

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0137.1
(630 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
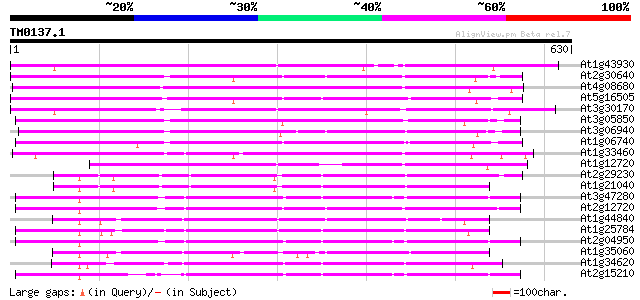
Score E
Sequences producing significant alignments: (bits) Value
At1g43930 hypothetical protein 264 1e-70
At2g30640 Mutator-like transposase 262 5e-70
At4g08680 putative MuDR-A-like transposon protein 255 6e-68
At5g16505 mutator-like transposase-like protein (MQK4.25) 249 3e-66
At3g30170 unknown protein 243 3e-64
At3g05850 unknown protein 242 4e-64
At3g06940 putative mudrA protein 216 2e-56
At1g06740 mudrA-like protein 215 5e-56
At1g33460 mutator transposase MUDRA, putative 215 7e-56
At1g12720 hypothetical protein 210 2e-54
At2g29230 Mutator-like transposase 202 4e-52
At1g21040 hypothetical protein 191 1e-48
At3g47280 putative protein 183 2e-46
At2g12720 putative protein 183 3e-46
At1g44840 hypothetical protein 182 5e-46
At1g25784 mutator-like transposase, putative 181 1e-45
At2g04950 putative transposase of transposon FARE2.9 (cds1) 180 2e-45
At1g35060 hypothetical protein 180 2e-45
At1g34620 Mutator-like protein 180 2e-45
At2g15210 putaive transposase of transposon FARE2.7 (cds1) 173 2e-43
>At1g43930 hypothetical protein
Length = 946
Score = 264 bits (674), Expect = 1e-70
Identities = 180/635 (28%), Positives = 292/635 (45%), Gaps = 30/635 (4%)
Query: 2 EFKFKLGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRR---RCGFLIFLSK 58
E +L F + + F+ A+ +S++ ++K ++ +RV A C C + + S
Sbjct: 234 EESLRLAKTFNNPEDFKIAVLRYSLKTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSY 293
Query: 59 VGGKHTYSIKTLIPEHTCSRVYNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYS 118
K +K H+C R ++ +A +E++ K E+VDE++ Y
Sbjct: 294 DKKKQKMQVKVYKDGHSCVRSGYSKMLKQRTIAWLFRERLRKNPKITKQEMVDEIKREYK 353
Query: 119 TGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQP 178
+T + +A+ + + + ++ + A++ R++PG +++ + S
Sbjct: 354 LTVTEDQCSKAKTMVMRERQATHQEHFARIWDYQAKIFRSNPGTTFEIETILGPTIGSLQ 413
Query: 179 RFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVE 238
RF R +IC + K + CRP IG+DG LK G LL A GRD +++ PIA+AVVE
Sbjct: 414 RFLRLFICFKSQKESWKQTCRPIIGIDGAFLKWDIKGHLLAATGRDGDNRIVPIAWAVVE 473
Query: 239 SECKESWKWFLELL--LADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANF 296
E ++W WF+ +L D+ R ISD+Q GL+ ++P EHR C RH+ N+
Sbjct: 474 IENDDNWDWFVRMLSRTLDLQDGRNVAIISDKQSGLVKAIHSVIPQAEHRQCARHIMENW 533
Query: 297 KKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLY 356
K+ + ++ L + ++ F MR LK N A + L+ +W + F +
Sbjct: 534 KRN-SHDMELQRLFWKIVRSYTEGEFGAHMRALKSYNASAFELLLKTLPVTWSRAFFKIG 592
Query: 357 PKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGR-------FAALNEKFQRYNH 409
C+ +NNLSE+FN TI AR KP+L M++ IR M R L +F + H
Sbjct: 593 SCCNDNLNNLSESFNRTIREARRKPLLDMLEDIRRQCMVRNEKRYVIADRLRTRFTKRAH 652
Query: 410 PFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGI 469
++++ ++ W+ + H K+E+K V D F VD+ +C C W + GI
Sbjct: 653 ----AEIEKMIAGSQVCQRWMAR---HNKHEIK-VGPVDSFTVDMNNNTCGCMKWQMTGI 704
Query: 470 PCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWPYTPDAPILPPLYK 529
PC HA + I E Y+ +Y ++ TY GI P+ GK +WP +LPP ++
Sbjct: 705 PCIHAASVIIGKRQKVEDYVSDWYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWR 764
Query: 530 R-KPGRPKKLRRR-------DPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTNPPTSQP 581
R PGRPK RR T E SR + C C GHN + C N + P
Sbjct: 765 RGNPGRPKNHARRKGVFESSTASSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPP 824
Query: 582 TEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQP 616
+ P P + P Q Q+P P
Sbjct: 825 AKR-PRGRPRKDKDPRMVFFGQGQTWQEPQAQGAP 858
>At2g30640 Mutator-like transposase
Length = 754
Score = 262 bits (669), Expect = 5e-70
Identities = 172/592 (29%), Positives = 286/592 (48%), Gaps = 32/592 (5%)
Query: 2 EFKFKLGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRRR-CGFLIFLSKVG 60
E +GMEF+ + R+ALRD ++ E++ +K+D R A C C + I +K+
Sbjct: 177 EHNLIVGMEFSDVLACRRALRDAAIALRFEMQTVKSDKTRFTAKCNSEGCPWRIHCAKLP 236
Query: 61 GKHTYSIKTLIPEHTCSRVYN--NRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYS 118
G T++I+T+ HTC + + + A+ +VA + E++ + + E+++++ +
Sbjct: 237 GLPTFTIRTIHGSHTCGGISHLGHHQASVQWVADAVTERLKVNPHCKPKEILEQIHQVHG 296
Query: 119 TGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQP 178
+T +AWR +++ +A V G Y L R+ +++R +PG+ H P
Sbjct: 297 ITLTYKQAWRGKERIMAAVRGSYEEDYRLLPRYCDQIRRTNPGSVAVVHGSPVDGS---- 352
Query: 179 RFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVE 238
F +++I + GF+ CRP IGLD LK++Y G LL A G D FP+AFA++
Sbjct: 353 -FQQFFISFQASICGFLNACRPLIGLDRTVLKSKYVGTLLLATGFDGEGAVFPLAFAIIS 411
Query: 239 SECKESWKWFL----ELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYA 294
E SW+WFL +LL + + + +S + + ++ D P H C+ L
Sbjct: 412 EENDSSWQWFLSELRQLLEVNSENMPKLTILSSRDQSIVDGVDTNFPTAFHGLCVHCLTE 471
Query: 295 NFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFT 354
+ + +F +++ +L+ +AAK FE KM E+ +++ EA W+ W + F
Sbjct: 472 SVRTQFNNSILV-NLVWEAAKCLTDFEFEGKMGEIAQISPEAASWIRNIQHSQWATYCFE 530
Query: 355 LYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPK 414
+ L N+SE+ N+ + A PI+ M++ IR LM F E +++ +P
Sbjct: 531 -GTRFGHLTANVSESLNSWVQDASGLPIIQMLESIRRQLMTLFNERRETSMQWSGMLVPS 589
Query: 415 PLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHA 474
+ + + + A++EV + ++IVD+R R+C C W+L G+PC HA
Sbjct: 590 AERHVLEAIEECRLYPVHKANEAQFEV--MTSEGKWIVDIRCRTCYCRGWELYGLPCSHA 647
Query: 475 VAAISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWPYTPDA----------PIL 524
VAA+ + + +SY+ Y+ TYA I P+ K W T A I
Sbjct: 648 VAALLACRQNVYRFTESYFTVANYRRTYAETIHPVPDKTEWKTTEPAGESGEDGEEIVIR 707
Query: 525 PPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTNP 576
PP R+P RPKK RR ED R RC RC GH TCT P
Sbjct: 708 PPRDLRQPPRPKK--RRSQGED----RGRQKRVVRCSRCNQAGHFRTTCTAP 753
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 255 bits (651), Expect = 6e-68
Identities = 159/594 (26%), Positives = 286/594 (47%), Gaps = 24/594 (4%)
Query: 4 KFKLGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRR--RCGFLIFLSKVGG 61
KF +G F + +F++A+ + ++++G + + + ++ C +C + ++ S
Sbjct: 108 KFVIGSTFFTGIEFKEAILEVTLKHGHNVVQDRWEKEKISFKCGMGGKCKWRVYCSYDPD 167
Query: 62 KHTYSIKTLIPEHTCSRVYNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYSTGI 121
+ + +KT H+CS + S +A +K+ + K+ ++ + ++ +
Sbjct: 168 RQLFVVKTSCTWHSCSPNGKCQILKSPVIARLFLDKLRLNEKFMPMDIQEYIKERWKMVS 227
Query: 122 TTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQPRFN 181
T + R R AL +++ + Q+ + + E+ +PG+ + + FN
Sbjct: 228 TIPQCQRGRLLALKMLKKEYEEQFAHIRGYVEEIHSQNPGSVAFIDTY--RNEKGEDVFN 285
Query: 182 RYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESEC 241
R+Y+C + + CRP IGLDG LK G+LL AVG DPN+Q +PIA+AVV+SE
Sbjct: 286 RFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGVLLTAVGHDPNNQIYPIAWAVVQSEN 345
Query: 242 KESWKWFLELLLADIG--GLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKK 299
E+W WF++ + D+ R++ +SD+ KGLLS + +P EHR C++H+ N KK
Sbjct: 346 AENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLSAVKQELPNAEHRMCVKHIVENLKKN 405
Query: 300 FGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKC 359
+++ L+ + A + + + + L+ +E ++ ++ +W + + L C
Sbjct: 406 HAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDEALYNDVLNEEPHTWSRCFYKLGSCC 465
Query: 360 DVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRL 419
+ + NN +E+FN+TI AR K ++ M++ IR M R N+K R+ F LK L
Sbjct: 466 EDVDNNATESFNSTITKARAKSLIPMLETIRRQGMTRIVKRNKKSLRHEGRFTKYALKML 525
Query: 420 AWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAIS 479
A E ++ C H +EV +D + VD+ + C+C W + GIPC H+ A+
Sbjct: 526 ALEKTDADRSKVYRCTHGVFEV--YIDENGHRVDIPKTQCSCGKWQISGIPCEHSYGAMI 583
Query: 480 FNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMW-------PYTPDAPILPPLYKRKP 532
G AE+YI ++ D ++ +Y P+ G + W P PILP K K
Sbjct: 584 EAGLDAENYISEFFSTDLWRDSYETATMPLRGPKYWLNSSYGLVTAPPEPILPGRKKEKS 643
Query: 533 GRPKKLRRRDPHEDDTDEGSRPNSRHRCGR---------CGTIGHNIRTCTNPP 577
+ K R + +E + + N + G+ CG GHN C P
Sbjct: 644 KKEKFARIKGKNESPKKKKRKKNEVEKLGKKGKIIHCKSCGEAGHNALRCKKFP 697
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 249 bits (636), Expect = 3e-66
Identities = 157/590 (26%), Positives = 279/590 (46%), Gaps = 30/590 (5%)
Query: 2 EFKFKLGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRRR-CGFLIFLSKVG 60
E +G EF ++ R+ L+D ++ +L+ +K+D R A C + C + I +K
Sbjct: 22 EHVLAVGKEFPDVETCRRTLKDLAIALHFDLRIVKSDRSRFIAKCSKEGCPWRIHAAKCP 81
Query: 61 GKHTYSIKTLIPEHTCSRV--YNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYS 118
G T++++TL EHTC V +++ A+ +VA ++ ++ +Y+ E++ ++R +
Sbjct: 82 GVQTFTVRTLNSEHTCEGVRDLHHQQASVGWVARSVEARIRDNPQYKPKEILQDIRDEHG 141
Query: 119 TGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQP 178
++ +AWR +++++A + G Y L + ++K +PG+ L P +
Sbjct: 142 VAVSYMQAWRGKERSMAALHGTYEEGYRFLPAYCEQIKLVNPGSFASVSALGP-----EN 196
Query: 179 RFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVE 238
F R +I C GF + CRP + LD HLK +Y G +LCA D +D FP+A A+V+
Sbjct: 197 CFQRLFIAYRACISGFFSSCRPLLELDRAHLKGKYLGAILCAAAVDADDGLFPLAIAIVD 256
Query: 239 SECKESWKWFL----ELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYA 294
+E E+W WFL +LL + + + +S++Q ++ + P H FCLR++
Sbjct: 257 NESDENWSWFLSELRKLLGMNTDSMPKLTILSERQSAVVEAVETHFPTAFHGFCLRYVSE 316
Query: 295 NFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFT 354
NF+ F ++ ++ A A FE K E+ E++++ W + W A+
Sbjct: 317 NFRDTFKNTKLV-NIFWSAVYALTPAEFETKSNEMIEISQDVVQWFELYLPHLWAV-AYF 374
Query: 355 LYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPK 414
+ ++E L + PI+ M++ IR + F E +N +P
Sbjct: 375 QGVRYGHFGLGITEVLYNWALECHELPIIQMMEHIRHQISSWFDNRRELSMGWNSILVPS 434
Query: 415 PLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHA 474
+R+ + + ++E+ S + IVD+R R C+C W + G+PC HA
Sbjct: 435 AERRITEAVADARCYQVLRANEVEFEIVSTERTN--IVDIRTRDCSCRRWQIYGLPCAHA 492
Query: 475 VAAISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWPYTPDAP--------ILPP 526
AA+ G + + + + +YQ TY+ I I + +W + I PP
Sbjct: 493 AAALISCGRNVHLFAEPCFTVSSYQQTYSQMIGEIPDRSLWKDEGEEAGGGVESLRIRPP 552
Query: 527 LYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTNP 576
+R PGRPKK R + RP +CGRC +GH+ + CT P
Sbjct: 553 KTRRPPGRPKKKVVR------VENLKRPKRIVQCGRCHLLGHSQKKCTQP 596
>At3g30170 unknown protein
Length = 995
Score = 243 bits (619), Expect = 3e-64
Identities = 176/633 (27%), Positives = 292/633 (45%), Gaps = 51/633 (8%)
Query: 2 EFKFKLGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRR------RCGFLIF 55
E +L + + F+ A+ +S++ ++K +++ V A C +C + ++
Sbjct: 223 EVLLELKKTYNTPDDFKLAVLRYSLKTRYDIKLFRSEARIVAAKCSYVDKDGVQCPWKVY 282
Query: 56 LSKVGGKHTYSIKTLIPEHTCSRVYNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRI 115
S H I+T + H C R +++ +A +E++ + K E+ E+
Sbjct: 283 CSYEKTMHKMQIRTYVNNHICVRSGHSKMLKRSSIAYLFEERLRVNPKLTKYEMAAEILR 342
Query: 116 HYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQI 175
Y+ +T + +A+ K + + + ++ + AE+ + +P ++ + TA
Sbjct: 343 EYNLEVTPDQCAKAKTKVVKARNASHEAHFARVWDYQAEVIKQNPWTEFE---IETTA-- 397
Query: 176 SQPRFNRYYICLEGCKRGFVAGC--RPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIA 233
G V G RPFIGLDG LK G LL AVGRD +++ PIA
Sbjct: 398 -----------------GAVIGAKQRPFIGLDGAFLKWDIKGHLLAAVGRDGDNKIVPIA 440
Query: 234 FAVVESECKESWKWFLELLLADIG--GLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRH 291
+AVVE E ++W WFL L A +G + +SD+Q GL+ ++P EHR C +H
Sbjct: 441 WAVVEIENDDNWDWFLRHLSASLGLCQMANIAILSDKQSGLVKAIHTILPHAEHRQCSKH 500
Query: 292 LYANFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKH 351
+ N+K+ + ++ L + A++ V F M EL++ N +A++ L +W +
Sbjct: 501 IMDNWKRD-SHDLELQRLFWKIARSYTVEEFNNHMMELQQYNRQAYESLQLTSPVTWSRA 559
Query: 352 AFTLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAA---LNEKFQRYN 408
F + C+ +NNLSE+FN TI AR KPIL M++ IR M R A + EK Q
Sbjct: 560 FFRIGTCCNDNLNNLSESFNRTIRQARRKPILDMLEDIRRQCMVRSAKRFLIAEKLQTRF 619
Query: 409 HPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVG 468
+ ++++ + ++ + H Y ++ + VD+ Q++C C +VG
Sbjct: 620 TKRAHEEIEKMIVGLRQCERYMARENLHEIY-----VNNVSYFVDMEQKTCDCRKCQMVG 674
Query: 469 IPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWPYTPDAPILPPLY 528
IPC HA I E Y+ YY + ++ TY GI P+ G +W T P+LPP +
Sbjct: 675 IPCIHATCVIIGKKEKVEDYVSDYYTKVRWRETYLKGIRPVQGMPLWCRTNRLPVLPPPW 734
Query: 529 KR-KPGRPKKLRRRDPHEDDTDEGSRPNSRHR------CGRCGTIGHNIRTCTNPPTSQP 581
+R GRP RR ++ S PN R C C GHN + C P P
Sbjct: 735 RRGNAGRPSNYARR-KGRNEAAAPSNPNKMSREKRIMTCSNCHQEGHNKQRCNIPTVLNP 793
Query: 582 TEDVPETEPTEAAQP-PQQPTEAAQPPQQPTEA 613
T+ P P + QP Q ++A Q P+ A
Sbjct: 794 TKR-PRGRPRKNQQPQSHQGSQADLESQGPSAA 825
>At3g05850 unknown protein
Length = 777
Score = 242 bits (618), Expect = 4e-64
Identities = 161/583 (27%), Positives = 276/583 (46%), Gaps = 34/583 (5%)
Query: 7 LGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRRR-CGFLIFLSKVGGKHTY 65
+G F ++ +FR+ALR +++ N ++ KND RV C+ C + I S++
Sbjct: 206 VGQRFKNVGEFREALRKYAIANQFGFRYKKNDSHRVTVKCKAEGCPWRIHASRLSTTQLI 265
Query: 66 SIKTLIPEHTCSRV--YNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYSTGITT 123
IK + P HTC N + +VAS +KEK+ + Y+ ++V +++ Y +
Sbjct: 266 CIKKMNPTHTCEGAGGINGLQTSRSWVASIIKEKLKVFPNYKPKDIVSDIKEEYGIQLNY 325
Query: 124 WRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQPRFNRY 183
++AWR ++ A ++G Y L F ++ +PG+ F ++ F+R
Sbjct: 326 FQAWRGKEIAREQLQGSYKDGYKQLPLFCEKIMETNPGSLATFTTKEDSS------FHRV 379
Query: 184 YICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESECKE 243
++ GF+ CRP + LD LK++Y G LL A D +D+ FP+AFAVV++E +
Sbjct: 380 FVSFHASVHGFLEACRPLVFLDSMQLKSKYQGTLLAATSVDGDDEVFPLAFAVVDAETDD 439
Query: 244 SWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKKFGGG 303
+W+WFL L + + F++D+QK L ++ H +CLR+L K G
Sbjct: 440 NWEWFLLQLRSLLSTPCYITFVADRQKNLQESIPKVFEKSFHAYCLRYLTDELIKDLKGP 499
Query: 304 V------VIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYP 357
+I D AA A FE + +K ++ EA+DW++ + +A+
Sbjct: 500 FSHEIKRLIVDDFYSAAYAPRADSFERHVENIKGLSPEAYDWIVQKSQPDHWANAYFRGA 559
Query: 358 KCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLK 417
+ + + ++ E F + A D PI M+D IR +MG N P
Sbjct: 560 RYNHMTSHSGEPFFSWASDANDLPITQMVDVIRGKIMGLIHVRRISANEANGNLTPSMEV 619
Query: 418 RLAWET-KSSNNWVPQACGHAKYEVKSVLDGDQF-IVDLRQRSCTCNWWDLVGIPCRHAV 475
+L E+ ++ V + + ++V+ G+ + +V++ + C+C W L G+PC HAV
Sbjct: 620 KLEKESLRAQTVHVAPSADNNLFQVR----GETYELVNMAECDCSCKGWQLTGLPCHHAV 675
Query: 476 AAISFNGGHAESYIDSYYGRDAYQATYAHGITPI---NGK--RMWPYTPDAPILPPLYKR 530
A I++ G + Y Y+ Y++TYA I P+ G+ R + PP +R
Sbjct: 676 AVINYYGRNPYDYCSKYFTVAYYRSTYAQSINPVPLLEGEMCRESSGGSAVTVTPPPTRR 735
Query: 531 KPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTC 573
PGRP K ++ P E+ + +C RC +GHN TC
Sbjct: 736 PPGRPPK--KKTPAEEVM------KRQLQCSRCKGLGHNKSTC 770
>At3g06940 putative mudrA protein
Length = 749
Score = 216 bits (551), Expect = 2e-56
Identities = 154/578 (26%), Positives = 263/578 (44%), Gaps = 32/578 (5%)
Query: 11 FTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRRR-CGFLIFLSKVGGKHTYSIKT 69
F + +FR AL +SV +G K+ KND RV C+ + C + I S++ IK
Sbjct: 186 FNTFLEFRDALHKYSVAHGFAYKYKKNDSHRVSVKCKAQGCPWRITASRLSTTQLICIKK 245
Query: 70 LIPEHTCSR--VYNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYSTGITTWRAW 127
+ P HTC R + AT +V + LKEK+ Y+ ++ ++++ Y + +AW
Sbjct: 246 MNPRHTCERAVIKPGYRATRGWVRTILKEKLKAFPDYKPKDIAEDIKKEYGIQLNYSQAW 305
Query: 128 RARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQPRFNRYYICL 187
RA++ A ++G Y+ L ++K +PG+ F ++ F+R +I
Sbjct: 306 RAKEIAREQLQGSYKEAYSQLPLICEKIKETNPGSIATFMTKEDSS------FHRLFISF 359
Query: 188 EGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESECKESWKW 247
GF G RP + LD L ++Y G++L A D D FP+AFA+V+SE +E+W W
Sbjct: 360 YASISGFKQGSRPLLFLDNAILNSKYQGVMLVATASDAEDGIFPVAFAIVDSETEENWLW 419
Query: 248 FLELLLADIGGLRRWIFISDQQKGLLSVFDELM-PGVEHRFCLRHLYANFKKKFGG---- 302
FLE L + R F++D Q GL + ++ H +CL L G
Sbjct: 420 FLEQLKTALSESRIITFVADFQNGLKNAIAQVFEKDAHHAYCLGQLAEKLNVDLKGQFSH 479
Query: 303 ---GVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKC 359
++ D A T V + + +K ++ +A++W++ W F +
Sbjct: 480 EARRYMLNDFYSVAYATTPVGYY-LALENIKSISPDAYNWVIESEPHHWANALFQ-GERY 537
Query: 360 DVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRL 419
+ + +N + F + + A + PI MID R+ +M K + + P ++L
Sbjct: 538 NKMNSNFGKDFYSWVSEAHEFPITQMIDEFRAKMMQSIYTRQVKSREWVTTLTPSNEEKL 597
Query: 420 AWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAIS 479
E + + + + ++V + IVD+ Q C C W L G+PC HA+A I
Sbjct: 598 QKEIELAQLLQVSSPEGSLFQVNGG-ESSVSIVDINQCDCDCKTWRLTGLPCSHAIAVIG 656
Query: 480 FNGGHAESYIDSYYGRDAYQATYAHGITPI-NGKRMWPYTPDAPIL---PPLYKRKPGRP 535
Y +Y ++++ YA I P+ N RM P ++ PP +R PGRP
Sbjct: 657 CIEKSPYEYCSTYLTVESHRLMYAESIQPVPNMDRMMLDDPPEGLVCVTPPPTRRTPGRP 716
Query: 536 KKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTC 573
K+++ +P + + +C +C +GHN +TC
Sbjct: 717 -KIKKVEPLD-------MMKRQLQCSKCKGLGHNKKTC 746
>At1g06740 mudrA-like protein
Length = 726
Score = 215 bits (548), Expect = 5e-56
Identities = 147/582 (25%), Positives = 272/582 (46%), Gaps = 26/582 (4%)
Query: 7 LGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICRRR-CGFLIFLSKVGGKHTY 65
+GMEF+ R+A+++ ++ E++ +K+D R A C + C + I +KV T+
Sbjct: 158 VGMEFSDAYACRRAIKNAAISLRFEMRTIKSDKTRFTAKCNSKGCPWRIHCAKVSNAPTF 217
Query: 66 SIKTLIPEHTCSRVYN--NRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYSTGITT 123
+I+T+ HTC + + ++ A+ +VA + EK+ ++ E+++E+ + ++
Sbjct: 218 TIRTIHGSHTCGGISHLGHQQASVQWVADVVAEKLKENPHFKPKEILEEIYRVHGISLSY 277
Query: 124 WRAWRARQKALAIVEGDAA-----SQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQP 178
+AWR +++ +A + G +Y L ++ E++R++PG+ H P
Sbjct: 278 KQAWRGKERIMATLRGSTLRGSFEEEYRLLPQYCDEIRRSNPGSVAVVHVNPIDGC---- 333
Query: 179 RFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVE 238
F +I + GF+ CRP I LD LK++Y G LL A G D + FP+AFA+V
Sbjct: 334 -FQHLFISFQASISGFLNACRPLIALDSTVLKSKYPGTLLLATGFDGDGAVFPLAFAIVN 392
Query: 239 SECKESWKWFLELLLADIG-GLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFK 297
E ++W FL L + + + +S ++ ++ + P H FCL +L F+
Sbjct: 393 EENDDNWHRFLSELRKILDENMPKLTILSSGERPVVDGVEANFPAAFHGFCLHYLTERFQ 452
Query: 298 KKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYP 357
++F V++ DL +AA V F+ K+ ++++++ EA W+ W F
Sbjct: 453 REFQSSVLV-DLFWEAAHCLTVLEFKSKINKIEQISPEASLWIQNKSPARWASSYFEGTR 511
Query: 358 KCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLK 417
+ N ++E+ + + PI+ ++ I +L+ E +++ +P K
Sbjct: 512 FGQLTANVITESLSNWVEDTSGLPIIQTMECIHRHLINMLKERRETSLHWSNVLVPSAEK 571
Query: 418 RLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAA 477
++ + S A++EV + +G+ +V++ SC C W + G+PC HAV A
Sbjct: 572 QMLAAIEQSRAHRVYRANEAEFEVMT-CEGN-VVVNIENCSCLCGRWQVYGLPCSHAVGA 629
Query: 478 ISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWPYTP---DAPILPPLYKRKPGR 534
+ Y +S + + Y+ YA + PI+ K W D+ + K G
Sbjct: 630 LLSCEEDVYRYTESCFTVENYRRAYAETLEPISDKVQWKENDSERDSENVIKTPKAMKGA 689
Query: 535 PKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTNP 576
P+K R R ++ R CGRC GH TCT P
Sbjct: 690 PRKRRVR------AEDRDRVRRVVHCGRCNQTGHFRTTCTAP 725
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 215 bits (547), Expect = 7e-56
Identities = 159/622 (25%), Positives = 281/622 (44%), Gaps = 46/622 (7%)
Query: 4 KFKLGMEFTSIQQFRQALRDHSVR---NGRELKFLKNDDVRVRAICRRRCGFLIFLSKVG 60
+ +G F + +F++ + ++++ N ++ ++ K D + R R+ C + ++ S
Sbjct: 175 RLAIGRTFFTGFEFKEVVLHYAMKHRINAKQNRWEK-DKISFRCAQRKECEWYVYASYSH 233
Query: 61 GKHTYSIKTLIPEHTCSRVYNNRSATSDFVASKLKEKVSIVRKYRLSEVVDEMRIHYSTG 120
+ + +KT +H+C+ + + +K+ + + ++ ++ +
Sbjct: 234 ERQLWVLKTKCLDHSCTSNGKCKLLKRRVIGRLFMDKLRLQPNFMPLDIQRHIKEQWKLV 293
Query: 121 ITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQPRF 180
T + R AL ++ + A Q+ L + AE+ + G++ + + F
Sbjct: 294 STIGQVQDGRLLALKWLKEEYAQQFAHLRGYVAEILSTNKGSTAIVDTIRDANE--NDVF 351
Query: 181 NRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESE 240
NR Y+C K F CRP IG+DG LK G LL A+ D N+Q +P+A+A V+ E
Sbjct: 352 NRIYVCFGAMKNAFYF-CRPLIGIDGTFLKHAVKGCLLTAIAHDANNQIYPVAWATVQFE 410
Query: 241 CKESWKWFL-----ELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYAN 295
E+W WFL +L L D G ++ ISD+ KG++S +P EHR C++H+ N
Sbjct: 411 NAENWLWFLNQLKHDLELKDGSG---YVVISDRCKGIISAVKNALPNAEHRPCVKHIVEN 467
Query: 296 FKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTL 355
KK+ G +++ + A + ++ + E++ ++ +M +WC+ F +
Sbjct: 468 LKKRHGSLDLLKKFVWNLAWSYSDTQYKANLNEMRAYIMSLYEDVMKEEPNTWCRSWFRI 527
Query: 356 YPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKP 415
C+ + NN +E+FNATI+ AR K ++ M++ IR M R + K ++
Sbjct: 528 GSYCEDVDNNATESFNATIVKARAKALVPMMETIRRQAMARISKRKAKIGKWKKKISEYV 587
Query: 416 LKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLR--QRSCTCNWWDLVGIPCRH 473
+ L E + + H K+EV DG+ V L+ + C+C W + GIPC H
Sbjct: 588 SEILKEEWELAVKCEVTKGTHEKFEV--WFDGNSNAVSLKTEEWDCSCCKWQVTGIPCEH 645
Query: 474 AVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWPY---TPDAPILPPLYK- 529
A AAI+ G E ++ + A++ Y G P+ G+ WP AP+ P+
Sbjct: 646 AYAAINDVGKDVEDFVIPMFYTIAWREQYDTGPDPVRGQMYWPTGLGLITAPLQDPVPPG 705
Query: 530 RKPGRPKKLRR-RDPHEDDTDEG------SRPNSRHRCGRCGTIGHNIRTCTNPP----- 577
RK G K R + P+E +G + C CG GHN C P
Sbjct: 706 RKKGEKKNYHRIKGPNESPKKKGPQRLKVGKKGVVMHCKSCGEAGHNAAGCKKFPKEKKG 765
Query: 578 -----------TSQPTEDVPET 588
TSQPT ++ ET
Sbjct: 766 RKKKNQEIKAGTSQPTMELQET 787
>At1g12720 hypothetical protein
Length = 585
Score = 210 bits (534), Expect = 2e-54
Identities = 140/500 (28%), Positives = 226/500 (45%), Gaps = 35/500 (7%)
Query: 90 VASKLKEKVSIVRKYRLSEVVDEMRIHYSTGITTWRAWRARQKALAIVEGDAASQYTTLF 149
+A+ +E++ + K E+V E++ Y +T + +A+ K L + ++ ++
Sbjct: 7 IAALFEERLRVNPKMTKYEMVAEIKREYKLEVTPDQCAKAKTKVLKARNASHDTHFSRIW 66
Query: 150 RFSAELKRASPGNSYKFHCLPPTAQISQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHL 209
+ AE+ +P + + T S+ RF R YIC K + CRP IG+DG L
Sbjct: 67 DYQAEVLNRNPNSDFDIETTARTFIGSKQRFFRLYICFNSQKVSWKQHCRPVIGIDGAFL 126
Query: 210 KTQYGGILLCAVGRDPNDQYFPIAFAVVESECKESWKWFLELLLADIG--GLRRWIFISD 267
K G LL AVGRD +++ P+A+AVVE E ++W WFL+ L +G + ISD
Sbjct: 127 KWDIKGHLLAAVGRDGDNRIVPLAWAVVEIENDDNWDWFLKKLSESLGLCEMVNLALISD 186
Query: 268 QQKGLLSVFDELMPGVEHRFCLRHLYANFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMR 327
+Q GL+ ++P EHR C +H+ N+K+ + ++ L + +++ + F M
Sbjct: 187 KQSGLVKAIHNVLPQAEHRQCSKHIMDNWKRD-SHDMELQRLFWKISRSYTIEEFNTHMA 245
Query: 328 ELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDKPILTMID 387
LK N +A+ L +W TI AR KP+L M++
Sbjct: 246 NLKSYNPQAYASLQLTSPMTW------------------------TIRQARRKPLLDMLE 281
Query: 388 WIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDG 447
IR M R A +R F P+ + S + +E+ ++
Sbjct: 282 DIRRQCMVRTAKRFIIAERLKSRFTPRAHAEIEKMIAGSAGCERHLARNNLHEI--YVND 339
Query: 448 DQFIVDLRQRSCTCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGIT 507
+ VD+ +++C C W++VGIPC H I E Y+ YY + ++ TY GI
Sbjct: 340 VGYFVDMDKKTCGCRKWEMVGIPCVHTPCVIIGRKEKVEDYVSDYYTKVRWRETYRDGIR 399
Query: 508 PINGKRMWPYTPDAPILPPLYKR-KPGRP-----KKLRRRDPHEDDTDEGSRPNSRHRCG 561
P+ G +WP P+LPP ++R PGR KK R + ++ SR N C
Sbjct: 400 PVQGMPLWPRMSRLPVLPPPWRRGNPGRQSNYARKKGRYETASSSNKNKMSRANRIMTCS 459
Query: 562 RCGTIGHNIRTCTNPPTSQP 581
C GHN +C N P
Sbjct: 460 NCKQEGHNKSSCKNATVLLP 479
>At2g29230 Mutator-like transposase
Length = 915
Score = 202 bits (515), Expect = 4e-52
Identities = 152/541 (28%), Positives = 256/541 (47%), Gaps = 30/541 (5%)
Query: 50 CGFLIFLSKVGGKHTYSIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSIVRKYRL 106
C + +++ K+ Y I++ EHTC+ R +R+AT+ + S ++ K + R
Sbjct: 390 CPWRVYIVKLEDSDNYQIRSANLEHTCTVEERSNYHRAATTRVIGSIIQSKYA--GNSRG 447
Query: 107 SEVVDEMRI---HYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNS 163
+D RI YS I+ W+AW++R+ A+ +G AA+ +T L + L+ A+PG+
Sbjct: 448 PRAIDLQRILLTDYSVRISYWKAWKSREIAMDSAQGSAANSFTLLPAYLHVLREANPGSI 507
Query: 164 YKFHCLPPTAQISQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGR 223
RF ++ +GF R + +DG HLK +YGG LL A G+
Sbjct: 508 VDLKT--EVDGKGNHRFKYMFLAFAASIQGFSCMKRVIV-IDGAHLKGKYGGCLLTASGQ 564
Query: 224 DPNDQYFPIAFAVVESECKESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGV 283
D N Q FPIAF VV+SE ++W+WF +L I F+SD+ + + + P
Sbjct: 565 DANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIPDGDNLTFVSDRHSSIYTGLRRVYPKA 624
Query: 284 EHRFCLRHLYAN----FKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDW 339
+H C+ HL N +KKK + + +AA+A + F E+ ++ +
Sbjct: 625 KHGACIVHLQRNIATSYKKKH-----LLFHVSRAARAFRICEFHTYFNEVIRLDPACARY 679
Query: 340 LMTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAA 399
L + W + A+ L + +V+ +N++E+ NA + AR+ PI++++++IR+ L+ FA
Sbjct: 680 LESVGFCHWTR-AYFLGKRYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAM 738
Query: 400 LNEKFQRYNHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSC 459
E + P PK + + + S + YE++ + V L +R+C
Sbjct: 739 RREAARTEASPLPPKMREVVHRNFEKSVRFAVHRLDRYDYEIREE-GASVYHVKLMERTC 797
Query: 460 TCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPIN--GKRMWPY 517
+C +DL+ +PC HA+AA G + + Y ++++ +Y I P+ G
Sbjct: 798 SCRAFDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALP 857
Query: 518 TPDAP--ILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTN 575
P A + PP +R GRPKK R E + RC RC GHN TC
Sbjct: 858 EPIASLRLFPPATRRPSGRPKKKRITSRGEFTCPQ----RQVTRCSRCTGAGHNRATCKM 913
Query: 576 P 576
P
Sbjct: 914 P 914
>At1g21040 hypothetical protein
Length = 904
Score = 191 bits (485), Expect = 1e-48
Identities = 141/504 (27%), Positives = 244/504 (47%), Gaps = 26/504 (5%)
Query: 50 CGFLIFLSKVGGKHTYSIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSIVRKYRL 106
C + +++ K+ Y I++ EHTC+ R +R+AT+ + S ++ K + R
Sbjct: 390 CPWRVYIVKLEDSDNYQIRSANLEHTCTVEERSNYHRAATTRVIGSIIQSKYA--GNSRG 447
Query: 107 SEVVDEMRI---HYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNS 163
+D RI YS I+ W+AW++R+ A+ +G AA+ +T L + L+ A+PG+
Sbjct: 448 PRAIDLQRILLTDYSVRISYWKAWKSREIAMDSAQGSAANSFTLLPAYLHVLREANPGSI 507
Query: 164 YKFHCLPPTAQISQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGR 223
RF ++ +GF R + +DG HLK +YGG LL A G+
Sbjct: 508 VDLKT--EVDGKGNHRFKYMFLAFAASIQGFSCMKRVIV-IDGAHLKGKYGGCLLTASGQ 564
Query: 224 DPNDQYFPIAFAVVESECKESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGV 283
D N Q FPIAF VV+SE ++W+WF +L I F+SD+ + + + P
Sbjct: 565 DANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIPDGDNLTFVSDRHSSIYTGLRRVYPKA 624
Query: 284 EHRFCLRHLYAN----FKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDW 339
+H C+ HL N +KKK + + +AA+A + F E+ ++ +
Sbjct: 625 KHGACIVHLQRNIATSYKKKH-----LLFHVSRAARAFRICEFHTYFNEVIRLDPACARY 679
Query: 340 LMTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAA 399
L + W + A+ L + +V+ +N++E+ NA + AR+ PI++++++IR+ L+ FA
Sbjct: 680 LESVGFCHWTR-AYFLGERYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAM 738
Query: 400 LNEKFQRYNHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSC 459
E + P PK + + + S + YE++ + V L +R+C
Sbjct: 739 RREAARTEASPLPPKMREVVHRNFEKSVRFAVHRLDRYDYEIREE-GASVYHVKLMERTC 797
Query: 460 TCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPIN--GKRMWPY 517
+C +DL+ +PC HA+AA G + + Y ++++ +Y I P+ G
Sbjct: 798 SCRAFDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALP 857
Query: 518 TPDAP--ILPPLYKRKPGRPKKLR 539
P A + PP +R GRPKK R
Sbjct: 858 EPIASLRLFPPATRRPSGRPKKKR 881
>At3g47280 putative protein
Length = 739
Score = 183 bits (465), Expect = 2e-46
Identities = 143/578 (24%), Positives = 257/578 (43%), Gaps = 26/578 (4%)
Query: 7 LGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAIC-RRRCGFLIFLSKVGGKHTY 65
+G F + LR +V++ + +K+D R C C + + ++ GG +Y
Sbjct: 173 IGKIFKDKDEMVFTLRKFAVKHSFKFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESY 232
Query: 66 SIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSIVR-KYRLSEVVDEMRIHYSTGI 121
I+ + H+C R ++R A++ + + + R ++++ R + GI
Sbjct: 233 VIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGI 292
Query: 122 TTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQPRFN 181
+AWR ++ A + G + L R+ ++ +PG+ F + S +F
Sbjct: 293 NYSKAWRVQKHAAELARGLPDDSFEVLPRWFHRVQVTNPGSITFFK------KDSANKFK 346
Query: 182 RYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESEC 241
++ RG+ R I +DG HL +++ G LL A +D N +PIAFA+V+SE
Sbjct: 347 YAFLAFGASIRGYKL-MRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSEN 405
Query: 242 KESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKKFG 301
SW WFL+ LL I +F+SD+ + S P H C HL N + F
Sbjct: 406 DASWDWFLKCLLNIIPDENDLVFVSDRAASIASRLSGNYPLAHHGLCTFHLQKNLETHFR 465
Query: 302 GGVVIRDLMMQAAKATYVHL-FEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCD 360
G +I + AA Y F+ E+ +++ +L + W + A++ + +
Sbjct: 466 GSSLIP--VYYAASRIYTKTEFDSLFWEITNSDKKLAQYLCEVDVRKWSR-AYSPSNRYN 522
Query: 361 VLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLA 420
++ +NL+E+ NA + R+ P++ + + IRS + F E+ ++ K++
Sbjct: 523 IMTSNLAESVNALLKQNREYPVVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMK 582
Query: 421 WETKSSNNWVPQAC--GHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAI 478
+S W+ + C ++EVK D +V+L +R+CTC +D+ PC H +A+
Sbjct: 583 ASYDTSTRWL-EVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 639
Query: 479 SFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMW--PYTPDAPI-LPPLYKRKPGRP 535
+ ++D ++ ++ Y+ I P W P T I LPP + G+
Sbjct: 640 KHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYWEIPETISEVICLPPSTRVPTGKR 699
Query: 536 KKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTC 573
KK +R P + H+C RCG GHN TC
Sbjct: 700 KK--KRIPSVWEHGRSQPKPKLHKCSRCGQSGHNKSTC 735
>At2g12720 putative protein
Length = 819
Score = 183 bits (464), Expect = 3e-46
Identities = 148/574 (25%), Positives = 251/574 (42%), Gaps = 20/574 (3%)
Query: 7 LGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAICR-RRCGFLIFLSKVGGKHTY 65
+G ++ S + L+ ++R+G + ++ V C RC + + + G +
Sbjct: 257 VGQQYRSKFELEYRLKLLAIRDGFDFDVPTSNKTTVSYECWVDRCLWRVRACRQGNDPNF 316
Query: 66 SIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSIVRK-YRLSEVVDEMRIHYSTGI 121
+ EHTCS R +R AT D + ++ + V + + + H+ +
Sbjct: 317 YVYIYDSEHTCSVRERSGRSRQATPDVLGVLYRDYLGDVGPDVKPKSIGIIITKHFRVKM 376
Query: 122 TTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQP-RF 180
+ ++++ + A + G S + L + ++RA+PG + QI + RF
Sbjct: 377 SYSKSYKTLRFARELTLGTPDSSFEELPSYLYMIRRANPGTVARL-------QIDESGRF 429
Query: 181 NRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESE 240
N +I GF R + +DG L Y G LL A+ +D N Q FP+AF VV++E
Sbjct: 430 NYMFIVFGASIAGFHY-MRRVVVVDGTFLHGSYKGTLLTALAQDGNFQIFPLAFGVVDTE 488
Query: 241 CKESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKKF 300
+SW+W L I ISD+ K + E+ P C HLY N KF
Sbjct: 489 NDDSWRWLFTQLKVVIPDATDLAIISDRHKSIGKAIGEVYPLAARGICTYHLYKNILVKF 548
Query: 301 GGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCD 360
+ L+ +AA+ ++ F E++E++ H +L + W + F + +
Sbjct: 549 KRKDLF-PLVKKAARCYRLNDFTNAFNEIEELDPLLHAYLQRAGVEMWARAHFP-GDRYN 606
Query: 361 VLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLA 420
++ N++E+ N + A++ PI+ M++ IR + FA + + + P K L
Sbjct: 607 LMTTNIAESMNRALSQAKNLPIVRMLEAIRQMMTRWFAERRDDASKQHTQLTPGVEKLLQ 666
Query: 421 WETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAISF 480
SS Q ++ +V + +V++ ++ CTC ++ +PC HA+AA
Sbjct: 667 TRVTSSRLLDVQTIDASRVQV--AYEASLHVVNVDEKQCTCRLFNKEKLPCIHAIAAAEH 724
Query: 481 NGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWP-YTPDAPILPPLYKRKPGRPKKLR 539
G S YY YA I P + + P D P LPP+ +PGRPKKLR
Sbjct: 725 MGVSRISLCSPYYKSSHLVNVYACAIMPSDTEVPVPQIVIDQPCLPPIVANQPGRPKKLR 784
Query: 540 RRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTC 573
+ E E RP H C RC GHN++TC
Sbjct: 785 MKSALEVAV-ETKRPRKEHACSRCKETGHNVKTC 817
>At1g44840 hypothetical protein
Length = 926
Score = 182 bits (462), Expect = 5e-46
Identities = 141/503 (28%), Positives = 229/503 (45%), Gaps = 21/503 (4%)
Query: 49 RCGFLIFLSKVGGKHTYSIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSI----- 100
+C + I + + G Y IK +HTCS R + ATS +AS K K S
Sbjct: 419 KCDWRILATLMKGTGYYEIKKASLDHTCSLDTRGQFMQKATSKVIASVFKAKYSDPTSGP 478
Query: 101 VRKYRLSEVVDEMRIHYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASP 160
V V++++R+ S + WRAR+ AL V G Y L + LK +P
Sbjct: 479 VPMDLQQLVLEDLRVSVSYS----KCWRARESALTSVAGSDEESYCYLAEYLHLLKLTNP 534
Query: 161 GNSYKFHCLPPTAQISQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCA 220
G S+ RF ++ GF R + +DG HLK +Y G+LL +
Sbjct: 535 GTITHIETERDVEDESKERFLYMFLAFGASIAGF-RHLRRILVVDGTHLKGKYKGVLLTS 593
Query: 221 VGRDPNDQYFPIAFAVVESECKESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELM 280
G+D N Q +P+ FAVV+SE ESW WF L I + +SD+ +L +
Sbjct: 594 SGQDANFQVYPLGFAVVDSENDESWTWFFTKLERIIADSKTLTILSDRHSSILVAVKRVF 653
Query: 281 PGVEHRFCLRHLYANFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWL 340
P H C+ HL N + K+ + L+ A A F+E +++ N+ +L
Sbjct: 654 PQANHGACIIHLCRNIQTKY-KNKALTQLVKNAGYAFTGTKFKEFYGQIETTNQNCGKYL 712
Query: 341 MTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAAL 400
+W +H F + +++ +N++E N + R I+ +I +IRS L F A
Sbjct: 713 HDIGMANWTRHYFR-GQRFNLMTSNIAETLNKALNKGRSSHIVELIRFIRSMLTRWFNAR 771
Query: 401 NEKFQRYNHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCT 460
+K ++ P P+ K++ ++N + YE+ +L G + +VDL ++ CT
Sbjct: 772 RKKSLKHKGPVPPEVDKQITKTMLTTNGSKVGRITNWSYEINGML-GGRNVVDLEKKQCT 830
Query: 461 CNWWDLVGIPCRHA-VAAISFNGGHAESYIDSYYGRDAYQATYAHGITPI---NGKRMWP 516
C +D + IPC HA VAA SF + ++ +D + A+ ++Y + P +++
Sbjct: 831 CKRYDKLKIPCSHALVAANSFKISY-KALVDDCFKPHAWVSSYPGAVFPEAIGRDEKILE 889
Query: 517 YTPDAPILPPLYKRKPGRPKKLR 539
+LPP +R GRPK R
Sbjct: 890 ELEHRSMLPPYTRRPSGRPKVAR 912
>At1g25784 mutator-like transposase, putative
Length = 1127
Score = 181 bits (458), Expect = 1e-45
Identities = 149/547 (27%), Positives = 244/547 (44%), Gaps = 26/547 (4%)
Query: 7 LGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAIC-RRRCGFLIFLSKVGGKHTY 65
+G F + + +Q + +++ K K+ ++ +C C + + V ++
Sbjct: 379 VGSVFKNRKVLQQTMSLQAIKQCFCFKQPKSCPKTLKMVCVDETCPWQLTARVVKDSESF 438
Query: 66 SIKTLIPEHTC---SRVYNNRSATSDFVASKLKEKVSIV----RKYRLSEVVDE---MRI 115
I + HTC SR Y N+ A + ++ + S R L +++ + +RI
Sbjct: 439 KITSYATTHTCNIDSRKYYNKHANYKLLGEVVRSRYSSTQGGPRAVDLPQLLLKDLNVRI 498
Query: 116 HYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQI 175
YST AWRA++ A+ V GD + Y L + L+ A+PG H P
Sbjct: 499 SYST------AWRAKEVAVENVRGDEIANYRFLPTYLYLLQLANPGTITHLHYTPEDD-- 550
Query: 176 SQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFA 235
+ RF ++ L +G + R + +DG L Y G LL A +D N Q FPIAF
Sbjct: 551 GKQRFKYVFVSLGASIKGLIY-MRKVVVVDGTQLVGPYKGCLLIACAQDGNFQIFPIAFG 609
Query: 236 VVESECKESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYAN 295
VV+ E SW WF E L + + +SD+ + + P H C HL N
Sbjct: 610 VVDGETDASWAWFFEKLAEIVPDSDDLMIVSDRHSSIYKGLSVVYPRAHHGACAVHLERN 669
Query: 296 FKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTL 355
+ G + L AAKA V FE+ L+E + + +L + W + A
Sbjct: 670 LSTYY-GKFGVSALFFSAAKAYRVRDFEKYFELLREKSAKCAKYLEDIGFEHWTR-AHCR 727
Query: 356 YPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKP 415
+ +++ +N SE+ N + A+ PI+ MI++IR LM FA+ +K R P+
Sbjct: 728 GERYNIMSSNNSESMNHVLTKAKTYPIVYMIEFIRDVLMRWFASRRKKVARCKSSVTPEV 787
Query: 416 LKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAV 475
+R E +S + + G Y+V S G+ F V L Q +CTC + + IPC HA+
Sbjct: 788 DERFLQELPASGKYAVKMSGPWSYQVTS-KSGEHFHVVLDQCTCTCLRYTKLRIPCEHAL 846
Query: 476 AAISFNGGHAESYIDSYYGRDAYQATYAHGITPINGKR---MWPYTPDAPILPPLYKRKP 532
AA +G +S + +YG + ++ I PI + + + D ++PP +R P
Sbjct: 847 AAAIEHGIDPKSCVGWWYGLQTFSDSFQEPILPIADPKDVVIPQHIVDLILIPPYSRRPP 906
Query: 533 GRPKKLR 539
GRP R
Sbjct: 907 GRPLSKR 913
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 180 bits (457), Expect = 2e-45
Identities = 143/578 (24%), Positives = 257/578 (43%), Gaps = 26/578 (4%)
Query: 7 LGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAIC-RRRCGFLIFLSKVGGKHTY 65
+G F + LR +V++ E +K+D + C C + + ++ GG +Y
Sbjct: 50 IGKFFKDKDEMVFTLRMSAVKHSFEFHTVKSDLTKYVLHCIDENCSWRLRATRAGGSESY 109
Query: 66 SIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSIVR-KYRLSEVVDEMRIHYSTGI 121
I+ + H+C R ++R A++ + + + R ++++ R + GI
Sbjct: 110 VIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGI 169
Query: 122 TTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQPRFN 181
+AWR ++ A + G + L R+ ++ +PG+ F + S +F
Sbjct: 170 NYSKAWRVQEHAAELARGLPDDSFEVLPRWFHRVQVTNPGSITFFK------KDSANKFK 223
Query: 182 RYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESEC 241
++ RG+ + I +DG HL +++ G LL A +D N +PIAFA+V+SE
Sbjct: 224 YAFLAFGASIRGYKL-MKKVISIDGAHLISKFKGTLLGASAQDGNFNLYPIAFAIVDSEN 282
Query: 242 KESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKKFG 301
SW WFL+ LL I +F+SD+ + S P + C HL N + F
Sbjct: 283 DASWDWFLKCLLNIIPDENDLVFVSDRAASIASGLSGNYPLAHNGLCTFHLQKNLETHFR 342
Query: 302 GGVVIRDLMMQAAKATYVHL-FEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCD 360
G +I + AA Y F+ E+ +++ +L + W + A++ + +
Sbjct: 343 GSSLIP--VYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWEVDVRKWSR-AYSPSNRYN 399
Query: 361 VLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLA 420
++ +NL+E+ NA + R+ PI+ + + IRS + F E+ ++ K++
Sbjct: 400 IMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMK 459
Query: 421 WETKSSNNWVPQAC--GHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAI 478
+S W+ + C ++EVK D +V+L +R+CTC +D+ PC H +A+
Sbjct: 460 ASYDTSTRWL-EVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 516
Query: 479 SFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMW--PYTPDAPI-LPPLYKRKPGRP 535
+ ++D ++ ++ Y+ I P W P T I LPP + GR
Sbjct: 517 KHINLNENMFVDEFHSTYKWRQAYSKSIHPNGDMEYWEIPETISEVICLPPSTRVPSGRR 576
Query: 536 KKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTC 573
KK +R P + H+C RCG GHN TC
Sbjct: 577 KK--KRIPSVWEHGRSQPKPKLHKCSRCGQSGHNNSTC 612
>At1g35060 hypothetical protein
Length = 873
Score = 180 bits (456), Expect = 2e-45
Identities = 146/512 (28%), Positives = 234/512 (45%), Gaps = 39/512 (7%)
Query: 49 RCGFLIFLSKVGGKHTYSIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSIVRKYR 105
+C + I + + G Y IK +HTCS R + ATS +AS K K S
Sbjct: 370 KCDWRILATVMNGTGYYEIKKAQLQHTCSVDTRRQYMKKATSKVIASVFKAKYSEASAGP 429
Query: 106 LSE-----VVDEMRIHYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASP 160
+ V++++R+ S + WRAR+ AL V G Y+ L + LK +P
Sbjct: 430 VPMDLQQLVLEDLRVSASYK----KCWRARESALTDVGGSDEESYSNLAEYLHLLKLTNP 485
Query: 161 GNSYKFHCLPPTAQISQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCA 220
G P + RF ++ +GF R + +DG HLK +Y G+LL A
Sbjct: 486 GTITHIETEPDIEDERKERFLYMFLAFGASIQGF-KHLRRVLVVDGTHLKGKYKGVLLTA 544
Query: 221 VGRDPNDQYFPIAFAVVESECKESWKWF---LELLLADIGGLRRWIFISDQQKGLLSVFD 277
G+D N Q +P+AFAVV+SE ++W WF LE ++AD L +SD+ + +
Sbjct: 545 SGQDANFQVYPLAFAVVDSENDDAWTWFFTKLERIIADNNTL---TILSDRHESIKVGVK 601
Query: 278 ELMPGVEHRFCLRHLYANFKKKFGGGVVIRDLMMQAAKATYVHL---FEEKMRELKEVN- 333
++ P H C+ HL N + +F R L A Y F+ ++ +N
Sbjct: 602 KVFPQAHHGACIIHLCRNIQARFKN----RGLTQLVKNAGYEFTSGKFKTLYNQINAINP 657
Query: 334 ---EEAHDWLMTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIR 390
+ HD M H W + F + +++ +N++E N + R I+ ++ +IR
Sbjct: 658 LCIKYLHDVGMAH----WTRLYFP-GQRFNLMTSNIAETLNKALFKGRSSHIVELLRFIR 712
Query: 391 SYLMGRFAALNEKFQRYNHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQF 450
S L F A +K Q ++ P P+ K+++ +S+ YEV L G
Sbjct: 713 SMLTRWFNAHRKKSQAHSGPVPPEVDKQISKNLTTSSGSKVGRVTSWSYEVVGKLGGSN- 771
Query: 451 IVDLRQRSCTCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITP-I 509
+VDL ++ CTC +D + IPC HA+ A + ++ +D Y+ ++ A+Y + P
Sbjct: 772 VVDLEKKQCTCKRYDKLKIPCGHALVAANSINLSYKALVDDYFKPHSWVASYKGAVFPEA 831
Query: 510 NGKR--MWPYTPDAPILPPLYKRKPGRPKKLR 539
NGK + P +LPP +R GRPK R
Sbjct: 832 NGKEEDIPEELPHRSMLPPYTRRPSGRPKVAR 863
>At1g34620 Mutator-like protein
Length = 1034
Score = 180 bits (456), Expect = 2e-45
Identities = 136/524 (25%), Positives = 252/524 (47%), Gaps = 33/524 (6%)
Query: 48 RRCGFLIFLSKVGGKHTYSIKTLIPEHTCS----RVYNNRSAT--------SDFVASKLK 95
+ C + ++ SKV ++ ++ HTC+ R Y+ + T S F+ K
Sbjct: 409 KTCPWRVYASKVDTSDSFQVRQANQRHTCTIDQRRRYHRLATTQVIGELMQSRFLGIKRG 468
Query: 96 EKVSIVRKYRLSEVVDEMRIHYSTGITTWRAWRARQKALAIVEGDAASQYTTLFRFSAEL 155
+++RK+ L + Y I+ W+AWRAR+ A+ G A Y + ++ L
Sbjct: 469 PNAAVIRKFLLDD--------YHVSISYWKAWRAREVAMEKSLGSMAGSYALIPAYAGLL 520
Query: 156 KRASPGN-SYKFHCLPPTAQISQPRFNRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYG 214
++A+PG+ + + PT RF +I +G+ A R I +DG +K +YG
Sbjct: 521 QQANPGSLCFTEYDDDPTGP---RRFKYQFIAFAASIKGY-AFMRKVIVVDGTSMKGRYG 576
Query: 215 GILLCAVGRDPNDQYFPIAFAVVESECKESWKWFLELLLADIGGLRRWIFISDQQKGLLS 274
G L+ A +D N Q FP+AF +V SE +++WF + L +FISD+ + + +
Sbjct: 577 GCLISACCQDGNFQIFPLAFGIVNSENDSAYEWFFQRLSIIAPDNPDLMFISDRHESIYT 636
Query: 275 VFDELMPGVEHRFCLRHLYANFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNE 334
++ H C HL+ N + + + R LM +AA+A +V F +++++N
Sbjct: 637 GLSKVYTQANHAACTVHLWRNIRHLYKPKSLCR-LMSEAAQAFHVTDFNRIFLKIQKLNP 695
Query: 335 EAHDWLMTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDKPILTMIDWIRSYLM 394
+L+ W + + + +++ +N+ E++N I AR+ P++ M+++IR+ LM
Sbjct: 696 GCAAYLVDLGFSEWTR-VHSKGRRFNIMDSNICESWNNVIREAREYPLICMLEYIRTTLM 754
Query: 395 GRFAALNEKFQRYNHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDL 454
FA + + P+ +R+ +S+ + + + +++V+ G+ FIV L
Sbjct: 755 DWFATRRAQAEDCPTTLAPRVQERVEENYQSAMSMSVKPICNFEFQVQE-RTGECFIVKL 813
Query: 455 RQRSCTCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPIN--GK 512
+ +C+C + +GIPC HA+AA + G +S + Y + + +Y I PI+ G
Sbjct: 814 DESTCSCLEFQGLGIPCAHAIAAAARIGVPTDSLAANGYFNELVKLSYEEKIYPIHSVGG 873
Query: 513 RMWPYT---PDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSR 553
+ P + PP +R PGRP+K+R E +G R
Sbjct: 874 EVAPEIAAGTTGELQPPFVRRPPGRPRKIRILSRGEFKARQGPR 917
>At2g15210 putaive transposase of transposon FARE2.7 (cds1)
Length = 580
Score = 173 bits (439), Expect = 2e-43
Identities = 146/578 (25%), Positives = 249/578 (42%), Gaps = 62/578 (10%)
Query: 7 LGMEFTSIQQFRQALRDHSVRNGRELKFLKNDDVRVRAIC-RRRCGFLIFLSKVGGKHTY 65
+G F + LR +V++ E +K+D R C C + + ++ GG +Y
Sbjct: 50 IGKNFKDKDEMVFTLRMFAVKHNFEFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESY 109
Query: 66 SIKTLIPEHTCS---RVYNNRSATSDFVASKLKEKVSIVR-KYRLSEVVDEMRIHYSTGI 121
I+ + H+C R ++R A++ + + + R ++++ R + GI
Sbjct: 110 VIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGI 169
Query: 122 TTWRAWRARQKALAIVEGDAASQYTTLFRFSAELKRASPGNSYKFHCLPPTAQISQPRFN 181
+AWR ++ A AEL R P +S F LP
Sbjct: 170 NYSKAWRVQEHA-------------------AELARGLPDDS--FEVLP----------- 197
Query: 182 RYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESEC 241
RG+ R I +DG HL +++ G LL A +D N +PIAFA+V+SE
Sbjct: 198 ----------RGYKL-MRKVISIDGAHLTSKFKGTLLGASAQDRNFNLYPIAFAIVDSEN 246
Query: 242 KESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFCLRHLYANFKKKFG 301
SW WFL+ LL I +F+S++ + S P H C HL N + F
Sbjct: 247 DASWDWFLKCLLNIIPDENDLVFVSERAASIASGLSGNYPLAHHGLCTFHLQKNLETHFR 306
Query: 302 GGVVIRDLMMQAAKATYVHL-FEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCD 360
G +I + AA Y F+ E+ +++ +L + W + A++ + +
Sbjct: 307 GSSLIP--VYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWEVHVRKWSR-AYSPSNRYN 363
Query: 361 VLMNNLSEAFNATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLA 420
++ +NL+E+ NA + R+ PI+ + + IRS + F E+ ++ K++
Sbjct: 364 IMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMK 423
Query: 421 WETKSSNNWVPQAC--GHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAI 478
+S W+ + C ++EVK D +V+L +R+CTC +D+ PC H +A+
Sbjct: 424 ASYDTSTRWL-EVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 480
Query: 479 SFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMW--PYTPDAPI-LPPLYKRKPGRP 535
+ + ++D ++ ++ Y+ I P W P T I LPP + GR
Sbjct: 481 NHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYWEIPETISEVICLPPSTRVPSGRR 540
Query: 536 KKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTC 573
KK +R P + H+C RCG GHN TC
Sbjct: 541 KK--KRIPSVWEHGRSQPKPKLHKCSRCGQSGHNKSTC 576
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,549,514
Number of Sequences: 26719
Number of extensions: 708609
Number of successful extensions: 5846
Number of sequences better than 10.0: 267
Number of HSP's better than 10.0 without gapping: 180
Number of HSP's successfully gapped in prelim test: 93
Number of HSP's that attempted gapping in prelim test: 2886
Number of HSP's gapped (non-prelim): 1591
length of query: 630
length of database: 11,318,596
effective HSP length: 105
effective length of query: 525
effective length of database: 8,513,101
effective search space: 4469378025
effective search space used: 4469378025
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0137.1


