

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0141.9
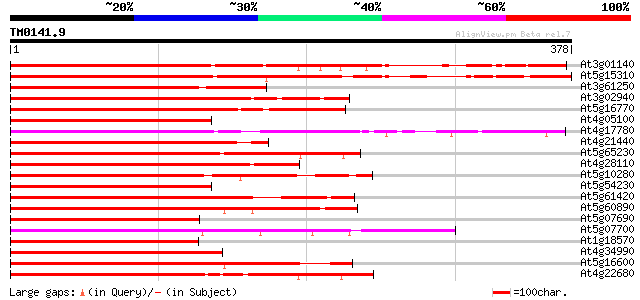
(378 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g01140 putative Myb-related transcription factor 368 e-102
At5g15310 myb-related protein - like 367 e-102
At3g61250 putative transcription factor (MYB17) 243 1e-64
At3g02940 MYB family transcription factor like protein 239 2e-63
At5g16770 putative transcription factor (MYB9) 232 3e-61
At4g05100 MYB - like protein 231 5e-61
At4g17780 MYB transcription factor like protein 230 8e-61
At4g21440 myb-related protein M4 229 1e-60
At5g65230 transcription factor-like protein 227 7e-60
At4g28110 putative transcription factor (MYB41) 223 1e-58
At5g10280 putative transcription factor MYB92 220 1e-57
At5g54230 Myb-related transcription factor-like protein 218 6e-57
At5g61420 putative transcription factor MYB28 215 4e-56
At5g60890 Myb transcription factor homolog (ATR1) 214 5e-56
At5g07690 transcription factor-like protein 212 3e-55
At5g07700 transcription factor (gb|AAD53097.1) 211 4e-55
At1g18570 unknown protein 210 9e-55
At4g34990 MYB-like protein 210 1e-54
At5g16600 transcription factor (gb|AAD53095.1) 209 3e-54
At4g22680 myb-like protein 208 3e-54
>At3g01140 putative Myb-related transcription factor
Length = 345
Score = 368 bits (944), Expect = e-102
Identities = 217/395 (54%), Positives = 250/395 (62%), Gaps = 74/395 (18%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+K GLKKGPWTPEEDQKLLAYIEEHGHGSWR+LP KAGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKAGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKF++QEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRLIKMG
Sbjct: 61 LRPDIKRGKFTVQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLIKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDPVTHK KN+ L SS G SK AA LSHMAQWESARLEAEARL RESKL HL +
Sbjct: 121 IDPVTHKHKNETLSSS--TGQSKNAATLSHMAQWESARLEAEARLARESKL-LHLQHYQN 177
Query: 181 ILPSPSSSSLNKH----DTVKAWNNGSSVVGD--LESPKSTLSFSEN-----------NN 223
S++ +H T W + GD LESP ST++FSEN +
Sbjct: 178 NNNLNKSAAPQQHCFTQKTSTNWTKPNQGNGDQQLESPTSTVTFSENLLMPLGIPTDSSR 237
Query: 224 NNHNDNNVAPIMIEFV---GTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMT 280
N +N+NN + MIE TS + +VKE E +W
Sbjct: 238 NRNNNNNESSAMIELAVSSSTSSDVSLVKEH-EHDW------------------------ 272
Query: 281 MEATWPPSDESLRTITTAATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGES 340
+R I G + EG T LL+ + R L P G E+
Sbjct: 273 -----------IRQIN-----------CGSGGIGEGFTSLLIGDS--VGRGL-PTGKNEA 307
Query: 341 NSGDGGDGCGSDFYEDNKNYWNNILNLVNSSPTNS 375
+G G + ++YEDNKNYWN+ILNLV+SSP++S
Sbjct: 308 TAGVGNES-EYNYYEDNKNYWNSILNLVDSSPSDS 341
>At5g15310 myb-related protein - like
Length = 326
Score = 367 bits (943), Expect = e-102
Identities = 211/380 (55%), Positives = 243/380 (63%), Gaps = 56/380 (14%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+K+GLKKGPWTPEEDQKLLAYIEEHGHGSWR+LP KAGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKLGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLHRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKF+LQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL+KMG
Sbjct: 61 LRPDIKRGKFNLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLVKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKL--RSHLGTS 178
IDPVTHKPKN+ SS+ G SK AA LSH AQWESARLEAEARL RESKL H T
Sbjct: 121 IDPVTHKPKNETPLSSL--GLSKNAAILSHTAQWESARLEAEARLARESKLLHLQHYQTK 178
Query: 179 SSILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVAPIMIEF 238
+S P +K LESP ST+SFSE + P IEF
Sbjct: 179 TSSQPHHHHGFTHKSLLPNWTTKPHEDQQQLESPTSTVSFSEMKES-------IPAKIEF 231
Query: 239 VGTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPPSDESLRTITTA 298
VG+S ++KE E +W ++T HE T
Sbjct: 232 VGSSTGVTLMKEP-EHDW-------------INSTMHEFETTQ----------------- 260
Query: 299 ATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDGCGSDFYEDNK 358
+G+ ++EG T LLL +S DRS S + + GGD ++YEDNK
Sbjct: 261 ---------MGEG-IEEGFTGLLLGGDS-IDRSFSGDKNETAGESSGGD---CNYYEDNK 306
Query: 359 NYWNNILNLVNSSPTNSPMF 378
NY ++I N V+ SP++SPMF
Sbjct: 307 NYLDSIFNFVDPSPSDSPMF 326
>At3g61250 putative transcription factor (MYB17)
Length = 299
Score = 243 bits (620), Expect = 1e-64
Identities = 116/173 (67%), Positives = 137/173 (79%), Gaps = 4/173 (2%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR+PCC+K+GLKKGPWTPEED+ L+A+I+++GHGSWR LP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRTPCCDKIGLKKGPWTPEEDEVLVAHIKKNGHGSWRTLPKLAGLLRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG F+ EE+ +IQLHA+LGNRW+AIA L RTDNEIKN WNTHLKKRL+ MG
Sbjct: 61 LRPDIKRGPFTADEEKLVIQLHAILGNRWAAIAAQLPGRTDNEIKNLWNTHLKKRLLSMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRS 173
+DP TH+P L S + + HMAQWESAR+EAEARL RES L S
Sbjct: 121 LDPRTHEP----LPSYGLAKQAPSSPTTRHMAQWESARVEAEARLSRESMLFS 169
>At3g02940 MYB family transcription factor like protein
Length = 321
Score = 239 bits (610), Expect = 2e-63
Identities = 121/230 (52%), Positives = 161/230 (69%), Gaps = 7/230 (3%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC++ GLKKGPWTPEEDQKL+ +I +HGHGSWRALP +AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDESGLKKGPWTPEEDQKLINHIRKHGHGSWRALPKQAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG F+ +EEQTII LH+LLGN+WS+IA HL RTDNEIKNYWNTH++K+LI+MG
Sbjct: 61 LRPDIKRGNFTAEEEQTIINLHSLLGNKWSSIAGHLPGRTDNEIKNYWNTHIRKKLIQMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQW-ESARLEAEARLVRESKLRSHLGTSS 179
IDPVTH+P+ D L+ AAN +++ ++ +L+A + + + L S + S
Sbjct: 121 IDPVTHRPRTDHLNVLAALPQLLAAANFNNLLNLNQNIQLDATS-VAKAQLLHSMIQVLS 179
Query: 180 SILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDN 229
+ + +SSS + H T SS + +L P + + +H D+
Sbjct: 180 N---NNTSSSFDIHHTTNNLFGQSSFLENL--PNIENPYDQTQGLSHIDD 224
>At5g16770 putative transcription factor (MYB9)
Length = 336
Score = 232 bits (591), Expect = 3e-61
Identities = 112/226 (49%), Positives = 159/226 (69%), Gaps = 4/226 (1%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC++ GLKKGPWT EED KL+ +I++HGHGSWRALP +AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDENGLKKGPWTQEEDDKLIDHIQKHGHGSWRALPKQAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG F+ +EEQTII LH+LLGN+WS+IA +L RTDNEIKNYWNTHL+K+L++MG
Sbjct: 61 LRPDIKRGNFTEEEEQTIINLHSLLGNKWSSIAGNLPGRTDNEIKNYWNTHLRKKLLQMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDPVTH+P+ D L+ AAN + + + ++ +A + +++L L T
Sbjct: 121 IDPVTHRPRTDHLNVLAALPQLIAAANFNSLLNL-NQNVQLDATTLAKAQL---LHTMIQ 176
Query: 181 ILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNH 226
+L + ++++ + N+ +++ G ++ F ++ N +H
Sbjct: 177 VLSTNNNTTNPSFSSSTMQNSNTNLFGQASYLENQNLFGQSQNFSH 222
>At4g05100 MYB - like protein
Length = 324
Score = 231 bits (589), Expect = 5e-61
Identities = 104/137 (75%), Positives = 119/137 (85%), Gaps = 1/137 (0%)
Query: 1 MGRSPCCEKV-GLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSN 59
MGRSPCCEK GLKKGPWTPEEDQKL+ YI HG+G+WR LP AGLQRCGKSCRLRW+N
Sbjct: 1 MGRSPCCEKKNGLKKGPWTPEEDQKLIDYINIHGYGNWRTLPKNAGLQRCGKSCRLRWTN 60
Query: 60 YLRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKM 119
YLRPDIKRG+FS +EE+TIIQLH+++GN+WSAIA L RTDNEIKNYWNTH++KRL+KM
Sbjct: 61 YLRPDIKRGRFSFEEEETIIQLHSIMGNKWSAIAARLPGRTDNEIKNYWNTHIRKRLLKM 120
Query: 120 GIDPVTHKPKNDALHSS 136
GIDPVTH P+ D L S
Sbjct: 121 GIDPVTHTPRLDLLDIS 137
>At4g17780 MYB transcription factor like protein
Length = 745
Score = 230 bits (587), Expect = 8e-61
Identities = 156/394 (39%), Positives = 206/394 (51%), Gaps = 57/394 (14%)
Query: 1 MGRSPCCEK-VGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSN 59
MGRSPCC++ G+KKGPW PEED KL AYI E+G+G+WR+LP AGL RCGKSCRLRW N
Sbjct: 1 MGRSPCCDQDKGVKKGPWLPEEDDKLTAYINENGYGNWRSLPKLAGLNRCGKSCRLRWMN 60
Query: 60 YLRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKM 119
YLRPDI+RGKFS EE TI++LHALLGN+WS IA HL RTDNEIKNYWNTH++K+L++M
Sbjct: 61 YLRPDIRRGKFSDGEESTIVRLHALLGNKWSKIAGHLPGRTDNEIKNYWNTHMRKKLLQM 120
Query: 120 GIDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSS 179
GIDPVTH+P+ + L +D S++ A + Q+ + + L ++
Sbjct: 121 GIDPVTHEPRTNDLSPILD--VSQMLAAAINNGQFGN------------NNLLNNNTALE 166
Query: 180 SILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVAPIMIEFV 239
IL + + T KA N SS +L +PK + N N+ N P + F+
Sbjct: 167 DILKLQLIHKMLQIITPKAIPNISSFKTNLLNPKPEPVVNSFNTNSVNPKPDPPAGL-FI 225
Query: 240 GTSLERGIVKEEG-------EQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPPSDESL 292
S GI E E W G++D N +P T S ESL
Sbjct: 226 NQS---GITPEAASDFIPSYENVWDGFED---NQLPGLVTV--------------SQESL 265
Query: 293 RTIT--TAATTAARAHV-LGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDGC 349
T T+ TT H+ G G D LL + S S+SPE ++
Sbjct: 266 NTAKPGTSTTTKVNDHIRTGMMPCYYG--DQLLETPSTGSVSVSPETTSLNHPSTAQHSS 323
Query: 350 GSDFYEDNKNY---------WNNILNLVNSSPTN 374
GSDF ED + + W + L ++ N
Sbjct: 324 GSDFLEDWEKFLDDETSDSCWKSFLEKIDKKSKN 357
>At4g21440 myb-related protein M4
Length = 350
Score = 229 bits (585), Expect = 1e-60
Identities = 107/174 (61%), Positives = 131/174 (74%), Gaps = 9/174 (5%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M RSPCCEK GLKKGPWT EEDQKL+ YI++HG+G+WR LP AGLQRCGKSCRLRW+NY
Sbjct: 1 MARSPCCEKNGLKKGPWTSEEDQKLVDYIQKHGYGNWRTLPKNAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG+FS +EE+TIIQLH+ LGN+WSAIA L RTDNEIKN+WNTH++K+L++MG
Sbjct: 61 LRPDIKRGRFSFEEEETIIQLHSFLGNKWSAIAARLPGRTDNEIKNFWNTHIRKKLLRMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSH 174
IDPVTH P+ D L S S ++ HM +RL+ ++ R H
Sbjct: 121 IDPVTHSPRLDLLDISSILASSLYNSSSHHMNM---------SRLMMDTNRRHH 165
>At5g65230 transcription factor-like protein
Length = 310
Score = 227 bits (579), Expect = 7e-60
Identities = 121/249 (48%), Positives = 158/249 (62%), Gaps = 15/249 (6%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSP ++ GLKKGPW PEED KL+ YI +HGH SW ALP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPSSDETGLKKGPWLPEEDDKLINYIHKHGHSSWSALPKLAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFS +EE+TI+ LHA+LGN+WS IA+HL RTDNEIKN+WNTHLKK+LI+MG
Sbjct: 61 LRPDIKRGKFSAEEEETILNLHAVLGNKWSMIASHLPGRTDNEIKNFWNTHLKKKLIQMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESA-RLEAEARLVRESKLRSHLGTSS 179
DP+TH+P+ D + SS+ S +NL + + +E +A L ++++
Sbjct: 121 FDPMTHQPRTDDIFSSLSQLMS--LSNLRGLVDLQQQFPMEDQALLNLQTEMAKLQLFQY 178
Query: 180 SILPSPSSSSLNKHD----TVKAWNNGSSVVGDLESPKSTLSFSENNN--------NNHN 227
+ PSP+ S+N + + N + DL S L NNN +NH
Sbjct: 179 LLQPSPAPMSINNINPNILNLLIKENSVTSNIDLGFLSSHLQDFNNNNLPSLKTLDDNHF 238
Query: 228 DNNVAPIMI 236
N +PI +
Sbjct: 239 SQNTSPIWL 247
>At4g28110 putative transcription factor (MYB41)
Length = 282
Score = 223 bits (568), Expect = 1e-58
Identities = 109/195 (55%), Positives = 137/195 (69%), Gaps = 2/195 (1%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+K G+KKGPWT EEDQKL+ YI HG G+WR LP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKNGVKKGPWTAEEDQKLIDYIRFHGPGNWRTLPKNAGLHRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG+FS +EE+TIIQLH+++GN+WSAIA L RTDNEIKN+WNTH++KRL++ G
Sbjct: 61 LRPDIKRGRFSFEEEETIIQLHSVMGNKWSAIAARLPGRTDNEIKNHWNTHIRKRLVRSG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDPVTH P+ D L S N S +A S+ L + ++R + L L +
Sbjct: 121 IDPVTHSPRLDLLDLSSLLSALFNQPNFSAVATHASSLLNPD--VLRLASLLLPLQNPNP 178
Query: 181 ILPSPSSSSLNKHDT 195
+ PS +L +T
Sbjct: 179 VYPSNLDQNLQTPNT 193
>At5g10280 putative transcription factor MYB92
Length = 334
Score = 220 bits (560), Expect = 1e-57
Identities = 116/249 (46%), Positives = 162/249 (64%), Gaps = 25/249 (10%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSP + GLKKGPWTP+ED+KL+ Y+++HGH SWRALP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPISDDSGLKKGPWTPDEDEKLVNYVQKHGHSSWRALPKLAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG+FS EEQTI+ LH++LGN+WS IA L RTDNEIKN+WNTHLKK+LI+MG
Sbjct: 61 LRPDIKRGRFSPDEEQTILNLHSVLGNKWSTIANQLPGRTDNEIKNFWNTHLKKKLIQMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQW----ESARLEAEARLVRESKLRSHLG 176
DP+TH+P+ D I +G S++ + S++ + + ++ E +++ + L
Sbjct: 121 FDPMTHRPRTD-----IFSGLSQLMSLSSNLRGFVDLQQQFPIDQEHTILKLQTEMAKLQ 175
Query: 177 TSSSIL-PSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVAPIM 235
+L PS S+++N +D L S SF E +NN ++N +
Sbjct: 176 LFQYLLQPSSMSNNVNPND-----------FDTLSLLNSIASFKETSNNTTSNN----LD 220
Query: 236 IEFVGTSLE 244
+ F+G+ L+
Sbjct: 221 LGFLGSYLQ 229
>At5g54230 Myb-related transcription factor-like protein
Length = 319
Score = 218 bits (554), Expect = 6e-57
Identities = 98/136 (72%), Positives = 115/136 (84%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MG+S E+ +KKGPWTPEED+KL+ YI+ HG G WR LP AGL+RCGKSCRLRW+NY
Sbjct: 1 MGKSSSSEESEVKKGPWTPEEDEKLVGYIQTHGPGKWRTLPKNAGLKRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG+FSLQEE+TIIQLH LLGN+WSAIA HL RTDNEIKNYWNTH+KK+L++MG
Sbjct: 61 LRPDIKRGEFSLQEEETIIQLHRLLGNKWSAIAIHLPGRTDNEIKNYWNTHIKKKLLRMG 120
Query: 121 IDPVTHKPKNDALHSS 136
IDPVTH P+ + L S
Sbjct: 121 IDPVTHCPRINLLQLS 136
>At5g61420 putative transcription factor MYB28
Length = 366
Score = 215 bits (547), Expect = 4e-56
Identities = 110/232 (47%), Positives = 146/232 (62%), Gaps = 20/232 (8%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M R PCC GLKKG WT EED+KL++YI +HG G WR +P KAGL+RCGKSCRLRW+NY
Sbjct: 1 MSRKPCCVGEGLKKGAWTTEEDKKLISYIHDHGEGGWRDIPQKAGLKRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
L+P+IKRG+FS +EEQ II LHA GN+WS IA HL +RTDNEIKNYWNTHLKKRL++ G
Sbjct: 61 LKPEIKRGEFSSEEEQIIIMLHASRGNKWSVIARHLPRRTDNEIKNYWNTHLKKRLMEQG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDPVTHKP + + ++D + A+ S S+ + +R
Sbjct: 121 IDPVTHKPLASSSNPTVDENLNSPNASSSDKQYSRSSSMPFLSR---------------- 164
Query: 181 ILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVA 232
P PSS ++ + + N+G+ + G S K F ++++ + N VA
Sbjct: 165 --PPPSSCNMVSKVSELSSNDGTPIQGSSLSCKK--RFKKSSSTSRLLNKVA 212
>At5g60890 Myb transcription factor homolog (ATR1)
Length = 295
Score = 214 bits (546), Expect = 5e-56
Identities = 112/242 (46%), Positives = 152/242 (62%), Gaps = 11/242 (4%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M R+PCC++ G+KKG WTPEEDQKL+AY+ HG G WR LP KAGL+RCGKSCRLRW+NY
Sbjct: 1 MVRTPCCKEEGIKKGAWTPEEDQKLIAYLHLHGEGGWRTLPEKAGLKRCGKSCRLRWANY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG+FS +E+ TII+LHAL GN+W+AIAT L+ RTDNEIKNYWNT+LKKRL + G
Sbjct: 61 LRPDIKRGEFSPEEDDTIIKLHALKGNKWAAIATSLAGRTDNEIKNYWNTNLKKRLKQKG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSK------VAANLSHMAQWESARLEAE--ARLVRESKLR 172
ID +THKP N + + +K A L+ +A + L + ++ +
Sbjct: 121 IDAITHKPINSTGQTGFEPKVNKPVYSSGSARLLNRVASKYAVELNRDLLTGIISGNSTV 180
Query: 173 SHLGTSSSILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVA 232
+ +S + SP+S+ LNK N ++ SF++ N N+ ++
Sbjct: 181 AEDSQNSGDVDSPTSTLLNKMAATSVLINTTTTYSGF---SDNCSFTDEFNEFFNNEEIS 237
Query: 233 PI 234
I
Sbjct: 238 DI 239
>At5g07690 transcription factor-like protein
Length = 336
Score = 212 bits (539), Expect = 3e-55
Identities = 95/128 (74%), Positives = 105/128 (81%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M R PCC GLKKG WT EED+KL++YI EHG G WR +P KAGL+RCGKSCRLRW+NY
Sbjct: 1 MSRKPCCVGEGLKKGAWTAEEDKKLISYIHEHGEGGWRDIPQKAGLKRCGKSCRLRWANY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
L+PDIKRG+FS +EEQ II LHA GN+WS IA HL KRTDNEIKNYWNTHLKK LI G
Sbjct: 61 LKPDIKRGEFSYEEEQIIIMLHASRGNKWSVIARHLPKRTDNEIKNYWNTHLKKLLIDKG 120
Query: 121 IDPVTHKP 128
IDPVTHKP
Sbjct: 121 IDPVTHKP 128
>At5g07700 transcription factor (gb|AAD53097.1)
Length = 338
Score = 211 bits (538), Expect = 4e-55
Identities = 129/322 (40%), Positives = 180/322 (55%), Gaps = 28/322 (8%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M + P C GLKKG WT EED+KL++YI +HG G WR +P KAGL+RCGKSCRLRW+NY
Sbjct: 1 MSKRPYCIGEGLKKGAWTTEEDKKLISYIHDHGEGGWRDIPEKAGLKRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
L+PDIKRG+FS +EEQ II LHA GN+WS IA HL KRTDNE+KNYWNTHLKKRLI G
Sbjct: 61 LKPDIKRGEFSYEEEQIIIMLHASRGNKWSVIARHLPKRTDNEVKNYWNTHLKKRLIDDG 120
Query: 121 IDPVTHKP------------KNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVR- 167
IDPVTHKP K D S + HS +++ + + S+ L + +
Sbjct: 121 IDPVTHKPLASSNPNPVEPMKFDFQKKSNQDEHSSQSSSTTPASLPLSSNLNSVKSKISS 180
Query: 168 -ESKLRS-HLGTSSSILPSPSSSSLNKHDTVKAWNNG----SSVVGDLESPKSTLSFSEN 221
E+++ S H+ S S+S L +A + G +S+ G L SP S+ ++
Sbjct: 181 GETQIESGHVSCKKRFGRSSSTSRLLNKVAARASSIGNILSTSIEGTLRSPASSSGLPDS 240
Query: 222 NNNNHN---DNNVAPIMIEFVGTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELT 278
+ ++ DN E +GTS++ I + + Q + + + + L
Sbjct: 241 FSQSYEYMIDNK------EDLGTSIDLNIPEYDFPQFLEQLINDDDENENIVGPEQDLLM 294
Query: 279 MTMEATWPPSDESLRTITTAAT 300
+T+ D+ L IT+ +T
Sbjct: 295 SDFPSTFVDEDDILGDITSWST 316
>At1g18570 unknown protein
Length = 352
Score = 210 bits (535), Expect = 9e-55
Identities = 96/128 (75%), Positives = 110/128 (85%), Gaps = 1/128 (0%)
Query: 1 MGRSPCCE-KVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSN 59
M R+PCC+ ++GLKKG WTPEEDQKLL+Y+ HG G WR LP KAGL+RCGKSCRLRW+N
Sbjct: 1 MVRTPCCKAELGLKKGAWTPEEDQKLLSYLNRHGEGGWRTLPEKAGLKRCGKSCRLRWAN 60
Query: 60 YLRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKM 119
YLRPDIKRG+F+ EE++II LHAL GN+WSAIA L RTDNEIKNYWNTH+KKRLIK
Sbjct: 61 YLRPDIKRGEFTEDEERSIISLHALHGNKWSAIARGLPGRTDNEIKNYWNTHIKKRLIKK 120
Query: 120 GIDPVTHK 127
GIDPVTHK
Sbjct: 121 GIDPVTHK 128
>At4g34990 MYB-like protein
Length = 274
Score = 210 bits (534), Expect = 1e-54
Identities = 93/143 (65%), Positives = 112/143 (78%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCCEK KG WT EED KL++YI+ HG G WR+LP AGLQRCGKSCRLRW NY
Sbjct: 1 MGRSPCCEKDHTNKGAWTKEEDDKLISYIKAHGEGCWRSLPRSAGLQRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPD+KRG F+L+E+ II+LH+LLGN+WS IAT L RTDNEIKNYWNTH+K++L++ G
Sbjct: 61 LRPDLKRGNFTLEEDDLIIKLHSLLGNKWSLIATRLPGRTDNEIKNYWNTHVKRKLLRKG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSK 143
IDP TH+P N+ S + SK
Sbjct: 121 IDPATHRPINETKTSQDSSDSSK 143
>At5g16600 transcription factor (gb|AAD53095.1)
Length = 327
Score = 209 bits (531), Expect = 3e-54
Identities = 109/233 (46%), Positives = 143/233 (60%), Gaps = 21/233 (9%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR PCC+KVGLKKGPWT EED+KL+ +I +GH WRALP +GL RCGKSCRLRW NY
Sbjct: 1 MGRQPCCDKVGLKKGPWTIEEDKKLINFILTNGHCCWRALPKLSGLLRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPD+KRG S EEQ +I LHA LGNRWS IA+HL RTDNEIKN+WNTH+KK+L KMG
Sbjct: 61 LRPDLKRGLLSEYEEQKVINLHAQLGNRWSKIASHLPGRTDNEIKNHWNTHIKKKLRKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSK--VAANLSHMAQWESARLEAEARLVRESKLRSHLGTS 178
IDP+THKP ++ S G K V + + Q + + E E + ++ +++ S
Sbjct: 121 IDPLTHKPLSEQEASQQAQGRKKSLVPHDDKNPKQDQQTKDEQEQHQLEQALEKNNTSVS 180
Query: 179 SSILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNV 231
LN H+ + + S +++++ ND+NV
Sbjct: 181 GDGFCIDEVPLLNPHEIL-------------------IDISSSHHHHSNDDNV 214
>At4g22680 myb-like protein
Length = 266
Score = 208 bits (530), Expect = 3e-54
Identities = 115/250 (46%), Positives = 156/250 (62%), Gaps = 10/250 (4%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR PCC+K+G+KKGPWT EED+KL+ +I +GH WRALP AGL+RCGKSCRLRW+NY
Sbjct: 1 MGRQPCCDKLGVKKGPWTVEEDKKLINFILTNGHCCWRALPKLAGLRRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPD+KRG S EEQ +I LHA LGN+WS IA+ L RTDNEIKN+WNTH+KK+L+KMG
Sbjct: 61 LRPDLKRGLLSHDEEQLVIDLHANLGNKWSKIASRLPGRTDNEIKNHWNTHIKKKLLKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDP+TH+P N S+IDN + + +N ++ + V S + +SS+
Sbjct: 121 IDPMTHQPLNQE-PSNIDNSKT-IPSNPDDVSVEPKT---TNTKYVEISVTTTEEESSST 175
Query: 181 ILPSPSSSSLNKH--DTVKAWNNGSSVVGDLESPKSTLSFSENN---NNNHNDNNVAPIM 235
+ SS H D + + S + E+ K S+S++N D+N++
Sbjct: 176 VTDQNSSMDNENHLIDNIYDDDELFSYLWSDETTKDEASWSDSNFGVGGTLYDHNISGAD 235
Query: 236 IEFVGTSLER 245
+F S ER
Sbjct: 236 ADFPIWSPER 245
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.128 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,655,520
Number of Sequences: 26719
Number of extensions: 449141
Number of successful extensions: 2284
Number of sequences better than 10.0: 207
Number of HSP's better than 10.0 without gapping: 170
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 1896
Number of HSP's gapped (non-prelim): 292
length of query: 378
length of database: 11,318,596
effective HSP length: 101
effective length of query: 277
effective length of database: 8,619,977
effective search space: 2387733629
effective search space used: 2387733629
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0141.9


