

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0137.11
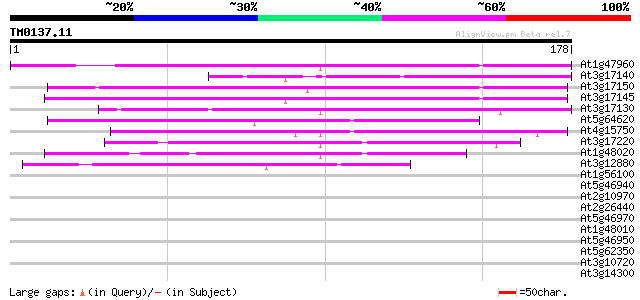
(178 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g47960 unknown protein 134 2e-32
At3g17140 hypothetical protein 79 1e-15
At3g17150 unknown protein 73 7e-14
At3g17145 unknown protein 70 6e-13
At3g17130 hypothetical protein 57 5e-09
At5g64620 invertase inhibitor homolog (emb|CAA73335.1) 54 3e-08
At4g15750 unknown protein 47 7e-06
At3g17220 unknown protein 45 2e-05
At1g48020 phosphatase-2C like protein 44 4e-05
At3g12880 unknown protein 43 1e-04
At1g56100 unknown protein 39 0.001
At5g46940 unknown protein 37 0.006
At2g10970 pseudogene 37 0.007
At2g26440 putative pectinesterase 36 0.010
At5g46970 unknown protein 35 0.021
At1g48010 hypothetical protein 35 0.028
At5g46950 unknown protein 33 0.081
At5g62350 ripening-related protein-like; contains similarity to ... 32 0.14
At3g10720 pectinesterase like protein 32 0.18
At3g14300 putative pectin methylesterase 32 0.24
>At1g47960 unknown protein
Length = 166
Score = 134 bits (338), Expect = 2e-32
Identities = 76/179 (42%), Positives = 106/179 (58%), Gaps = 14/179 (7%)
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPR 60
M+ +K + LI + +V+ +S + ++ TCK TP + C+ L +DPR
Sbjct: 1 MKMMKVMMLIVMMMMVMVMVS------------EGSIIEPTCKETPDFNLCVSLLNSDPR 48
Query: 61 SSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGD-KPALNSCQGRYNAILKADIPQA 119
SSAD +GLALI++D I+ LN+I+ L K + K AL+ C RY IL AD+P+A
Sbjct: 49 GSSADTSGLALILIDKIKGLATKTLNEINGLYKKRPELKRALDECSRRYKTILNADVPEA 108
Query: 120 TQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNLL 178
+A+ G PKF EDGV DAGVEA+ C+ GF +G SPLTS M + V RAI+R LL
Sbjct: 109 IEAISKGVPKFGEDGVIDAGVEASVCQGGF-NGSSPLTSLTKSMQKISNVTRAIVRMLL 166
>At3g17140 hypothetical protein
Length = 112
Score = 79.3 bits (194), Expect = 1e-15
Identities = 54/118 (45%), Positives = 70/118 (58%), Gaps = 10/118 (8%)
Query: 64 ADVTGLALIMVDVIQAKTNGVLN---KISQLLKGGGDKPALNSCQGRYNAILKADIPQAT 120
A V G+A+ + + A+T G +N K +LK KP L+ C RY I+ D+ A
Sbjct: 2 AMVVGVAMGNI-ALGAETVGQINEAYKTKPMLK----KP-LDECSMRYKTIVDVDVHTAI 55
Query: 121 QALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNLL 178
++K GNPKFAE V DAGVEA+ CE GF+ G+SPLTS M +V RAIIR LL
Sbjct: 56 ISIK-GNPKFAEGAVVDAGVEASICEGGFTKGQSPLTSLTQRMEKICDVTRAIIRMLL 112
>At3g17150 unknown protein
Length = 175
Score = 73.2 bits (178), Expect = 7e-14
Identities = 48/168 (28%), Positives = 78/168 (45%), Gaps = 5/168 (2%)
Query: 13 IFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALI 72
IF+V++ I +++ + P + LV + CK Y + C+ L DPRS ++++ GLA I
Sbjct: 10 IFVVLSQIQIALSQT-IQSPPGSSLVQRLCKRNRYQALCISTLNVDPRSKTSNLQGLASI 68
Query: 73 MVDVIQAKTNGVLNKISQLLK---GGGDKPALNSCQGRYNAILKADIPQATQALKTGNPK 129
+D K N L +LK G D +C Y A + +P LK
Sbjct: 69 SLDATTKKFNVTLTYYISVLKNVRGRVDFERYGTCIEEYGAAVDRFLPAVKADLKAKKYP 128
Query: 130 FAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNL 177
A + D + CE F +G+SP+T+ N +H ++ II+ L
Sbjct: 129 EAMSEMKDVVAKPGYCEDQF-AGESPVTARNKAVHDIADMTADIIKTL 175
>At3g17145 unknown protein
Length = 177
Score = 70.1 bits (170), Expect = 6e-13
Identities = 47/169 (27%), Positives = 76/169 (44%), Gaps = 4/169 (2%)
Query: 12 NIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLAL 71
+IF+V I +++ + ++ + + + CK PS C+ L DPRS ++++ LA
Sbjct: 6 SIFVVFMQIQVALSSQPIRYATEPKPIQELCKFNINPSLCVSTLNLDPRSKNSNLRELAW 65
Query: 72 IMVDVIQAKTNGVLN---KISQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGNP 128
I +D K N +LN +S+ +K D +C Y + +P A LK G
Sbjct: 66 ISIDATSNKVNKMLNYLISVSKNIKDREDLKKYKTCIDDYGTAARRFLPAALDDLKAGFF 125
Query: 129 KFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNL 177
A+ + + CE+ F G SPLT N H + IIR L
Sbjct: 126 SLAKSDMESVVSIPDHCEAQF-GGSSPLTGRNKATHDIANMTADIIRYL 173
>At3g17130 hypothetical protein
Length = 183
Score = 57.0 bits (136), Expect = 5e-09
Identities = 44/159 (27%), Positives = 76/159 (47%), Gaps = 11/159 (6%)
Query: 29 VLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKI 88
V+Q SD L+ K C+ TP+ C L+ S S D L M V+ L I
Sbjct: 24 VVQSSD-DLIDKICQATPFCDLCEASLRPFSPSPS-DPKSLGAAMASVVLGNMTDTLGYI 81
Query: 89 SQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
L+K D + AL C Y ++K +IPQA +A++ G FA + DA + ++C
Sbjct: 82 QSLIKHAHDPAAERALAQCAELYRPVVKFNIPQAMEAMQGGKFGFAIYVLGDAEKQTDSC 141
Query: 146 ESGFSSGKS------PLTSENNGMHIATEVARAIIRNLL 178
+ G ++ + +T+ N + +VA +++++L+
Sbjct: 142 QKGITNAGADDESSVAVTARNKLVKNLCDVAISVLKSLM 180
>At5g64620 invertase inhibitor homolog (emb|CAA73335.1)
Length = 180
Score = 54.3 bits (129), Expect = 3e-08
Identities = 40/141 (28%), Positives = 62/141 (43%), Gaps = 5/141 (3%)
Query: 13 IFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALI 72
IF+++ T++ S + + ++ TCK T Y C+ L++DPRS +AD GLA I
Sbjct: 6 IFLLLVTLTFSASTLISAKSNTTTIIESTCKTTNYYKFCVSALKSDPRSPTADTKGLASI 65
Query: 73 MVDV----IQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGNP 128
MV V + N + +S +K K L C +Y A+ + Q L
Sbjct: 66 MVGVGMTNATSTANYIAGNLSATVKDTVLKKVLQDCSEKY-ALAADSLRLTIQDLDDEAY 124
Query: 129 KFAEDGVADAGVEANTCESGF 149
+A V A N C + F
Sbjct: 125 DYASMHVLAAQDYPNVCRNIF 145
>At4g15750 unknown protein
Length = 171
Score = 46.6 bits (109), Expect = 7e-06
Identities = 43/153 (28%), Positives = 73/153 (47%), Gaps = 9/153 (5%)
Query: 33 SDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKIS--Q 90
+D +L+ C +T Y + CL L+ADP S + D GL I++ + ++ + ++N ++ +
Sbjct: 20 ADEELIKTECNHTEYQNVCLFCLEADPISFNIDRAGLVNIIIHCLGSQLDVLINTVTSLK 79
Query: 91 LLKGGGD--KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESG 148
L+KG G+ + L C + AI + + A L T N A + V A TCE
Sbjct: 80 LMKGEGEANENVLKDCVTGF-AIAQLRLQGANIDLITLNYDKAYELVKTALNYPRTCEEN 138
Query: 149 FSSGKSPLTSENNGMHIA----TEVARAIIRNL 177
K +S+ +A T VA+ +I L
Sbjct: 139 LQKLKFKDSSDVYDDILAYSQLTSVAKTLIHRL 171
>At3g17220 unknown protein
Length = 173
Score = 45.1 bits (105), Expect = 2e-05
Identities = 36/136 (26%), Positives = 63/136 (45%), Gaps = 8/136 (5%)
Query: 31 QPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQ 90
Q +D + + KN + C +++++P++S AD+ LA I Q + KI
Sbjct: 26 QVADIKAICGKAKNQSF---CTSYMKSNPKTSGADLQTLANITFGSAQTSASEGFRKIQS 82
Query: 91 LLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCES 147
L+K + K A SC Y + + + + A Q+L +G+ K V+ A +TCE
Sbjct: 83 LVKTATNPTMKKAYTSCVQHYKSAI-SSLNDAKQSLASGDGKGLNIKVSAAMEGPSTCEQ 141
Query: 148 GFSSGK-SPLTSENNG 162
+ K P +N+G
Sbjct: 142 DMADFKVDPSAVKNSG 157
>At1g48020 phosphatase-2C like protein
Length = 176
Score = 44.3 bits (103), Expect = 4e-05
Identities = 37/137 (27%), Positives = 62/137 (45%), Gaps = 9/137 (6%)
Query: 12 NIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLAL 71
N F+ + IG + S+ + C T PS CL+FL + +S ++ LA
Sbjct: 8 NAFLSSLMFLLLIGSSYAITSSEMSTI---CDKTLNPSFCLKFLNT--KFASPNLQALAK 62
Query: 72 IMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNP 128
+D QA+ L K+ ++ GG D K A SC Y + + ++ +A + L +G+
Sbjct: 63 TTLDSTQARATQTLKKLQSIIDGGVDPRSKLAYRSCVDEYESAI-GNLEEAFEHLASGDG 121
Query: 129 KFAEDGVADAGVEANTC 145
V+ A A+TC
Sbjct: 122 MGMNMKVSAALDGADTC 138
>At3g12880 unknown protein
Length = 179
Score = 42.7 bits (99), Expect = 1e-04
Identities = 33/127 (25%), Positives = 58/127 (44%), Gaps = 9/127 (7%)
Query: 5 KSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSA 64
K ++L+ + I+ ++ V+ ++ +L+ + C N+ P+ CL+ L +DP S A
Sbjct: 3 KKMALLAKLLILTTLVTT----ISVISRANEELMMQQCHNSDNPTLCLRCLSSDPGSHEA 58
Query: 65 DVTGLALIMVDVIQAK----TNGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQAT 120
D GLA I++ I + T SQ + AL C G + K + +A
Sbjct: 59 DKVGLARIILKCINSHLLVLTKNASTLGSQHYQNPNTAAALKQC-GLGFSTAKHGVGEAD 117
Query: 121 QALKTGN 127
L TG+
Sbjct: 118 TQLITGD 124
>At1g56100 unknown protein
Length = 232
Score = 38.9 bits (89), Expect = 0.001
Identities = 17/62 (27%), Positives = 35/62 (56%)
Query: 33 SDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLL 92
++ L+ + C N P+ C+Q L++DP S AD G+A I++ + ++ + + + +L
Sbjct: 20 AEKDLMKEECHNAQVPTICMQCLESDPTSVHADRVGIAEIIIHCLDSRLDIITKQKGELQ 79
Query: 93 KG 94
G
Sbjct: 80 IG 81
>At5g46940 unknown protein
Length = 176
Score = 37.0 bits (84), Expect = 0.006
Identities = 32/141 (22%), Positives = 61/141 (42%), Gaps = 14/141 (9%)
Query: 37 LVAKTCKNTPYPSA------CLQFLQADPRSSSA-DVTGLALIMVDVIQAKTNGVLNKIS 89
L+ +CK S C+ L+ +P++ +A D+ GL + K + +
Sbjct: 23 LIRNSCKKATATSPKFKYNLCVTSLETNPQAKTAKDLAGLVMASTKNAVTKATTLKGTVD 82
Query: 90 QLLKGGGDKPA----LNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
+++KG L C Y + + +A +K+ N + ++ A +TC
Sbjct: 83 KIIKGKKVNKMTAMPLRDCLQLYTDAI-GSLNEALAGVKSRNYPTVKTVLSAAMDTPSTC 141
Query: 146 ESGFSSGK--SPLTSENNGMH 164
E+GF K SP+T EN+ ++
Sbjct: 142 ETGFKERKAPSPVTKENDNLY 162
>At2g10970 pseudogene
Length = 194
Score = 36.6 bits (83), Expect = 0.007
Identities = 29/120 (24%), Positives = 55/120 (45%), Gaps = 12/120 (10%)
Query: 32 PSDAQLVAKTCKNTPYPSA----CLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNK 87
P + L++K CK T S+ C++ L D R+ SA ++++ +A + + K
Sbjct: 45 PYPSLLISKACKGTASFSSLEEECIKSLTLDHRTKSAST------VLELAKAALSLAMEK 98
Query: 88 I--SQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
+Q L G KP SC Y + + +A +++ G+ +D ++ A A+ C
Sbjct: 99 AEHTQFLIGSPKKPCFKSCMENYKDSVVEGLMKAQRSMGNGDVDETDDDLSLARDAADYC 158
>At2g26440 putative pectinesterase
Length = 547
Score = 36.2 bits (82), Expect = 0.010
Identities = 42/169 (24%), Positives = 72/169 (41%), Gaps = 23/169 (13%)
Query: 4 LKSLSLICNIFIVVATISM-SIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSS 62
L S +L +F++ T S+ S + L P + + CKNTPYP AC L+ S
Sbjct: 3 LSSFNLSSLLFLLFFTPSVFSYSYQPSLNPHETSATS-FCKNTPYPDACFTSLKL---SI 58
Query: 63 SADVT-GLALIMVDVIQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKA-DIPQAT 120
S +++ + ++ +Q + K++ LL G G + N +G+ ++ D+ T
Sbjct: 59 SINISPNILSFLLQTLQTALSEA-GKLTDLLSGAG--VSNNLVEGQRGSLQDCKDLHHIT 115
Query: 121 QALKTGNPKFAEDGVADAGVEAN-------------TCESGFSSGKSPL 156
+ + +DGV D+ A+ TC G S PL
Sbjct: 116 SSFLKRSISKIQDGVNDSRKLADARAYLSAALTNKITCLEGLESASGPL 164
>At5g46970 unknown protein
Length = 164
Score = 35.0 bits (79), Expect = 0.021
Identities = 29/114 (25%), Positives = 54/114 (46%), Gaps = 8/114 (7%)
Query: 51 CLQFLQADPRSSSA-DVTGLALIMVDVIQAKTNGVLNKISQLLKGG-----GDKPALNSC 104
C++ L+ +P+S +A + L ++ +KT + + ++LK +KP L C
Sbjct: 43 CVKSLEENPQSKTARSLDRLVMLSTKNAVSKTTSMKGIVDKILKENRFEMYSEKP-LRDC 101
Query: 105 QGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTS 158
Y+ + + +A +K+ + K A ++ A +CE GF GK PL S
Sbjct: 102 LELYSDATNS-LKEALTIIKSRDYKTANVVISAAMGAPPSCEIGFKEGKKPLKS 154
>At1g48010 hypothetical protein
Length = 178
Score = 34.7 bits (78), Expect = 0.028
Identities = 24/99 (24%), Positives = 42/99 (42%), Gaps = 4/99 (4%)
Query: 52 LQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRY 108
++F DP+ D+ G+A ++ Q N++ L D K + SC Y
Sbjct: 44 MEFSALDPKIGKLDLPGVAKCLIHYAQQNALDTHNQLQLLANSTTDNRTKESYISCSKYY 103
Query: 109 NAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCES 147
+ L + +A + LK NP + + A AN C++
Sbjct: 104 DKALYS-FDEALKDLKVNNPDYLNIEITAARQNANDCKT 141
>At5g46950 unknown protein
Length = 174
Score = 33.1 bits (74), Expect = 0.081
Identities = 33/117 (28%), Positives = 50/117 (42%), Gaps = 7/117 (5%)
Query: 51 CLQFLQADPRSSSAD-VTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKPA---LNSCQG 106
C+Q L DP+S +A + LAL + AK + ++Q LK + L C G
Sbjct: 43 CVQSLTQDPQSKAATTLEDLALASTKNVAAKITNLKGIVAQDLKDQRYQDIVEDLKLCLG 102
Query: 107 RYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSG--KSPLTSENN 161
Y + A +K+ + A ++ A CE F KSP+T+ENN
Sbjct: 103 FYKDA-NDSLKTALANIKSRDYDGANSNLSAALNAPGDCEDDFKEAEKKSPITNENN 158
>At5g62350 ripening-related protein-like; contains similarity to
pectinesterase (MMI9.18)
Length = 202
Score = 32.3 bits (72), Expect = 0.14
Identities = 36/162 (22%), Positives = 62/162 (38%), Gaps = 20/162 (12%)
Query: 4 LKSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSS 63
L S+S + ++ + AT + G + + +CK T YP+ C+ L
Sbjct: 9 LLSISYLLSLELTAATAASQTGASK----KAINFIQSSCKTTTYPALCVHSLSVYANDIQ 64
Query: 64 ADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDK----PALNSCQGRYNAILKADIPQA 119
LA + V ++ +S+L + G K A+ C N + + ++
Sbjct: 65 TSPKRLAETAIAVTLSRAQSTKLFVSRLTRMKGLKKREVEAIKDCVEEMNDTVDR-LTKS 123
Query: 120 TQALK-TGNPK----------FAEDGVADAGVEANTCESGFS 150
Q LK G+ K A+ + A + NTC GFS
Sbjct: 124 VQELKLCGSAKDQDQFAYHMSNAQTWTSAALTDENTCSDGFS 165
>At3g10720 pectinesterase like protein
Length = 619
Score = 32.0 bits (71), Expect = 0.18
Identities = 15/34 (44%), Positives = 18/34 (52%)
Query: 30 LQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSS 63
L PS +Q + CK+TPYP C L A S S
Sbjct: 71 LPPSQSQSPSLACKSTPYPKLCRTILNAVKSSPS 104
>At3g14300 putative pectin methylesterase
Length = 968
Score = 31.6 bits (70), Expect = 0.24
Identities = 25/119 (21%), Positives = 49/119 (41%), Gaps = 15/119 (12%)
Query: 6 SLSLICNIFIVVATISMSIGHCRVLQPSD------------AQLVAKTCKNTPYPSACLQ 53
SL+++ I ++ + I R+L S A + C T YP++C+
Sbjct: 227 SLAIVAKILSTISDFGIPIHGRRLLNSSPHATPISVPKLTPAASLRNVCSVTRYPASCVS 286
Query: 54 FLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRYN 109
+ P S++ D L + + V+ + N + +L + D K +L+ C +N
Sbjct: 287 SISKLPSSNTTDPEALFRLSLQVVINELNSIAGLPKKLAEETDDERLKSSLSVCGDVFN 345
Score = 27.3 bits (59), Expect = 4.4
Identities = 19/77 (24%), Positives = 37/77 (47%), Gaps = 4/77 (5%)
Query: 32 PSDAQLVAKTCKNTPYPSACLQFLQADPRS-SSADVTGLALIMVDVIQAKTN---GVLNK 87
P+ + ++ C T YP++C+ + P S ++ D L + + V + N G+ K
Sbjct: 453 PTPSSVLRTVCNVTNYPASCISSISKLPLSKTTTDPKVLFRLSLQVTFDELNSIVGLPKK 512
Query: 88 ISQLLKGGGDKPALNSC 104
+++ G K AL+ C
Sbjct: 513 LAEETNDEGLKSALSVC 529
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,641,261
Number of Sequences: 26719
Number of extensions: 134540
Number of successful extensions: 381
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 353
Number of HSP's gapped (non-prelim): 39
length of query: 178
length of database: 11,318,596
effective HSP length: 93
effective length of query: 85
effective length of database: 8,833,729
effective search space: 750866965
effective search space used: 750866965
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0137.11


