

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0137.2
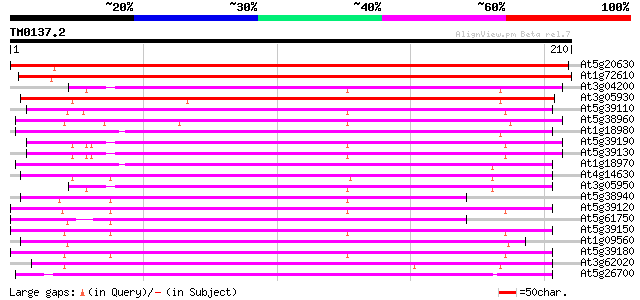
(210 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g20630 germin-like protein (GLP3b) 297 3e-81
At1g72610 germin-like protein 275 2e-74
At3g04200 germin-like protein 149 1e-36
At3g05930 germin-like protein 146 9e-36
At5g39110 germin -like protein 145 1e-35
At5g38960 germin - like protein 145 1e-35
At1g18980 unknown protein 144 3e-35
At5g39190 germin-like protein (GLP2a) copy2 143 7e-35
At5g39130 germin - like protein 142 2e-34
At1g18970 germin-like protein (GLP4) 140 5e-34
At4g14630 germin precursor oxalate oxidase 140 6e-34
At3g05950 germin-like protein 139 8e-34
At5g38940 germin - like protein 138 2e-33
At5g39120 germin - like protein 137 3e-33
At5g61750 germin-like protein - like 137 5e-33
At5g39150 germin - like protein 137 5e-33
At1g09560 germin-like protein 137 5e-33
At5g39180 germin-like protein 136 7e-33
At3g62020 germin-like protein (GLP10) 136 7e-33
At5g26700 nectarin - like protein 135 1e-32
>At5g20630 germin-like protein (GLP3b)
Length = 211
Score = 297 bits (760), Expect = 3e-81
Identities = 145/210 (69%), Positives = 174/210 (82%), Gaps = 1/210 (0%)
Query: 1 MKMVLTILFILSLLS-LSHASVVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIA 59
MKM++ I FI+SL+S +S ASV DFCV D P P+GY+CK P +VT +DFA++GLG A
Sbjct: 1 MKMIIQIFFIISLISTISFASVQDFCVADPKGPQSPSGYSCKNPDQVTENDFAFTGLGTA 60
Query: 60 GNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTIC 119
GNTSNIIKAAVTPAF + G+NGLG+SLARLDLA GGVIPLHTHPGASE LVV+QGTIC
Sbjct: 61 GNTSNIIKAAVTPAFAPAYAGINGLGVSLARLDLAGGGVIPLHTHPGASEVLVVIQGTIC 120
Query: 120 AGFVASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALF 179
AGF++S N VYLKTL +GD MVFPQGLLHFQ+N G ALAFV+F S++PGLQIL FALF
Sbjct: 121 AGFISSANKVYLKTLNRGDSMVFPQGLLHFQLNSGKGPALAFVAFGSSSPGLQILPFALF 180
Query: 180 KSDFPTDLIAATTFLDAAQIKKLKGVLGGS 209
+D P++L+ ATTFL A++KKLKGVLGG+
Sbjct: 181 ANDLPSELVEATTFLSDAEVKKLKGVLGGT 210
>At1g72610 germin-like protein
Length = 208
Score = 275 bits (702), Expect = 2e-74
Identities = 141/208 (67%), Positives = 165/208 (78%), Gaps = 1/208 (0%)
Query: 4 VLTILFILSLL-SLSHASVVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNT 62
+L +F+LSLL +LS+ASV DFCV +L PAGY C +P V A DF +SGLG GNT
Sbjct: 1 MLRTIFLLSLLFALSNASVQDFCVANLKRAETPAGYPCIRPIHVKATDFVFSGLGTPGNT 60
Query: 63 SNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGF 122
+NII AAVTPAF AQFPG+NGLG+S ARLDLA GVIP+HTHPGASE L V+ G+I AGF
Sbjct: 61 TNIINAAVTPAFAAQFPGLNGLGLSTARLDLAPKGVIPMHTHPGASEVLFVLTGSITAGF 120
Query: 123 VASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALFKSD 182
V+S N VY++TLK G VMVFPQGLLHFQIN G SSA A V+F+SANPGLQILDFALF +
Sbjct: 121 VSSANAVYVQTLKPGQVMVFPQGLLHFQINAGKSSASAVVTFNSANPGLQILDFALFANS 180
Query: 183 FPTDLIAATTFLDAAQIKKLKGVLGGSG 210
PT+L+ TTFLDA +KKLKGVLGG+G
Sbjct: 181 LPTELVVGTTFLDATTVKKLKGVLGGTG 208
>At3g04200 germin-like protein
Length = 227
Score = 149 bits (375), Expect = 1e-36
Identities = 88/194 (45%), Positives = 117/194 (59%), Gaps = 12/194 (6%)
Query: 23 DFCVG----DLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNTSNIIKAAVTPAFDAQF 78
DFCV + F NG CK P VTA+DF+YSGL IA NT+N + + VT +
Sbjct: 33 DFCVAASETNRVFVNGKF---CKDPKSVTANDFSYSGLNIARNTTNFLGSNVTTVDVNKI 89
Query: 79 PGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGFVAS---DNTVYLKTLK 135
PG+N LG+SLARLD A GG P H HP A+E LVV +G + GFV+S +N ++ K LK
Sbjct: 90 PGLNTLGVSLARLDFAQGGQNPPHIHPRATEILVVTKGKLLVGFVSSNQDNNRLFYKVLK 149
Query: 136 KGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALFKSD--FPTDLIAATTF 193
+GDV VFP GL+HFQ+N + A+AF F S NPG + A+F S+ P +++A
Sbjct: 150 RGDVFVFPIGLIHFQMNVRRTRAVAFAGFGSQNPGTIRIADAVFGSNPSIPQEVLAKAFQ 209
Query: 194 LDAAQIKKLKGVLG 207
LD ++ L V G
Sbjct: 210 LDVKLVRFLHIVFG 223
>At3g05930 germin-like protein
Length = 219
Score = 146 bits (368), Expect = 9e-36
Identities = 82/204 (40%), Positives = 125/204 (61%), Gaps = 4/204 (1%)
Query: 5 LTILFILSLLSLSHASVV-DFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNTS 63
+T + + + ++L+ +++ DFCV DL+ GY CK P+KVT +DF + GL A T+
Sbjct: 10 VTFMLVAAHMALADTNMLQDFCVADLSNGLKVNGYPCKDPAKVTPEDFYFIGLATAAATA 69
Query: 64 NI-IKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGF 122
N + +AVT A + PG+N LG+S++R+D A GG+ P H HP ASEA+ V++G + GF
Sbjct: 70 NSSMGSAVTGANVEKVPGLNTLGVSISRIDYAPGGLNPPHLHPRASEAIFVLEGRLFVGF 129
Query: 123 VASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALFKSD 182
+ + + K + KGDV VFP+ LLHFQ N + A +F S PG Q++ +LF S+
Sbjct: 130 LTTTGKLISKHVNKGDVFVFPKALLHFQQNPNKAPASVLAAFDSQLPGTQVVGPSLFGSN 189
Query: 183 --FPTDLIAATTFLDAAQIKKLKG 204
P DL+A A +I+K+KG
Sbjct: 190 PPIPDDLLAKAFGAAAPEIQKIKG 213
>At5g39110 germin -like protein
Length = 222
Score = 145 bits (367), Expect = 1e-35
Identities = 92/210 (43%), Positives = 121/210 (56%), Gaps = 13/210 (6%)
Query: 7 ILFILSLLSLSHAS------VVDFCV--GDLTFPNGPAGYACKKPSKVTADDFAYSGLGI 58
IL LS L +S A + DFCV GDL G CK P + A+DF YSGL
Sbjct: 8 ILITLSALVISFAEANDPSPLQDFCVAIGDLKNGVFVNGKFCKDPKQAKAEDFFYSGLNQ 67
Query: 59 AGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTI 118
AG T+N +K+ VT Q PG+N LGISL R+D A G P HTHP A+E LV+V+GT+
Sbjct: 68 AGTTNNKVKSNVTTVNVDQIPGLNTLGISLVRIDYAPYGQNPPHTHPRATEILVLVEGTL 127
Query: 119 CAGFVAS---DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILD 175
GFV+S +N ++ K L GDV VFP G++HFQ+N G + A+AF SS N G+ +
Sbjct: 128 YVGFVSSNQDNNRLFAKVLNPGDVFVFPIGMIHFQVNIGKTPAVAFAGLSSQNAGVITIA 187
Query: 176 FALFKSDFP--TDLIAATTFLDAAQIKKLK 203
+F S P D++A LD +K L+
Sbjct: 188 DTVFGSTPPINPDILAQAFQLDVNVVKDLE 217
>At5g38960 germin - like protein
Length = 221
Score = 145 bits (367), Expect = 1e-35
Identities = 95/218 (43%), Positives = 126/218 (57%), Gaps = 13/218 (5%)
Query: 3 MVLTILFILSLLSLSHA------SVVDFCVGDLTFPNG--PAGYACKKPSKVTADDFAYS 54
+ L IL++L+ +L A + DFC+G T N G CK P VTADDF +S
Sbjct: 4 LYLAILYLLAASTLPFAIASDPSPLQDFCIGVNTPANALFVNGKFCKDPKLVTADDFYFS 63
Query: 55 GLGIAGNT-SNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVV 113
GL A T S+ + + VT Q PG+N LGISL R+D G P HTHP A+E L+V
Sbjct: 64 GLDKARTTESSPVGSNVTTVNVNQIPGLNTLGISLVRIDYGINGQNPPHTHPRATEILLV 123
Query: 114 VQGTICAGFVAS--DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGL 171
+GT+ GF +S +N ++ KTL KGDV VFP+GL+HFQ+N G A+AF S SS NPG+
Sbjct: 124 QEGTLFVGFFSSFPENRLFNKTLNKGDVFVFPEGLIHFQVNIGKQPAVAFASLSSQNPGV 183
Query: 172 QILDFALFKSDFPTD--LIAATTFLDAAQIKKLKGVLG 207
I+ LF S P D ++A LD I +L+ G
Sbjct: 184 IIIGNTLFGSKPPIDPNVLAKAFQLDPKVIIQLQKKFG 221
>At1g18980 unknown protein
Length = 220
Score = 144 bits (363), Expect = 3e-35
Identities = 82/203 (40%), Positives = 119/203 (58%), Gaps = 4/203 (1%)
Query: 3 MVLTILFILSLLSLSHASVVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNT 62
+++ +F++ LS + DFCVGDL G+ CK S V+A DF +SGLG NT
Sbjct: 15 LLVICVFVIPSLSSDSDPLQDFCVGDLKASPSINGFPCK--SSVSASDFFFSGLGGPLNT 72
Query: 63 SNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGF 122
S AV+PA FPG+N LG+S+ ++ A GGV P H+HP A+EA VV++G++ GF
Sbjct: 73 STPNGVAVSPANVLTFPGLNTLGLSMNNVEFAPGGVNPPHSHPRATEAGVVIEGSVFVGF 132
Query: 123 VASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALFKSD 182
+ ++NT++ K L G++ V P+GL+HFQ N G A SF+S PG +L LF S+
Sbjct: 133 LTTNNTLFSKVLNAGEMFVVPRGLVHFQWNVGKVKARLITSFNSQLPGSAVLPSTLFGSN 192
Query: 183 --FPTDLIAATTFLDAAQIKKLK 203
P ++ T D + KLK
Sbjct: 193 PTIPNAVLTKTFRTDDVTVNKLK 215
>At5g39190 germin-like protein (GLP2a) copy2
Length = 222
Score = 143 bits (360), Expect = 7e-35
Identities = 90/211 (42%), Positives = 124/211 (58%), Gaps = 13/211 (6%)
Query: 7 ILFILSLLSLSHASVV-DFCVG--DL--TFPNGPAGYACKKPSKVTADDFAYSGLGIAGN 61
I +LS ++ S + DFCV DL F NG CK P +V A DF +SGL + GN
Sbjct: 13 IALVLSFVNAYDPSPLQDFCVAIDDLKGVFVNGRF---CKDPKRVDAKDFFFSGLNVPGN 69
Query: 62 TSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAG 121
T+N + + VT Q PG+N +GISL R+D A G P HTHP SE LV+V+GT+ G
Sbjct: 70 TNNQVGSNVTTVNVDQIPGLNTMGISLVRIDYAPHGQNPPHTHPRGSEILVLVEGTLYVG 129
Query: 122 FVAS---DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFAL 178
FV+S +N ++ K L GDV VFP G++HFQ+N G A+AF SS N G+ + +
Sbjct: 130 FVSSNQDNNRLFAKVLHPGDVFVFPIGMIHFQVNVGKIPAVAFAGLSSQNAGVITIANTV 189
Query: 179 FKSDFP--TDLIAATTFLDAAQIKKLKGVLG 207
F S+ P +L+A LDA+ +K+L+ G
Sbjct: 190 FGSNPPIYPELLARAFQLDASVVKELQAKFG 220
>At5g39130 germin - like protein
Length = 222
Score = 142 bits (357), Expect = 2e-34
Identities = 90/211 (42%), Positives = 123/211 (57%), Gaps = 13/211 (6%)
Query: 7 ILFILSLLSLSHASVV-DFCVG--DL--TFPNGPAGYACKKPSKVTADDFAYSGLGIAGN 61
I +LS ++ S + DFCV DL F NG CK P +V A DF +SGL + GN
Sbjct: 13 IALVLSFVNAYDPSPLQDFCVAIDDLKGVFVNGRF---CKDPERVDAKDFFFSGLNVPGN 69
Query: 62 TSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAG 121
T+N + + VT Q PG+N +GISL R+D A G P HTHP SE LV+V+GT+ G
Sbjct: 70 TNNQVGSNVTTVNVDQIPGLNTMGISLVRIDYAPHGQNPPHTHPRGSEILVLVEGTLYVG 129
Query: 122 FVAS---DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFAL 178
FV+S +N ++ K L GDV VFP G++HFQ+N G A+AF SS N G+ + +
Sbjct: 130 FVSSNQDNNRLFAKVLHPGDVFVFPIGMIHFQLNIGKIPAIAFAGLSSQNAGVITIANTV 189
Query: 179 FKSDFP--TDLIAATTFLDAAQIKKLKGVLG 207
F S+ P +L+A LDA +K+L+ G
Sbjct: 190 FGSNPPIYPELLARAFQLDANVVKELQAKFG 220
>At1g18970 germin-like protein (GLP4)
Length = 220
Score = 140 bits (353), Expect = 5e-34
Identities = 80/203 (39%), Positives = 115/203 (56%), Gaps = 4/203 (1%)
Query: 3 MVLTILFILSLLSLSHASVVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNT 62
+++ LF++ LS + DFCVGDL G+ CK S V+A DF YSGLG +T
Sbjct: 15 LLMFCLFVIPSLSSDSDPLQDFCVGDLKASASINGFPCK--SAVSASDFFYSGLGGPLDT 72
Query: 63 SNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGF 122
SN V PA FPG+N LGIS+ ++LA GGV P H HP A+E V++G++ GF
Sbjct: 73 SNPNGVTVAPANVLTFPGLNTLGISMNNVELAPGGVNPPHLHPRATEVGTVIEGSVFVGF 132
Query: 123 VASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALF--K 180
++++NT++ K L G+ V P+GL+HFQ N G A +F+S PG +L LF K
Sbjct: 133 LSTNNTLFSKVLNAGEAFVIPRGLVHFQWNVGQVKARMITAFNSQLPGAVVLPSTLFGSK 192
Query: 181 SDFPTDLIAATTFLDAAQIKKLK 203
+ P ++ D ++ LK
Sbjct: 193 PEIPNAVLTRAFRTDDTTVQNLK 215
>At4g14630 germin precursor oxalate oxidase
Length = 222
Score = 140 bits (352), Expect = 6e-34
Identities = 88/214 (41%), Positives = 121/214 (56%), Gaps = 15/214 (7%)
Query: 5 LTILFILSLLSLSHASVV--------DFCVGDLTFPNGPA--GYACKKPSKVTADDFAYS 54
L+ L LSL +L+ V+ DFCVG T +G G CK P V ADDF +S
Sbjct: 6 LSFLAALSLFALTLPLVIASDPSPLQDFCVGVNTPADGVFVNGKFCKDPRIVFADDFFFS 65
Query: 55 GLGIAGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVV 114
L GNT+N + + VT G+N LGISL R+D A G P HTHP A+E LVV
Sbjct: 66 SLNRPGNTNNAVGSNVTTVNVNNLGGLNTLGISLVRIDYAPNGQNPPHTHPRATEILVVQ 125
Query: 115 QGTICAGFVASD---NTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGL 171
QGT+ GF++S+ N ++ KTL GDV VFP+GL+HFQ N GG+ A+A + SS N G+
Sbjct: 126 QGTLLVGFISSNQDGNRLFAKTLNVGDVFVFPEGLIHFQFNLGGTPAVAIAALSSQNAGV 185
Query: 172 QILDFALF--KSDFPTDLIAATTFLDAAQIKKLK 203
+ +F K D +++A +D ++ L+
Sbjct: 186 ITIANTIFGSKPDVDPNVLARAFQMDVNAVRNLQ 219
>At3g05950 germin-like protein
Length = 229
Score = 139 bits (351), Expect = 8e-34
Identities = 84/190 (44%), Positives = 110/190 (57%), Gaps = 12/190 (6%)
Query: 23 DFCVG----DLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNTSNIIKAAVTPAFDAQF 78
DFCV F NG CK P V A+DF SGL IAGNT N + + VT +
Sbjct: 32 DFCVAVDDASGVFVNGKF---CKDPKYVKAEDFFTSGLNIAGNTINRVGSNVTNVNVDKI 88
Query: 79 PGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGFVAS---DNTVYLKTLK 135
PG+N LG+SL R+D A GG P HTHP A+E LVVV+GT+ GFV S +N ++ K L
Sbjct: 89 PGLNTLGVSLVRIDFAPGGQNPPHTHPRATEILVVVEGTLLVGFVTSNQDNNRLFSKVLY 148
Query: 136 KGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALF--KSDFPTDLIAATTF 193
GDV VFP G++HFQ+N G ++A+AF S NPG + A+F K +++A
Sbjct: 149 PGDVFVFPIGMIHFQVNVGRTNAVAFAGLGSQNPGTITIADAVFGSKPSIMPEILAKAFQ 208
Query: 194 LDAAQIKKLK 203
LD +K L+
Sbjct: 209 LDVNVVKYLE 218
>At5g38940 germin - like protein
Length = 221
Score = 138 bits (347), Expect = 2e-33
Identities = 83/179 (46%), Positives = 107/179 (59%), Gaps = 12/179 (6%)
Query: 5 LTILFILSLLSLS--------HASVVDFCVGDLTFPNGPA--GYACKKPSKVTADDFAYS 54
L+ L +LSLL+L+ + + DFCV T NG G CK P VTADDF +S
Sbjct: 4 LSFLAVLSLLALTLPLAIASDPSQLQDFCVSANTSANGVFVNGKFCKDPKLVTADDFFFS 63
Query: 55 GLGIAGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVV 114
GL A ++ + + VT G+N LGISL R+D A G P HTHP A+E LVV
Sbjct: 64 GLQTARPITSPVGSTVTAVNVNNLLGLNTLGISLVRIDYAVNGQNPPHTHPRATEILVVE 123
Query: 115 QGTICAGFVAS--DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGL 171
QGT+ GFV S DN ++ K L +GDV VFP+GL+HFQ N G + A+AF + SS NPG+
Sbjct: 124 QGTLLVGFVTSNPDNRLFSKVLNEGDVFVFPEGLIHFQANIGKAPAVAFAALSSQNPGV 182
>At5g39120 germin - like protein
Length = 221
Score = 137 bits (346), Expect = 3e-33
Identities = 86/212 (40%), Positives = 122/212 (56%), Gaps = 9/212 (4%)
Query: 1 MKMVLTILFILSLLSLSH--ASVVDFCVGDLTFPNGPA--GYACKKPSKVTADDFAYSGL 56
M ++L + L ++ ++ + + DFCV NG G CK P + A+DF SGL
Sbjct: 5 MSLILITFWALVTIAKAYDPSPLQDFCVAIDDPKNGVFVNGKFCKDPKQAKAEDFFSSGL 64
Query: 57 GIAGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQG 116
AG T+N +K+ VT Q PG+N LGISL R+D A G P HTHP A+E LV+V+G
Sbjct: 65 NQAGITNNKVKSNVTTVNVDQIPGLNTLGISLVRIDYAPYGQNPPHTHPRATEILVLVEG 124
Query: 117 TICAGFVAS---DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQI 173
T+ GFV+S +N ++ K L GDV VFP G++HFQ+N G + A+AF SS N G+
Sbjct: 125 TLYVGFVSSNQDNNRLFAKVLNPGDVFVFPIGMIHFQVNIGKTPAVAFAGLSSQNAGVIT 184
Query: 174 LDFALFKSDFP--TDLIAATTFLDAAQIKKLK 203
+ +F S P D++A LD +K L+
Sbjct: 185 IADTVFGSTPPINPDILAQAFQLDVNVVKDLE 216
>At5g61750 germin-like protein - like
Length = 210
Score = 137 bits (344), Expect = 5e-33
Identities = 76/181 (41%), Positives = 108/181 (58%), Gaps = 16/181 (8%)
Query: 1 MKMVLTILFILSLLSLSHAS--VVDFCVGDLTFPNGPA--------GYACKKPSKVTADD 50
MK + I+F LS+S S + D C P P GY CK P+K+TA D
Sbjct: 1 MKFFVVIVFCAIFLSVSGDSDNMQDTC------PTAPGEQSIFFINGYPCKNPTKITAQD 54
Query: 51 FAYSGLGIAGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEA 110
F + L AG+T N +++ VT +FPG+N LG+S++R DL G +P H+HP +SE
Sbjct: 55 FKSTKLTEAGDTDNYLQSNVTLLTALEFPGLNTLGLSVSRTDLERDGSVPFHSHPRSSEM 114
Query: 111 LVVVQGTICAGFVASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPG 170
L VV+G + AGFV ++N ++ L+KGDV VFP+GLLHF ++ G A AF ++S NPG
Sbjct: 115 LFVVKGVVFAGFVDTNNKIFQTVLQKGDVFVFPKGLLHFCLSGGFEPATAFSFYNSQNPG 174
Query: 171 L 171
+
Sbjct: 175 V 175
>At5g39150 germin - like protein
Length = 221
Score = 137 bits (344), Expect = 5e-33
Identities = 86/214 (40%), Positives = 124/214 (57%), Gaps = 11/214 (5%)
Query: 1 MKMVLTILFILSLLSLSHA----SVVDFCVGDLTFPNGPA--GYACKKPSKVTADDFAYS 54
+ M L ++ + +L++++ A + DFCV NG G CK P + A+DF S
Sbjct: 3 VSMSLILITLSALVTIAKAYDPSPLQDFCVAIDDPKNGVFVNGKFCKDPKQAKAEDFFSS 62
Query: 55 GLGIAGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVV 114
GL AG T+N +++ VT Q PG+N LGISL R+D A G P HTHP A+E LV+V
Sbjct: 63 GLNQAGITNNKVQSNVTTVNVDQIPGLNTLGISLVRIDYAPYGQNPPHTHPRATEILVLV 122
Query: 115 QGTICAGFVAS---DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGL 171
+GT+ GFV+S +N ++ K L GDV VFP G++HFQ+N G + A+AF SS N G+
Sbjct: 123 EGTLYVGFVSSNQDNNRLFAKVLNPGDVFVFPIGMIHFQVNIGKTPAVAFAGLSSQNAGV 182
Query: 172 QILDFALFKSDFP--TDLIAATTFLDAAQIKKLK 203
+ +F S P D++A LD +K L+
Sbjct: 183 ITIADTVFGSTPPINPDILAQAFQLDVNVVKDLE 216
>At1g09560 germin-like protein
Length = 219
Score = 137 bits (344), Expect = 5e-33
Identities = 82/196 (41%), Positives = 112/196 (56%), Gaps = 7/196 (3%)
Query: 5 LTILFILSLLSLSHAS------VVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGI 58
LT+L +L+ +S +S + D CV DL G+ CK + VT+ DF GL
Sbjct: 6 LTLLLLLTTVSFFISSSADPDMLQDLCVADLPSGIKINGFPCKDAATVTSADFFSQGLAK 65
Query: 59 AGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTI 118
G T+N A VT A PG+N LG+SL+R+D A GG+ P HTHP A+E + V++GT+
Sbjct: 66 PGLTNNTFGALVTGANVMTIPGLNTLGVSLSRIDYAPGGLNPPHTHPRATEVVFVLEGTL 125
Query: 119 CAGFVASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFAL 178
GF+ + N + ++LKKGDV FP+GL+HFQ N+G A +F+S PG Q L L
Sbjct: 126 DVGFLTTANKLISQSLKKGDVFAFPKGLVHFQKNNGDVPASVIAAFNSQLPGTQSLGATL 185
Query: 179 FKSDFPT-DLIAATTF 193
F S P D I A F
Sbjct: 186 FGSTPPVPDNILAQAF 201
>At5g39180 germin-like protein
Length = 221
Score = 136 bits (343), Expect = 7e-33
Identities = 86/214 (40%), Positives = 124/214 (57%), Gaps = 11/214 (5%)
Query: 1 MKMVLTILFILSLLSLSHA----SVVDFCVGDLTFPNGPA--GYACKKPSKVTADDFAYS 54
+ M L ++ + +L++++ A + DFCV NG G CK P + A+DF S
Sbjct: 3 VSMSLILITLSALVTIAKAYDPSPLQDFCVAIDDPKNGVFVNGKFCKDPKQAKAEDFFSS 62
Query: 55 GLGIAGNTSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVV 114
GL AG T+N +++ VT Q PG+N LGISL R+D A G P HTHP A+E LV+V
Sbjct: 63 GLNQAGITNNKVQSNVTTVNVDQIPGLNTLGISLVRIDYAPYGQNPPHTHPRATEILVLV 122
Query: 115 QGTICAGFVAS---DNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGL 171
+GT+ GFV+S +N ++ K L GDV VFP G++HFQ+N G + A+AF SS N G+
Sbjct: 123 EGTLYVGFVSSNQDNNRLFAKVLNPGDVFVFPIGMIHFQVNIGKTPAVAFAGLSSQNAGV 182
Query: 172 QILDFALFKSDFP--TDLIAATTFLDAAQIKKLK 203
+ +F S P D++A LD +K L+
Sbjct: 183 ITIADIVFGSTPPINPDILAQAFQLDVNVVKDLE 216
>At3g62020 germin-like protein (GLP10)
Length = 220
Score = 136 bits (343), Expect = 7e-33
Identities = 77/205 (37%), Positives = 118/205 (57%), Gaps = 10/205 (4%)
Query: 9 FILSLLSLSHA-------SVVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGN 61
F +LLSL+ ++ D CV D T G+ CK S +TA DF ++G+G
Sbjct: 8 FFFTLLSLNVIVLAYDPDTLQDLCVADRTSGIKVNGFTCKPESNITASDFFFAGIGKPAV 67
Query: 62 TSNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAG 121
+N + +AVT A + G+N LG+SLAR+D A GG+ P HTHP A+E + V++G + G
Sbjct: 68 VNNTVGSAVTGANVEKIAGLNTLGVSLARIDYAPGGLNPPHTHPRATEVIFVLEGELDVG 127
Query: 122 FVASDNTVYLKTLKKGDVMVFPQGLLHFQ-INDGGSSALAFVSFSSANPGLQILDFALFK 180
F+ + N ++ KT+KKG+V VFP+GL+H+Q ND A +F+S PG Q + LF
Sbjct: 128 FITTANKLFAKTVKKGEVFVFPRGLIHYQKNNDKAKPASVISAFNSQLPGTQSIAATLFT 187
Query: 181 SD--FPTDLIAATTFLDAAQIKKLK 203
+ P ++ T + +I+K+K
Sbjct: 188 ATPAIPDHVLTTTFQIGTKEIEKIK 212
>At5g26700 nectarin - like protein
Length = 213
Score = 135 bits (341), Expect = 1e-32
Identities = 77/201 (38%), Positives = 113/201 (55%), Gaps = 4/201 (1%)
Query: 3 MVLTILFILSLLSLSHASVVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNT 62
+V+T+LF+ S + D CV DL+ GY CK +++T +DF + GL T
Sbjct: 10 VVVTMLFVAMA---SAEMLQDVCVADLSNAVKVNGYTCKDSTQITPEDFYFKGLANIAAT 66
Query: 63 SNIIKAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGF 122
+ + VT A + PG+N LG+S++R+D A G+ P H HP ASE + V++G + GF
Sbjct: 67 NTSTGSVVTGANVEKLPGLNTLGLSMSRIDYAPNGLNPPHVHPRASEIIFVLEGQLYVGF 126
Query: 123 VASDNTVYLKTLKKGDVMVFPQGLLHFQINDGGSSALAFVSFSSANPGLQILDFALFKSD 182
V + + K L KGDV FP+GL+HFQ N S A +F S PG Q L +LF +
Sbjct: 127 VTTAGKLIAKNLNKGDVFTFPKGLIHFQKNIANSPASVLAAFDSQLPGTQSLVASLFGA- 185
Query: 183 FPTDLIAATTFLDAAQIKKLK 203
P D++A + L Q+KK+K
Sbjct: 186 LPDDILAKSFQLKHKQVKKIK 206
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,522,025
Number of Sequences: 26719
Number of extensions: 177339
Number of successful extensions: 433
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 355
Number of HSP's gapped (non-prelim): 44
length of query: 210
length of database: 11,318,596
effective HSP length: 95
effective length of query: 115
effective length of database: 8,780,291
effective search space: 1009733465
effective search space used: 1009733465
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0137.2


