypothetical protein 160 1e-39
At4g07850 putative polyprotein 138 4e-33
At2g14640 putative retroelement pol polyprotein 130 8e-31
At4g07830 putative reverse transcriptase 123 1e-28
At3g31970 hypothetical protein 121 7e-28
At4g16910 retrotransposon like protein 118 4e-27
At2g14650 putative retroelement pol polyprotein 117 1e-26
At2g10780 pseudogene 116 2e-26
At2g07660 putative retroelement pol polyprotein 114 8e-26
At2g04670 putative retroelement pol polyprotein 104 6e-23
At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein 96 4e-20
At3g31480 hypothetical protein 79 5e-15
At1g36120 putative reverse transcriptase gb|AAD22339.1 79 5e-15
At2g05610 putative retroelement pol polyprotein 77 1e-14
At2g13060 pseudogene 68 9e-12
At3g29490 hypothetical protein 67 1e-11
At2g06320 putative retroelement pol polyprotein 64 1e-10
At4g10580 putative reverse-transcriptase -like protein 60 2e-09
>At1g36590 hypothetical protein
Length = 1499
Score = 166 bits (419), Expect = 2e-41
Identities = 83/160 (51%), Positives = 105/160 (64%), Gaps = 1/160 (0%)
Query: 4 PTTVWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIR 63
P T+W + MD I GLP S GK I+VVVDRL+K AHF+ L H +SA VA ++ + +
Sbjct: 1149 PDTIWSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFK 1208
Query: 64 LHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRL 122
LHG P++IVSD D F F G LK +SAYHPQ+DGQTEVVNRCLE YLR +
Sbjct: 1209 LHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCM 1268
Query: 123 IWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQDP 162
P+ WS LA AE+W+NTNY++S M+PF + GQ P
Sbjct: 1269 CHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVP 1308
Score = 38.1 bits (87), Expect = 0.007
Identities = 14/28 (50%), Positives = 23/28 (82%)
Query: 218 VFLKLQPYFRRCLARRINEKLSPRFYGP 245
V++KLQPY ++ + R N+KLSP+++GP
Sbjct: 1368 VYVKLQPYRQQSVVMRANQKLSPKYFGP 1395
>At3g11970 hypothetical protein
Length = 1499
Score = 165 bits (418), Expect = 3e-41
Identities = 83/160 (51%), Positives = 105/160 (64%), Gaps = 1/160 (0%)
Query: 4 PTTVWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIR 63
P T+W + MD I GLP S GK I+VVVDRL+K AHF+ L H +SA VA ++ + +
Sbjct: 1149 PDTIWSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFK 1208
Query: 64 LHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRL 122
LHG P++IVSD D F F G LK +SAYHPQ+DGQTEVVNRCLE YLR +
Sbjct: 1209 LHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCM 1268
Query: 123 IWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQDP 162
P+ WS LA AE+W+NTNY++S M+PF + GQ P
Sbjct: 1269 CHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVP 1308
Score = 38.1 bits (87), Expect = 0.007
Identities = 14/28 (50%), Positives = 23/28 (82%)
Query: 218 VFLKLQPYFRRCLARRINEKLSPRFYGP 245
V++KLQPY ++ + R N+KLSP+++GP
Sbjct: 1368 VYVKLQPYRQQSVVMRANQKLSPKYFGP 1395
>At1g35370 hypothetical protein
Length = 1447
Score = 160 bits (404), Expect = 1e-39
Identities = 80/160 (50%), Positives = 105/160 (65%), Gaps = 1/160 (0%)
Query: 4 PTTVWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIR 63
P +W ++ MD I GLP S GK I+VVVDRL+K AHF+ L H +SA VA F+ + +
Sbjct: 1120 PDKIWCDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYK 1179
Query: 64 LHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRL 122
HG P++IVSD D F F + + G +L+ SSAYHPQ+DGQTEVVNRCLE YLR +
Sbjct: 1180 HHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCM 1239
Query: 123 IWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQDP 162
P W+ L AE+W+NTNY++S M+PF + GQ P
Sbjct: 1240 CHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAP 1279
Score = 39.3 bits (90), Expect = 0.003
Identities = 14/28 (50%), Positives = 24/28 (85%)
Query: 218 VFLKLQPYFRRCLARRINEKLSPRFYGP 245
V++KLQPY ++ + R+N+KLSP+++GP
Sbjct: 1339 VYVKLQPYRQQSVVLRVNQKLSPKYFGP 1366
>At4g07850 putative polyprotein
Length = 1138
Score = 138 bits (348), Expect = 4e-33
Identities = 75/165 (45%), Positives = 102/165 (61%), Gaps = 2/165 (1%)
Query: 4 PTTVWENIYMDSIGGLPKSK-GKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEII 62
P W +I MD + GLP+++ GK++I VVVDR +K AHF+P A +A F +E++
Sbjct: 826 PLHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFIPCHKTDDAMHIANLFFREVV 885
Query: 63 RLHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRR 121
RLHG P TIVSD D+ F F + L GT+L FS+ HPQTDGQTEVVNR L LR
Sbjct: 886 RLHGMPKTIVSDRDTKFLSYFWKTLWSKLGTKLLFSTTCHPQTDGQTEVVNRTLSTLLRA 945
Query: 122 LIWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQDPLSLL 166
LI K W +CL EF +N + +++ SPF + G +P++ L
Sbjct: 946 LIKKNLKTWEDCLPHVEFAYNHSVHSATKFSPFQIVYGFNPITPL 990
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 130 bits (328), Expect = 8e-31
Identities = 81/204 (39%), Positives = 113/204 (54%), Gaps = 13/204 (6%)
Query: 53 VAAGFIQEIIRLHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVV 111
VAA F+ +++LHG P +IVSD D F F + + + T+L S+AYHPQTDGQTEVV
Sbjct: 634 VAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTDGQTEVV 693
Query: 112 NRCLEAYLRRLIWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQDPL-------- 163
NRC+E +LR + PK WS+ + WAE+W+NT ++ S M+PF AL G+ P
Sbjct: 694 NRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRPPSPIPAYELG 753
Query: 164 SLLKGLSYHQGWKR*IS*YVVEMSCWLI*GKIC*HDAFFC**A*ARCGVCSW*LVFLKLQ 223
S++ G Q R + E+ L+ C V W V L++Q
Sbjct: 754 SVVCGELNEQMAAR--DELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDW--VLLRIQ 809
Query: 224 PYFRRCLARRINEKLSPRFYGPLQ 247
PY ++ L RR ++KLS RFYGP Q
Sbjct: 810 PYRQKTLFRRSSQKLSHRFYGPFQ 833
>At4g07830 putative reverse transcriptase
Length = 611
Score = 123 bits (309), Expect = 1e-28
Identities = 67/158 (42%), Positives = 95/158 (59%), Gaps = 1/158 (0%)
Query: 4 PTTVWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIR 63
P W+ I MD + GLP S+ K+ I V+VDRLTK AHFL + A+ +A ++ EI++
Sbjct: 238 PEWKWDFITMDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVK 297
Query: 64 LHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRL 122
LHG P +IVSD DS F F GT+++ S+AYHPQTDGQ+E + LE LR
Sbjct: 298 LHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMC 357
Query: 123 IWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQ 160
+ +W++ L+ EF +N +Y S M+PF L G+
Sbjct: 358 VLDWRGHWADHLSLVEFAYNNSYQASIGMAPFEVLYGR 395
>At3g31970 hypothetical protein
Length = 1329
Score = 121 bits (303), Expect = 7e-28
Identities = 67/154 (43%), Positives = 93/154 (59%), Gaps = 1/154 (0%)
Query: 8 WENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIRLHGF 67
W+ I MD + GLP S+ K+ I V+VDRLTK AHFL + A +A ++ EI+ LHG
Sbjct: 975 WDFITMDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAVLLAKKYVSEIVELHGV 1034
Query: 68 PSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRLIWTC 126
P +IVSD DS F F GT+++ S+AYHPQTDGQ+E + LE LR +
Sbjct: 1035 PVSIVSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDR 1094
Query: 127 PKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQ 160
+W++ L+ EF +N +Y S M+PF AL G+
Sbjct: 1095 GGHWADHLSLVEFAYNNSYQASIRMAPFEALYGR 1128
>At4g16910 retrotransposon like protein
Length = 687
Score = 118 bits (296), Expect = 4e-27
Identities = 69/161 (42%), Positives = 94/161 (57%), Gaps = 5/161 (3%)
Query: 4 PTTVWENIYMDSIGGLP---KSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQE 60
P W++I MD + GLP KSK + VVVDRLTK AHF+ + +A +A +I E
Sbjct: 312 PEWKWDHIMMDFVTGLPTGIKSK-HNAVWVVVDRLTKSAHFMAISDKDAAEIIAEKYIDE 370
Query: 61 IIRLHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYL 119
I+RLHG P +IVSD D+ F F + + GT++ S+AYHPQTDGQ+E + LE L
Sbjct: 371 IVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNLSTAYHPQTDGQSERTIQTLEDML 430
Query: 120 RRLIWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQ 160
R + NW L EF +N ++ S MSP+ AL G+
Sbjct: 431 RACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGR 471
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 117 bits (293), Expect = 1e-26
Identities = 65/155 (41%), Positives = 93/155 (59%), Gaps = 1/155 (0%)
Query: 4 PTTVWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIR 63
P W+ I +D + GLP S+ K+ I V+VDRLTK AHFL + A+ +A ++ EI++
Sbjct: 973 PEWKWDFITIDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVK 1032
Query: 64 LHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRL 122
LHG P +IVSD DS F F GT+++ S+AYHPQT GQ+E + LE LR
Sbjct: 1033 LHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMC 1092
Query: 123 IWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNAL 157
+ +W++ L+ EF +N +Y S M+PF AL
Sbjct: 1093 VLDWGGHWADHLSLVEFAYNNSYPASIGMAPFEAL 1127
>At2g10780 pseudogene
Length = 1611
Score = 116 bits (291), Expect = 2e-26
Identities = 68/161 (42%), Positives = 92/161 (56%), Gaps = 5/161 (3%)
Query: 4 PTTVWENIYMDSIGGLP---KSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQE 60
P W++I MD + GLP KSK + VVVDRLTK AHF+ + A +A +I E
Sbjct: 1228 PEWKWDHITMDFVTGLPTGIKSK-HNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDE 1286
Query: 61 IIRLHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYL 119
I+RLHG P +IVSD D+ F F + + GT++ S+AYHPQTD Q+E + LE L
Sbjct: 1287 IVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQTLEDML 1346
Query: 120 RRLIWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQ 160
R + NW L EF +N ++ S MSP+ AL G+
Sbjct: 1347 RACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGR 1387
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 114 bits (285), Expect = 8e-26
Identities = 67/157 (42%), Positives = 90/157 (56%), Gaps = 5/157 (3%)
Query: 8 WENIYMDSIGGLP---KSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIRL 64
W++I MD + LP KSK + VVVDRLTK AHF+ + A +A +I EI+RL
Sbjct: 624 WDHITMDFVTRLPTGIKSK-HNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRL 682
Query: 65 HGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRLI 123
HG P +IVSD D+ F F + GT++ S+AYHPQTDGQ+E + LE LR +
Sbjct: 683 HGIPVSIVSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACV 742
Query: 124 WTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQ 160
NW L EF +N ++ S MSP+ AL G+
Sbjct: 743 LDWGGNWEKYLRLIEFAYNNSFQASIGMSPYEALYGR 779
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 104 bits (260), Expect = 6e-23
Identities = 57/136 (41%), Positives = 83/136 (60%), Gaps = 1/136 (0%)
Query: 26 ETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIRLHGFPSTIVSDHDS-FPE*FLE 84
+ I V++DRLTK AHFL + A+ +A ++ EI++LHG P +IVSD DS F F
Sbjct: 1075 DAIWVIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWR 1134
Query: 85 VLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRLIWTCPKNWSNCLAWAEFWFNTN 144
GT+++ S+AYHPQTDGQ+E + LE LR + +W++ L+ EF +N +
Sbjct: 1135 AFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNS 1194
Query: 145 YNTSRNMSPFNALSGQ 160
Y S M+PF AL G+
Sbjct: 1195 YQASIGMAPFEALYGR 1210
>At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 280
Score = 95.5 bits (236), Expect = 4e-20
Identities = 55/133 (41%), Positives = 75/133 (56%), Gaps = 2/133 (1%)
Query: 13 MDSIGGLPKS-KGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIRLHGFPSTI 71
MD + GLP++ +G +++ VVVDR +K HF+ AS +A F +E++RLHG P +I
Sbjct: 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADASNIAKLFFKEVVRLHGVPKSI 60
Query: 72 VSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRLIWTCPKNW 130
SD D+ F F L + GT L SS H Q DGQTEV NR L +R + PK W
Sbjct: 61 TSDRDTKFLSHFWSTLWRMFGTALNRSSTPHTQIDGQTEVTNRTLGNMVRSICGDNPKQW 120
Query: 131 SNCLAWAEFWFNT 143
L EF +N+
Sbjct: 121 DLALPQIEFAYNS 133
>At3g31480 hypothetical protein
Length = 338
Score = 78.6 bits (192), Expect = 5e-15
Identities = 48/116 (41%), Positives = 67/116 (57%), Gaps = 3/116 (2%)
Query: 13 MDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIRLHGFPSTIV 72
MD + GLP S+ K+ I V+ DRLT A FL + + +A ++ EI++LHG +IV
Sbjct: 1 MDFVVGLPMSRTKDAIWVIGDRLTNSADFLAIRKTDGVAVLANKYVSEIVKLHGVHVSIV 60
Query: 73 SDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRR--LIWT 125
SD DS F F GT+++ S+AYHP+TDG+ E + LE LR L WT
Sbjct: 61 SDKDSKFTSAFWIAFQAEMGTKVQISTAYHPKTDGKFEKTIQTLEDMLRMTPLCWT 116
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 78.6 bits (192), Expect = 5e-15
Identities = 42/111 (37%), Positives = 66/111 (58%), Gaps = 1/111 (0%)
Query: 57 FIQEIIRLHGFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCL 115
++ EI++LHG P +I+S DS F F GT+++ S+AYHPQTDGQ+E + L
Sbjct: 959 YVSEIVKLHGVPVSILSHRDSKFTSAFWRAFQVEMGTKVQMSTAYHPQTDGQSERTIQTL 1018
Query: 116 EAYLRRLIWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQDPLSLL 166
E L+ + +W++ L+ +F +N +Y S M+PF AL G+ +LL
Sbjct: 1019 EDMLQMCVLDWGGHWADHLSLVKFAYNNSYQASIGMAPFEALYGRPCRTLL 1069
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 77.4 bits (189), Expect = 1e-14
Identities = 35/69 (50%), Positives = 47/69 (67%)
Query: 4 PTTVWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIR 63
P +W ++ MD I GLP S GK I+VVVDRL+K AHF+ L H +SA VA ++ + +
Sbjct: 540 PDRIWSDVSMDFIDGLPLSNGKTVIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVFK 599
Query: 64 LHGFPSTIV 72
LHG PS+IV
Sbjct: 600 LHGCPSSIV 608
Score = 36.6 bits (83), Expect = 0.021
Identities = 14/28 (50%), Positives = 22/28 (78%)
Query: 218 VFLKLQPYFRRCLARRINEKLSPRFYGP 245
VF+KLQPY ++ + R +KLSP+++GP
Sbjct: 696 VFVKLQPYRQQSVVMRSTQKLSPKYFGP 723
>At2g13060 pseudogene
Length = 930
Score = 67.8 bits (164), Expect = 9e-12
Identities = 50/156 (32%), Positives = 76/156 (48%), Gaps = 4/156 (2%)
Query: 7 VWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEII-RLH 65
V++ +D +G P S ILV VD ++K+ + L + S V + II
Sbjct: 266 VFDRWGIDFMGPFPPSNKNLYILVDVDYVSKWVE--AIASLKNDSAVVMKLFKSIIFPRF 323
Query: 66 GFPSTIVSDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRLIW 124
G P ++SD D F LE L+ G Q + ++ YHPQT GQ EV NR ++ L + +
Sbjct: 324 GVPRIVISDGDKHFINKILEKLLLQYGVQHRVATPYHPQTSGQVEVSNRQIKEILEKTVG 383
Query: 125 TCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQ 160
K WS L A + + T + T +PF+ L G+
Sbjct: 384 KAKKEWSYKLYDALWAYKTAFKTPLGTTPFHLLYGK 419
>At3g29490 hypothetical protein
Length = 438
Score = 67.0 bits (162), Expect = 1e-11
Identities = 38/120 (31%), Positives = 61/120 (50%), Gaps = 19/120 (15%)
Query: 41 FLPLCHLFSASEVAAGFIQEIIRLHGFPSTIVSDHDSFPE*FLEVLIQGAGTQLKFSSAY 100
FL + A+ +A ++ EI++LHG P+ + GT+++ S+ Y
Sbjct: 40 FLAIRKTDGAAVLAKKYVSEIVKLHGVPAEM-------------------GTKVQMSTPY 80
Query: 101 HPQTDGQTEVVNRCLEAYLRRLIWTCPKNWSNCLAWAEFWFNTNYNTSRNMSPFNALSGQ 160
HPQTDGQ E + LE LR + +W++ L+ EF +N +Y M+PF AL G+
Sbjct: 81 HPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQAGIGMAPFEALYGR 140
>At2g06320 putative retroelement pol polyprotein
Length = 466
Score = 64.3 bits (155), Expect = 1e-10
Identities = 46/146 (31%), Positives = 70/146 (47%), Gaps = 2/146 (1%)
Query: 13 MDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIRLHGFPSTIV 72
+D +G P S G + ILVVVD ++K+ + A V F I G ++
Sbjct: 173 IDFMGPFPSSYGNKYILVVVDYVSKWVEAIA-SPTNDAKVVLKLFKTIIFPRFGVSWVVI 231
Query: 73 SDHDS-FPE*FLEVLIQGAGTQLKFSSAYHPQTDGQTEVVNRCLEAYLRRLIWTCPKNWS 131
SD F E L++ G + K ++ YHPQT GQ E+ NR ++ L + + K+WS
Sbjct: 232 SDGGKHFINKVFENLLKKHGVKHKVATPYHPQTSGQVEISNREIKTILEKTVGITRKDWS 291
Query: 132 NCLAWAEFWFNTNYNTSRNMSPFNAL 157
L A + + T + T +PFN L
Sbjct: 292 TKLDDALWAYKTAFKTPIGTTPFNLL 317
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 60.1 bits (144), Expect = 2e-09
Identities = 31/73 (42%), Positives = 43/73 (58%)
Query: 4 PTTVWENIYMDSIGGLPKSKGKETILVVVDRLTKFAHFLPLCHLFSASEVAAGFIQEIIR 63
P W+ I MD + GL S+ K+ I V+VDRLTK AHFL + A+ +A F+ EI++
Sbjct: 1008 PEWKWDFITMDLVVGLRVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVK 1067
Query: 64 LHGFPSTIVSDHD 76
LHG P + D
Sbjct: 1068 LHGVPLNMKEAQD 1080
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.349 0.154 0.574
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,720,592
Number of Sequences: 26719
Number of extensions: 300664
Number of successful extensions: 1096
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1019
Number of HSP's gapped (non-prelim): 53
length of query: 354
length of database: 11,318,596
effective HSP length: 100
effective length of query: 254
effective length of database: 8,646,696
effective search space: 2196260784
effective search space used: 2196260784
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0147a.4


