

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.9
(645 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
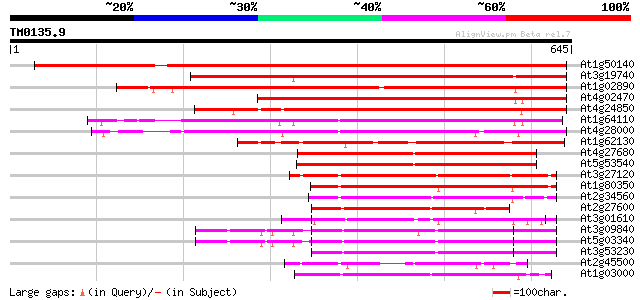
Score E
Sequences producing significant alignments: (bits) Value
At1g50140 hypothetical protein, 5' partial 859 0.0
At3g19740 ATPase, putative 675 0.0
At1g02890 hypothetical protein 456 e-128
At4g02470 407 e-113
At4g24850 unknown protein (At4g24850) 397 e-111
At1g64110 unknown protein 396 e-110
At4g28000 putative protein 395 e-110
At1g62130 hypothetical protein 305 7e-83
At4g27680 unknown protein 275 4e-74
At5g53540 26S proteasome regulatory particle chain RPT6-like pro... 273 3e-73
At3g27120 spastin protein, putative 229 3e-60
At1g80350 CAD ATPase (AAA1) 221 7e-58
At2g34560 katanin like protein 207 1e-53
At2g27600 putative AAA-type ATPase 194 2e-49
At3g01610 putative cell division control protein 178 7e-45
At3g09840 putative transitional endoplasmic reticulum ATPase 175 8e-44
At5g03340 transitional endoplasmic reticulum ATPase 172 5e-43
At3g53230 CDC48 - like protein 169 4e-42
At2g45500 hypothetical protein 168 1e-41
At1g03000 putative peroxisome assembly factor-2 161 1e-39
>At1g50140 hypothetical protein, 5' partial
Length = 601
Score = 859 bits (2220), Expect = 0.0
Identities = 432/614 (70%), Positives = 515/614 (83%), Gaps = 15/614 (2%)
Query: 29 PLANGQRGEVYEVNGDRVAVILDISEDRENEVELESVNNDGRKPPVYWINVKNIEHDLDA 88
PL++GQRGEVYEV G+RVAVI + +D+ +E + + P++W++VK++++DLD
Sbjct: 1 PLSSGQRGEVYEVIGNRVAVIFEYGDDKTSEGSEKKPAEQPQMLPIHWLDVKDLKYDLDM 60
Query: 89 QSQDCYLAMEALREVLNSKQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSGPI 148
Q+ D Y+AMEAL EVL S QPLIVYFPD +QWL ++VPK+ R EF KV+EMFD+LSGPI
Sbjct: 61 QAVDGYIAMEALNEVLQSIQPLIVYFPDSTQWLSRAVPKTRRKEFVDKVKEMFDKLSGPI 120
Query: 149 VLICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTD--KGQKTSDDDEINKLFSN 206
V+ICGQNK ++G+KE+ +F PL LK LT+ G+ S+++EI KLF+N
Sbjct: 121 VMICGQNKIETGSKEREKF-------------PLPLKGLTEGFTGRGKSEENEIYKLFTN 167
Query: 207 VVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVIL 266
V+ + PPK+E+ L FKKQL EDR+IVISRSN+NEL K LEEH+L C DL +++D VIL
Sbjct: 168 VMRLHPPKEEDTLRLFKKQLGEDRRIVISRSNINELLKALEEHELLCTDLYQVNTDGVIL 227
Query: 267 TKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKN 326
TKQKAEK +GWAKNHYL+SC +P VKG +L LPRESLEI+I+RL+ +E S +PSQ+LKN
Sbjct: 228 TKQKAEKAIGWAKNHYLASCPVPLVKGGRLSLPRESLEISIARLRKLEDNSLKPSQNLKN 287
Query: 327 LAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPV 386
+AKDE+E NF+SAVV PGEIGVKF+DIGALEDVKKALNELVILPMRRPELF GNLLRP
Sbjct: 288 IAKDEYERNFVSAVVAPGEIGVKFEDIGALEDVKKALNELVILPMRRPELFARGNLLRPC 347
Query: 387 KGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAP 446
KGILLFGPPGTGKTL+AKALATEAGANFIS++GSTLTSKWFGDAEKLTKALFSFA+KLAP
Sbjct: 348 KGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFATKLAP 407
Query: 447 VIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDD 506
VIIFVDE+DSLLGARGG+ EHEATRRMRNEFMAAWDGLRSK++QRILILGATNRPFDLDD
Sbjct: 408 VIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRPFDLDD 467
Query: 507 AVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCIT 566
AVIRRLPRRIYVDLPDA+NR KIL+I L ENL+ DFQF+KLA T+GYSGSDLKNLCI
Sbjct: 468 AVIRRLPRRIYVDLPDAENRLKILKIFLTPENLESDFQFEKLAKETEGYSGSDLKNLCIA 527
Query: 567 AAYRPVQELLEEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEM 626
AAYRPVQELL+EE+ G + LR L+LDDF+QSK+KV PSVAYDA ++NELRKWNE
Sbjct: 528 AAYRPVQELLQEEQKGARAEASPGLRSLSLDDFIQSKAKVSPSVAYDATTMNELRKWNEQ 587
Query: 627 YGEGGTRTKSPFGF 640
YGEGG+RTKSPFGF
Sbjct: 588 YGEGGSRTKSPFGF 601
>At3g19740 ATPase, putative
Length = 439
Score = 675 bits (1741), Expect = 0.0
Identities = 341/441 (77%), Positives = 387/441 (87%), Gaps = 10/441 (2%)
Query: 208 VSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILT 267
+++ PPK+E L F KQL EDR+IV+SRSNLNEL K LEE++L C DL +++D VILT
Sbjct: 1 MNLVPPKEEENLIVFNKQLGEDRRIVMSRSNLNELLKALEENELLCTDLYQVNTDGVILT 60
Query: 268 KQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLK-- 325
KQ+AEKV+GWA+NHYLSSC P +K +L LPRES+EI++ RLK E +SR+P+Q+LK
Sbjct: 61 KQRAEKVIGWARNHYLSSCPSPSIKEGRLILPRESIEISVKRLKAQEDISRKPTQNLKRF 120
Query: 326 ----NLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGN 381
N+AKDEFE+NF+SAVV PGEIGVKFDDIGALE VKK LNELVILPMRRPELFT GN
Sbjct: 121 LFLQNIAKDEFETNFVSAVVAPGEIGVKFDDIGALEHVKKTLNELVILPMRRPELFTRGN 180
Query: 382 LLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFA 441
LLRP KGILLFGPPGTGKTL+AKALATEAGANFIS++GSTLTSKWFGDAEKLTKALFSFA
Sbjct: 181 LLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFA 240
Query: 442 SKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRP 501
SKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSK++QRILILGATNRP
Sbjct: 241 SKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRP 300
Query: 502 FDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLK 561
FDLDDAVIRRLPRRIYVDLPDA+NR KIL+I L ENL+ F+FDKLA T+GYSGSDLK
Sbjct: 301 FDLDDAVIRRLPRRIYVDLPDAENRLKILKIFLTPENLETGFEFDKLAKETEGYSGSDLK 360
Query: 562 NLCITAAYRPVQELLEEEKAGDNNITNVA--LRPLNLDDFVQSKSKVGPSVAYDAVSVNE 619
NLCI AAYRPVQELL+EE +++TN + LRPL+LDDF+QSK+KV PSVAYDA ++NE
Sbjct: 361 NLCIAAAYRPVQELLQEE--NKDSVTNASPDLRPLSLDDFIQSKAKVSPSVAYDATTMNE 418
Query: 620 LRKWNEMYGEGGTRTKSPFGF 640
LRKWNE YGEGGTRTKSPFGF
Sbjct: 419 LRKWNEQYGEGGTRTKSPFGF 439
>At1g02890 hypothetical protein
Length = 1240
Score = 456 bits (1172), Expect = e-128
Identities = 252/541 (46%), Positives = 356/541 (65%), Gaps = 28/541 (5%)
Query: 123 KSVPKSI--RSEFFHKVEEMFDQLSGPIVLICGQNKAQSGTKEK---GQFTMILPNFARL 177
K + KS+ ++ + ++ + L IV+I Q + + KEK G F +
Sbjct: 703 KDIEKSVSGNTDVYITLKSKLENLPENIVVIASQTQLDN-RKEKSHPGGFLFTKFGSNQT 761
Query: 178 AKLPLSLK-----RLTDKGQKTSDD-DEINKLFSNVVSIQPPKDENLLATFKKQLEEDRK 231
A L L+ RL D+ + +I +LF N V+IQ P+DE L +K +LE D +
Sbjct: 762 ALLDLAFPDTFGGRLQDRNTEMPKAVKQITRLFPNKVTIQLPEDEASLVDWKDKLERDTE 821
Query: 232 IVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCV 291
I+ +++N+ +R VL ++QL C D+ L D L EKVVG+A NH+L +C P V
Sbjct: 822 ILKAQANITSIRAVLSKNQLVCPDIEILCIKDQTLPSDSVEKVVGFAFNHHLMNCSEPTV 881
Query: 292 KGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPGEIGVKF 350
K +KL + ES+ + L ++ ++ +SLK++ ++EFE +S V+PP +IGV F
Sbjct: 882 KDNKLIISAESITYGLQLLHEIQNENKSTKKSLKDVVTENEFEKKLLSDVIPPSDIGVSF 941
Query: 351 DDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEA 410
DIGALE+VK L ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AKA+ATEA
Sbjct: 942 SDIGALENVKDTLKELVMLPLQRPELFGKGQLTKPTKGILLFGPPGTGKTMLAKAVATEA 1001
Query: 411 GANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEAT 470
GANFI++S S++TSK EK KA+FS ASK+AP +IFVDEVDS+LG R EHEA
Sbjct: 1002 GANFINISMSSITSK----GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAM 1057
Query: 471 RRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKIL 530
R+M+NEFM WDGLR+K+ +R+L+L ATNRPFDLD+AVIRRLPRR+ V+LPD+ NR KIL
Sbjct: 1058 RKMKNEFMINWDGLRTKDKERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDSANRSKIL 1117
Query: 531 RIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK---------- 580
+IL E + D + +AN+TDGYSGSDLKNLC+TAA+ P++E+LE+EK
Sbjct: 1118 SVILAKEEMAEDVDLEAIANMTDGYSGSDLKNLCVTAAHLPIREILEKEKKERSVAQAEN 1177
Query: 581 -AGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFG 639
A ++ +RPLN++DF + +V SVA D+ ++NEL++WNE+YGEGG+R K+
Sbjct: 1178 RAMPQLYSSTDVRPLNMNDFKTAHDQVCASVASDSSNMNELQQWNELYGEGGSRKKTSLS 1237
Query: 640 F 640
+
Sbjct: 1238 Y 1238
>At4g02470
Length = 371
Score = 407 bits (1045), Expect = e-113
Identities = 201/367 (54%), Positives = 272/367 (73%), Gaps = 12/367 (3%)
Query: 286 CLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPG 344
C P VK +KL + ES+ + L ++ ++ +SLK++ ++EFE +S V+PP
Sbjct: 3 CTEPIVKDNKLVISAESISYGLQTLHDIQNENKSLKKSLKDVVTENEFEKKLLSDVIPPS 62
Query: 345 EIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAK 404
+IGV FDDIGALE+VK+ L ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AK
Sbjct: 63 DIGVSFDDIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAK 122
Query: 405 ALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGA 464
A+ATEAGANFI++S S++TSKWFG+ EK KA+FS ASK+AP +IFVDEVDS+LG R
Sbjct: 123 AVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP 182
Query: 465 FEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAD 524
EHEA R+M+NEFM WDGLR+K+ +R+L+L ATNRPFDLD+AVIRRLPRR+ V+LPDA
Sbjct: 183 GEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDAT 242
Query: 525 NRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK---- 580
NR KIL +IL E + PD + +AN+TDGYSGSDLKNLC+TAA+ P++E+LE+EK
Sbjct: 243 NRSKILSVILAKEEIAPDVDLEAIANMTDGYSGSDLKNLCVTAAHFPIREILEKEKKEKT 302
Query: 581 --AGDNNITN-----VALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTR 633
+N T +R L ++DF + +V SV+ D+ ++NEL++WNE+YGEGG+R
Sbjct: 303 AAQAENRPTPPLYSCTDVRSLTMNDFKAAHDQVCASVSSDSSNMNELQQWNELYGEGGSR 362
Query: 634 TKSPFGF 640
K+ +
Sbjct: 363 KKTSLSY 369
>At4g24850 unknown protein (At4g24850)
Length = 442
Score = 397 bits (1021), Expect = e-111
Identities = 209/443 (47%), Positives = 292/443 (65%), Gaps = 19/443 (4%)
Query: 213 PKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDL----LHLDSDDVILTK 268
P+DE L +K Q++ D + +SN N LR VL L C L + D+ L +
Sbjct: 2 PQDEKRLTLWKHQMDRDAETSKVKSNFNHLRMVLRRRGLGCEGLETTWSRMCLKDLTLQR 61
Query: 269 QKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLA 328
EK++GWA +++S P K+ L RES+E I L+ + S +
Sbjct: 62 DSVEKIIGWAFGNHISKN--PDTDPAKVTLSRESIEFGIGLLQN--DLKGSTSSKKDIVV 117
Query: 329 KDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKG 388
++ FE +S V+ P +I V FDDIGALE VK L ELV+LP++RPELF G L +P KG
Sbjct: 118 ENVFEKRLLSDVILPSDIDVTFDDIGALEKVKDILKELVMLPLQRPELFCKGELTKPCKG 177
Query: 389 ILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVI 448
ILLFGPPGTGKT++AKA+A EA ANFI++S S++TSKWFG+ EK KA+FS ASK++P +
Sbjct: 178 ILLFGPPGTGKTMLAKAVAKEADANFINISMSSITSKWFGEGEKYVKAVFSLASKMSPSV 237
Query: 449 IFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAV 508
IFVDEVDS+LG R EHEA+R+++NEFM WDGL ++E +R+L+L ATNRPFDLD+AV
Sbjct: 238 IFVDEVDSMLGRREHPREHEASRKIKNEFMMHWDGLTTQERERVLVLAATNRPFDLDEAV 297
Query: 509 IRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAA 568
IRRLPRR+ V LPD NR IL++IL E+L PD ++A++T+GYSGSDLKNLC+TAA
Sbjct: 298 IRRLPRRLMVGLPDTSNRAFILKVILAKEDLSPDLDIGEIASMTNGYSGSDLKNLCVTAA 357
Query: 569 YRPVQELLEEEKAGDNNIT-----------NVALRPLNLDDFVQSKSKVGPSVAYDAVSV 617
+RP++E+LE+EK + + LR LN++DF + V SV+ ++ ++
Sbjct: 358 HRPIKEILEKEKRERDAALAQGKVPPPLSGSSDLRALNVEDFRDAHKWVSASVSSESATM 417
Query: 618 NELRKWNEMYGEGGTRTKSPFGF 640
L++WN+++GEGG+ + F F
Sbjct: 418 TALQQWNKLHGEGGSGKQQSFSF 440
>At1g64110 unknown protein
Length = 824
Score = 396 bits (1017), Expect = e-110
Identities = 239/613 (38%), Positives = 352/613 (56%), Gaps = 104/613 (16%)
Query: 90 SQDCYLAMEALREVL---NSKQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSG 146
S D L +++L +VL + P+++Y D +L +S ++ +++ +LSG
Sbjct: 242 SFDEKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRS------QRTYNLFQKLLQKLSG 295
Query: 147 PIVLICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSN 206
P VLI G +++ + D++++ +F
Sbjct: 296 P-VLILGSRIVDLSSEDAQEI-----------------------------DEKLSAVFPY 325
Query: 207 VVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVIL 266
+ I+PP+DE L ++K QLE D ++ ++ N N + +VL E+ L C DL + +D +
Sbjct: 326 NIDIRPPEDETHLVSWKSQLERDMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKV 385
Query: 267 TKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAIS------------------ 308
E++V A +++L + P + KL + SL S
Sbjct: 386 LSNYIEEIVVSALSYHLMNNKDPEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTK 445
Query: 309 ----------------RLKGVETMS--RQPSQSLK---------NLAKD-EFESNFISAV 340
+ + V T+S +P + K +A D EFE V
Sbjct: 446 EESSKEVKAESIKPETKTESVTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEV 505
Query: 341 VPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKT 400
+P EI V F DIGAL+++K++L ELV+LP+RRP+LFT G LL+P +GILLFGPPGTGKT
Sbjct: 506 IPAEEINVTFKDIGALDEIKESLQELVMLPLRRPDLFT-GGLLKPCRGILLFGPPGTGKT 564
Query: 401 LMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGA 460
++AKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ ASK++P IIFVDEVDS+LG
Sbjct: 565 MLAKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQ 624
Query: 461 RGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDL 520
R EHEA R+++NEFM+ WDGL +K +RIL+L ATNRPFDLD+A+IRR RRI V L
Sbjct: 625 RTRVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGL 684
Query: 521 PDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE- 579
P +NR+KILR +L E +D + + +LA +T+GY+GSDLKNLC TAAYRPV+EL+++E
Sbjct: 685 PAVENREKILRTLLAKEKVDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQER 744
Query: 580 -------------KAGDNNITN----VALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRK 622
KAG+ + + LRPLN DF ++K++V S A + + EL++
Sbjct: 745 IKDTEKKKQREPTKAGEEDEGKEERVITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQ 804
Query: 623 WNEMYGEGGTRTK 635
WNE+YGEGG+R K
Sbjct: 805 WNELYGEGGSRKK 817
>At4g28000 putative protein
Length = 726
Score = 395 bits (1016), Expect = e-110
Identities = 238/582 (40%), Positives = 343/582 (58%), Gaps = 80/582 (13%)
Query: 95 LAMEALREVLNS---KQPLIVYFPDGSQWLHKSVPKSIRSEFFHKV-EEMFDQLSGPIVL 150
L +++L +VL S P+I+Y D V K +SE F+K+ + + +LSGP+++
Sbjct: 187 LFLQSLYKVLVSISETNPIIIYLRD--------VEKLCQSERFYKLFQRLLTKLSGPVLV 238
Query: 151 ICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSNVVSI 210
+ + L D Q+ + I+ LF + I
Sbjct: 239 LGSR-----------------------------LLEPEDDCQEVGEG--ISALFPYNIEI 267
Query: 211 QPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQK 270
+PP+DEN L ++K + E+D K++ + N N + +VL + L C DL + D +
Sbjct: 268 RPPEDENQLMSWKTRFEDDMKVIQFQDNKNHIAEVLAANDLECDDLGSICHADTMFLSSH 327
Query: 271 AEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKG-----VETMSRQPSQSLK 325
E++V A +++L + P K +L + SL ++ L+ +++ + K
Sbjct: 328 IEEIVVSAISYHLMNNKEPEYKNGRLVISSNSLSHGLNILQEGQGCFEDSLKLDTNIDSK 387
Query: 326 NLAKD-EFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLR 384
+A D EFE V+P EIGV F DIG+L++ K++L ELV+LP+RRP+LF G LL+
Sbjct: 388 EVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLFK-GGLLK 446
Query: 385 PVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKL 444
P +GILLFGPPGTGKT+MAKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ A+K+
Sbjct: 447 PCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKV 506
Query: 445 APVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDL 504
+P IIFVDEVDS+LG R EHEA R+++NEFM WDGL S RIL+L ATNRPFDL
Sbjct: 507 SPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFDL 566
Query: 505 DDAVIRRLPRRIYVDLPDADNRKKILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLK 561
D+A+IRR RRI V LP ++R+KILR +L K ENLD F +LA +TDGYSGSDLK
Sbjct: 567 DEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTENLD----FQELAQMTDGYSGSDLK 622
Query: 562 NLCITAAYRPVQELLEEEKAGD-----------------------NNITNVALRPLNLDD 598
N C TAAYRPV+EL+++E D + + LRPL+++D
Sbjct: 623 NFCTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGITLRPLSMED 682
Query: 599 FVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFGF 640
+KS+V S A + +NEL++WN++YGEGG+R K +
Sbjct: 683 MKVAKSQVAASFAAEGAGMNELKQWNDLYGEGGSRKKEQLSY 724
>At1g62130 hypothetical protein
Length = 372
Score = 305 bits (780), Expect = 7e-83
Identities = 177/398 (44%), Positives = 243/398 (60%), Gaps = 54/398 (13%)
Query: 263 DVILTKQKAEKVVGWAKNHYLSSCLLPCVKGD-KLCLPRESLEIAISRLKGVETMSRQPS 321
D+ L + AEK++GWA +H++ S P D ++ L ESL+ G+E + +
Sbjct: 3 DLTLRRDSAEKIIGWALSHHIKSN--PGADPDVRVILSLESLKC------GIELLEIESK 54
Query: 322 QSLKNLAKDE-FESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHG 380
+SLK++ + FE IS ++PP EIGV FDDIGALE+VK L ELV+LP + PELF G
Sbjct: 55 KSLKDIVTENTFE---ISDIIPPSEIGVTFDDIGALENVKDTLKELVMLPFQWPELFCKG 111
Query: 381 NLLR--------------------PVKGILLFGPPGTGKTLMAKALATEAGANFISVSGS 420
L + P GILLFGP GTGKT++AKA+ATEAGAN I++S
Sbjct: 112 QLTKMLTLWIGGFLISLLLYFSTQPCNGILLFGPSGTGKTMLAKAVATEAGANLINMS-- 169
Query: 421 TLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAA 480
S+WF + EK KA+FS ASK++P IIF+DEV+S+L H + +NEF+
Sbjct: 170 --MSRWFSEGEKYVKAVFSLASKISPSIIFLDEVESML--------HRYRLKTKNEFIIN 219
Query: 481 WDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLD 540
WDGLR+ E +R+L+L ATNRPFDLD+AVIRRLP R+ V LPDA +R KIL++IL E+L
Sbjct: 220 WDGLRTNEKERVLVLAATNRPFDLDEAVIRRLPHRLMVGLPDARSRSKILKVILSKEDLS 279
Query: 541 PDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLDDFV 600
PDF D++A++T+GYSG+DLK A E + LR L ++DF
Sbjct: 280 PDFDIDEVASMTNGYSGNDLKERDAAVA---------EGRVPPAGSGGSDLRVLKMEDFR 330
Query: 601 QSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPF 638
+ V S++ +V++ LR+WNE YGEGG+R F
Sbjct: 331 NALELVSMSISSKSVNMTALRQWNEDYGEGGSRRNESF 368
>At4g27680 unknown protein
Length = 398
Score = 275 bits (704), Expect = 4e-74
Identities = 136/274 (49%), Positives = 188/274 (67%), Gaps = 1/274 (0%)
Query: 332 FESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILL 391
+E V+ P I V+F IG LE +K+AL ELVILP++RPELF +G LL P KG+LL
Sbjct: 65 YEDVIACDVINPDHIDVEFGSIGGLETIKQALYELVILPLKRPELFAYGKLLGPQKGVLL 124
Query: 392 FGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFV 451
+GPPGTGKT++AKA+A E+GA FI+V S L SKWFGDA+KL A+FS A KL P IIF+
Sbjct: 125 YGPPGTGKTMLAKAIAKESGAVFINVRVSNLMSKWFGDAQKLVSAVFSLAYKLQPAIIFI 184
Query: 452 DEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRR 511
DEV+S LG R +HEA M+ EFMA WDG + + R+++L ATNRP +LD+A++RR
Sbjct: 185 DEVESFLGQRRST-DHEAMANMKTEFMALWDGFSTDPHARVMVLAATNRPSELDEAILRR 243
Query: 512 LPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRP 571
LP+ + +PD R +IL++ LK E ++PD FD +A L +GY+GSD+ LC AAY P
Sbjct: 244 LPQAFEIGIPDRRERAEILKVTLKGERVEPDIDFDHIARLCEGYTGSDIFELCKKAAYFP 303
Query: 572 VQELLEEEKAGDNNITNVALRPLNLDDFVQSKSK 605
++E+L+ E+ G + L L+L+ + + K
Sbjct: 304 IREILDAERKGKPCLDPRPLSQLDLEKVLATSKK 337
>At5g53540 26S proteasome regulatory particle chain RPT6-like
protein
Length = 403
Score = 273 bits (697), Expect = 3e-73
Identities = 134/276 (48%), Positives = 189/276 (67%), Gaps = 1/276 (0%)
Query: 330 DEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGI 389
+++E V+ P I V+F IG LE +K+AL ELVILP++RPELF +G LL P KG+
Sbjct: 66 NQYEDVIACDVINPLHIDVEFGSIGGLESIKQALYELVILPLKRPELFAYGKLLGPQKGV 125
Query: 390 LLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVII 449
LL+GPPGTGKT++AKA+A E+ A FI+V S L SKWFGDA+KL A+FS A KL P II
Sbjct: 126 LLYGPPGTGKTMLAKAIARESEAVFINVKVSNLMSKWFGDAQKLVSAVFSLAYKLQPAII 185
Query: 450 FVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVI 509
F+DEVDS LG R ++EA M+ EFMA WDG + +N R+++L ATNRP +LD+A++
Sbjct: 186 FIDEVDSFLGQRRST-DNEAMSNMKTEFMALWDGFTTDQNARVMVLAATNRPSELDEAIL 244
Query: 510 RRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAY 569
RR P+ + +PD R +IL+++LK E+++ D +D++A L + Y+GSD+ LC AAY
Sbjct: 245 RRFPQSFEIGMPDCQERAQILKVVLKGESVESDINYDRIARLCEDYTGSDIFELCKKAAY 304
Query: 570 RPVQELLEEEKAGDNNITNVALRPLNLDDFVQSKSK 605
P++E+LE EK G L L+L+ + + K
Sbjct: 305 FPIREILEAEKEGKRVSVPRPLTQLDLEKVLATSKK 340
>At3g27120 spastin protein, putative
Length = 327
Score = 229 bits (585), Expect = 3e-60
Identities = 131/309 (42%), Positives = 190/309 (61%), Gaps = 10/309 (3%)
Query: 322 QSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGN 381
+ L+NL E +S + + V++DDI LE KK + E+VI P+ RP++F
Sbjct: 23 EKLRNLEPRLIEH--VSNEIMDRDPNVRWDDIAGLEHAKKCVTEMVIWPLLRPDIFK--G 78
Query: 382 LLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFA 441
P KG+LLFGPPGTGKT++ KA+A EA A F +S S+LTSKW G+ EKL +ALF A
Sbjct: 79 CRSPGKGLLLFGPPGTGKTMIGKAIAGEAKATFFYISASSLTSKWIGEGEKLVRALFGVA 138
Query: 442 SKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRP 501
S P +IFVDE+DSLL R EHE++RR++ +F+ +G S Q IL++GATNRP
Sbjct: 139 SCRQPAVIFVDEIDSLLSQRKSDGEHESSRRLKTQFLIEMEGFDSGSEQ-ILLIGATNRP 197
Query: 502 FDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLD--PDFQFDKLANLTDGYSGSD 559
+LD+A RRL +R+Y+ LP ++ R I++ +LK + L D + + NLT+GYSGSD
Sbjct: 198 QELDEAARRRLTKRLYIPLPSSEARAWIIQNLLKKDGLFTLSDDDMNIICNLTEGYSGSD 257
Query: 560 LKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNE 619
+KNL A P++E L + N+T +R + L DF + +V PSV+ + + + E
Sbjct: 258 MKNLVKDATMGPLREAL-KRGIDITNLTKDDMRLVTLQDFKDALQEVRPSVSQNELGIYE 316
Query: 620 LRKWNEMYG 628
WN +G
Sbjct: 317 --NWNNQFG 323
>At1g80350 CAD ATPase (AAA1)
Length = 523
Score = 221 bits (564), Expect = 7e-58
Identities = 114/291 (39%), Positives = 183/291 (62%), Gaps = 13/291 (4%)
Query: 347 GVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKAL 406
GV++DD+ L + K+ L E V+LP+ PE F + RP KG+L+FGPPGTGKTL+AKA+
Sbjct: 235 GVRWDDVAGLSEAKRLLEEAVVLPLWMPEYFQ--GIRRPWKGVLMFGPPGTGKTLLAKAV 292
Query: 407 ATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFE 466
ATE G F +VS +TL SKW G++E++ + LF A AP IF+DE+DSL +RGG+ E
Sbjct: 293 ATECGTTFFNVSSATLASKWRGESERMVRCLFDLARAYAPSTIFIDEIDSLCNSRGGSGE 352
Query: 467 HEATRRMRNEFMAAWDGLRSKENQR------ILILGATNRPFDLDDAVIRRLPRRIYVDL 520
HE++RR+++E + DG+ + +++L ATN P+D+D+A+ RRL +RIY+ L
Sbjct: 353 HESSRRVKSELLVQVDGVSNTATNEDGSRKIVMVLAATNFPWDIDEALRRRLEKRIYIPL 412
Query: 521 PDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELL---E 577
PD ++RK ++ I L+ + D + +A T+GYSG DL N+C A+ ++ +
Sbjct: 413 PDFESRKALININLRTVEVASDVNIEDVARRTEGYSGDDLTNVCRDASMNGMRRKIAGKT 472
Query: 578 EEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
++ + + +++ P+ + DF ++ KV PSV+ + +E KW +G
Sbjct: 473 RDEIKNMSKDDISNDPVAMCDFEEAIRKVQPSVSSSDIEKHE--KWLSEFG 521
>At2g34560 katanin like protein
Length = 384
Score = 207 bits (527), Expect = 1e-53
Identities = 116/292 (39%), Positives = 177/292 (59%), Gaps = 15/292 (5%)
Query: 344 GEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMA 403
G +K++ I LE+ KK L E V++P++ P F LL P KGILLFGPPGTGKT++A
Sbjct: 96 GNPNIKWESIKGLENAKKLLKEAVVMPIKYPTYFN--GLLTPWKGILLFGPPGTGKTMLA 153
Query: 404 KALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARG- 462
KA+ATE F ++S S++ SKW GD+EKL + LF A AP IF+DE+D+++ RG
Sbjct: 154 KAVATECNTTFFNISASSVVSKWRGDSEKLIRVLFDLARHHAPSTIFLDEIDAIISQRGG 213
Query: 463 -GAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLP 521
G EHEA+RR++ E + DGL+ K N+ + +L ATN P++LD A++RRL +RI V LP
Sbjct: 214 EGRSEHEASRRLKTELLIQMDGLQ-KTNELVFVLAATNLPWELDAAMLRRLEKRILVPLP 272
Query: 522 DADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELL----- 576
D + R+ + +++ + D D L ++GYSGSD++ LC AA +P++ L
Sbjct: 273 DPEARRGMFEMLIPSQPGDEPLPHDVLVEKSEGYSGSDIRILCKEAAMQPLRRTLAILED 332
Query: 577 EEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
E+ ++ + + P+ +D ++ S PS A + K+N+ YG
Sbjct: 333 REDVVPEDELPKIG--PILPEDIDRALSNTRPSAHLHA---HLYDKFNDDYG 379
>At2g27600 putative AAA-type ATPase
Length = 435
Score = 194 bits (492), Expect = 2e-49
Identities = 102/230 (44%), Positives = 147/230 (63%), Gaps = 8/230 (3%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
+K+ D+ LE K+AL E VILP++ P+ FT RP + LL+GPPGTGK+ +AKA+A
Sbjct: 129 IKWSDVAGLESAKQALQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 186
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
TEA + F SVS S L SKW G++EKL LF A + AP IIFVDE+DSL G RG E
Sbjct: 187 TEADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESAPSIIFVDEIDSLCGTRGEGNES 246
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRK 527
EA+RR++ E + G+ ++++L+L ATN P+ LD A+ RR +RIY+ LP+A R+
Sbjct: 247 EASRRIKTELLVQMQGV-GHNDEKVLVLAATNTPYALDQAIRRRFDKRIYIPLPEAKARQ 305
Query: 528 KILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQE 574
+ ++ L H +PDF++ L T+G+SGSD+ + PV++
Sbjct: 306 HMFKVHLGDTPHNLTEPDFEY--LGQKTEGFSGSDVSVCVKDVLFEPVRK 353
>At3g01610 putative cell division control protein
Length = 703
Score = 178 bits (452), Expect = 7e-45
Identities = 103/280 (36%), Positives = 167/280 (58%), Gaps = 18/280 (6%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
VK+DD+G L+ ++ N ++ P+++P+++ + G LL+GPPG GKTL+AKA A
Sbjct: 408 VKWDDVGGLDHLRLQFNRYIVRPIKKPDIYKAFGVDLET-GFLLYGPPGCGKTLIAKAAA 466
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARG--GAF 465
EAGANF+ + G+ L +K+ G++E + LF A AP +IF DEVD+L +RG GA+
Sbjct: 467 NEAGANFMHIKGAELLNKYVGESELAIRTLFQRARTCAPCVIFFDEVDALTTSRGKEGAW 526
Query: 466 EHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
R+ N+F+ DG E + + ++GATNRP +D A +R R +YV LP+A
Sbjct: 527 ---VVERLLNQFLVELDG---GERRNVYVIGATNRPDVVDPAFLRPGRFGNLLYVPLPNA 580
Query: 524 DNRKKILRIILKHENLDPDFQFDKLA-NLTDGYSGSDLKNLCITAAYRPVQELLEEEKAG 582
D R IL+ I + + +DP D +A N +G+SG+DL +L A ++ V+E++ ++
Sbjct: 581 DERASILKAIARKKPIDPSVDLDGIAKNNCEGFSGADLAHLVQKATFQAVEEMIGSSESS 640
Query: 583 DNNITNVALRPLNLDDFVQSKSKVGPSV------AYDAVS 616
++++T++ + F Q+ S V PSV YDA+S
Sbjct: 641 EDDVTDITQCTIKTRHFEQALSLVSPSVNKQQRRHYDALS 680
Score = 140 bits (352), Expect = 3e-33
Identities = 101/348 (29%), Positives = 175/348 (50%), Gaps = 34/348 (9%)
Query: 313 VETMSRQPSQSLKNLA--KDEFESNFISAVVPPGEIGVK------FDDIGALEDVKKALN 364
VET+S + L + K+ S +S G++ V+ F D G ++ + L
Sbjct: 70 VETVSNKGRSKLATMGARKEAKVSLSLSGATGNGDLEVEGTKGPTFKDFGGIKKILDELE 129
Query: 365 ELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTS 424
V+ P+ PE F + +P GIL GPPG GKT +A A+A EAG F +S + + S
Sbjct: 130 MNVLFPILNPEPFKKIGV-KPPSGILFHGPPGCGKTKLANAIANEAGVPFYKISATEVIS 188
Query: 425 KWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGL 484
G +E+ + LFS A + AP I+F+DE+D+ +G++ + E +R+ + + DG
Sbjct: 189 GVSGASEENIRELFSKAYRTAPSIVFIDEIDA-IGSKRENQQREMEKRIVTQLLTCMDGP 247
Query: 485 RSKENQR--------ILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADNRKKILRIIL 534
+K ++ +L++GATNRP LD A+ R R I + PD D R +IL ++
Sbjct: 248 GNKGDKNAPDSSAGFVLVIGATNRPDALDPALRRSGRFETEIALTAPDEDARAEILSVVA 307
Query: 535 KHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLE---EEKAGDNNITNVAL 591
+ L+ F ++A LT G+ G+DL+++ A + ++ +L+ E++GD L
Sbjct: 308 QKLRLEGPFDKKRIARLTPGFVGADLESVAYLAGRKAIKRILDSRKSEQSGDGEDDKSWL 367
Query: 592 RP-----------LNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
R + + DF ++ + V S+ + S+ KW+++ G
Sbjct: 368 RMPWPEEELEKLFVKMSDFEEAVNLVQASLTREGFSIVPDVKWDDVGG 415
>At3g09840 putative transitional endoplasmic reticulum ATPase
Length = 809
Score = 175 bits (443), Expect = 8e-44
Identities = 95/237 (40%), Positives = 142/237 (59%), Gaps = 8/237 (3%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LE+VK+ L E V P+ PE F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 477 VSWNDIGGLENVKRELQETVQYPVEHPEKFEKFGM-SPSKGVLFYGPPGCGKTLLAKAIA 535
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFISV G L + WFG++E + +F A + AP ++F DE+DS+ RGG
Sbjct: 536 NECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGGGSGG 595
Query: 468 E---ATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPD 522
+ A R+ N+ + DG+ +K+ + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 596 DGGGAADRVLNQLLTEMDGMNAKKT--VFIIGATNRPDIIDSALLRPGRLDQLIYIPLPD 653
Query: 523 ADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE 579
D+R I + L+ + D LA T G+SG+D+ +C A ++E +E++
Sbjct: 654 EDSRLNIFKAALRKSPIAKDVDIGALAKYTQGFSGADITEICQRACKYAIRENIEKD 710
Score = 166 bits (420), Expect = 4e-41
Identities = 137/432 (31%), Positives = 208/432 (47%), Gaps = 33/432 (7%)
Query: 214 KDENLLATFKKQLEED--RKIVISRSNLN-ELRKVLEEHQLSCVDLLHLDSDDVILTKQK 270
KD +A + EE R + RSNL L V+ HQ C D+ + ++
Sbjct: 69 KDTVCIALADETCEEPKIRMNKVVRSNLRVRLGDVISVHQ--CPDVKYGKRVHILPVDDT 126
Query: 271 AEKVVGWAKNHYLSSCLL----PCVKGDKLCLPR---ESLEIAISRLKGVETMSRQPSQS 323
E V G + YL L P KGD L L R S+E + E P
Sbjct: 127 VEGVTGNLFDAYLKPYFLEAYRPVRKGD-LFLVRGGMRSVEFKVIETDPAEYCVVAPDTE 185
Query: 324 L---KNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHG 380
+ K E E V +DD+G + + ELV LP+R P+LF
Sbjct: 186 IFCEGEPVKREDEERLDD---------VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSI 236
Query: 381 NLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSF 440
+ +P KGILL+GPPG+GKTL+A+A+A E GA F ++G + SK G++E + F
Sbjct: 237 GV-KPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEIMSKLAGESESNLRKAFEE 295
Query: 441 ASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNR 500
A K AP IIF+DE+DS+ R E RR+ ++ + DGL+S+ + ++++GATNR
Sbjct: 296 AEKNAPSIIFIDEIDSIAPKREKT-NGEVERRIVSQLLTLMDGLKSRAH--VIVMGATNR 352
Query: 501 PFDLDDAVIR--RLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGS 558
P +D A+ R R R I + +PD R ++LRI K+ L D ++++ T GY G+
Sbjct: 353 PNSIDPALRRFGRFDREIDIGVPDEIGRLEVLRIHTKNMKLAEDVDLERISKDTHGYVGA 412
Query: 559 DLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNL--DDFVQSKSKVGPSVAYDAVS 616
DL LC AA + ++E ++ D++I L + + + F + PS + V
Sbjct: 413 DLAALCTEAALQCIREKMDVIDLEDDSIDAEILNSMAVTNEHFHTALGNSNPSALRETVV 472
Query: 617 VNELRKWNEMYG 628
WN++ G
Sbjct: 473 EVPNVSWNDIGG 484
>At5g03340 transitional endoplasmic reticulum ATPase
Length = 810
Score = 172 bits (436), Expect = 5e-43
Identities = 94/236 (39%), Positives = 141/236 (58%), Gaps = 7/236 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LE+VK+ L E V P+ PE F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 477 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGM-SPSKGVLFYGPPGCGKTLLAKAIA 535
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFISV G L + WFG++E + +F A + AP ++F DE+DS+ RG +
Sbjct: 536 NECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSAGD 595
Query: 468 E--ATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ +K+ + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 596 AGGAADRVLNQLLTEMDGMNAKKT--VFIIGATNRPDIIDSALLRPGRLDQLIYIPLPDE 653
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE 579
D+R I + L+ + D LA T G+SG+D+ +C A ++E +E++
Sbjct: 654 DSRLNIFKACLRKSPVAKDVDVTALAKYTQGFSGADITEICQRACKYAIRENIEKD 709
Score = 165 bits (417), Expect = 8e-41
Identities = 137/432 (31%), Positives = 209/432 (47%), Gaps = 33/432 (7%)
Query: 214 KDENLLATFKKQLEED--RKIVISRSNLN-ELRKVLEEHQLSCVDLLHLDSDDVILTKQK 270
KD +A + EE R + RSNL L V+ HQ C D+ + ++
Sbjct: 69 KDTVCIALADETCEEPKIRMNKVVRSNLRVRLGDVISVHQ--CPDVKYGKRVHILPVDDT 126
Query: 271 AEKVVGWAKNHYLSSCLL----PCVKGDKLCLPR---ESLEIAISRLKGVETMSRQPSQS 323
E V G + YL L P KGD L L R S+E + E P
Sbjct: 127 VEGVTGNLFDAYLKPYFLEAYRPVRKGD-LFLVRGGMRSVEFKVIETDPAEYCVVAPDTE 185
Query: 324 L---KNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHG 380
+ K E E E+G +DD+G + + ELV LP+R P+LF
Sbjct: 186 IFCEGEPVKREDEERL-------DEVG--YDDVGGVRKQMAQIRELVELPLRHPQLFKSI 236
Query: 381 NLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSF 440
+ +P KGILL+GPPG+GKTL+A+A+A E GA F ++G + SK G++E + F
Sbjct: 237 GV-KPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEIMSKLAGESESNLRKAFEE 295
Query: 441 ASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNR 500
A K AP IIF+DE+DS+ R E RR+ ++ + DGL+S+ + ++++GATNR
Sbjct: 296 AEKNAPSIIFIDEIDSIAPKREKT-NGEVERRIVSQLLTLMDGLKSRAH--VIVMGATNR 352
Query: 501 PFDLDDAVIR--RLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGS 558
P +D A+ R R R I + +PD R ++LRI K+ L D ++++ T GY G+
Sbjct: 353 PNSIDPALRRFGRFDREIDIGVPDEIGRLEVLRIHTKNMKLAEDVDLERISKDTHGYVGA 412
Query: 559 DLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNL--DDFVQSKSKVGPSVAYDAVS 616
DL LC AA + ++E ++ D++I L + + + F + PS + V
Sbjct: 413 DLAALCTEAALQCIREKMDVIDLEDDSIDAEILNSMAVSNEHFHTALGNSNPSALRETVV 472
Query: 617 VNELRKWNEMYG 628
W ++ G
Sbjct: 473 EVPNVSWEDIGG 484
>At3g53230 CDC48 - like protein
Length = 815
Score = 169 bits (428), Expect = 4e-42
Identities = 91/236 (38%), Positives = 142/236 (59%), Gaps = 7/236 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LE+VK+ L E V P+ PE F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 478 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGM-SPSKGVLFYGPPGCGKTLLAKAIA 536
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFIS+ G L + WFG++E + +F A + AP ++F DE+DS+ RG +
Sbjct: 537 NECQANFISIKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSVGD 596
Query: 468 E--ATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ +K+ + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 597 AGGAADRVLNQLLTEMDGMNAKKT--VFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDE 654
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE 579
++R +I + L+ + D LA T G+SG+D+ +C + ++E +E++
Sbjct: 655 ESRYQIFKSCLRKSPVAKDVDLRALAKYTQGFSGADITEICQRSCKYAIRENIEKD 710
Score = 164 bits (416), Expect = 1e-40
Identities = 101/285 (35%), Positives = 157/285 (54%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DD+G + + ELV LP+R P+LF + +P KGILL+GPPG+GKTL+A+A+A
Sbjct: 205 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGV-KPPKGILLYGPPGSGKTLIARAVA 263
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+DS+ R
Sbjct: 264 NETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HG 322
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL+S+ + ++++GATNRP +D A+ R R R I + +PD
Sbjct: 323 EVERRIVSQLLTLMDGLKSRAH--VIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIG 380
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R ++LRI K+ L D ++++ T GY G+DL LC AA + ++E ++ D
Sbjct: 381 RLEVLRIHTKNMKLAEDVDLERVSKDTHGYVGADLAALCTEAALQCIREKMDVIDLDDEE 440
Query: 586 ITNVALRPLNL--DDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I L + + D F + PS + V W ++ G
Sbjct: 441 IDAEILNSMAVSNDHFQTALGNSNPSALRETVVEVPNVSWEDIGG 485
>At2g45500 hypothetical protein
Length = 481
Score = 168 bits (425), Expect = 1e-41
Identities = 115/307 (37%), Positives = 167/307 (53%), Gaps = 67/307 (21%)
Query: 317 SRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPEL 376
S +P + N+ D+ I+ + VK+DD+ L K+AL E+VILP +R +L
Sbjct: 183 SPKPVKESGNVYDDKLVE-MINTTIVDRSPSVKWDDVAGLNGAKQALLEMVILPAKRRDL 241
Query: 377 FTHGNLLRPVK-----GILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAE 431
FT L RP + G+LLFGPPG GKT++AKA+A+E+ A F +VS S+LTSKW
Sbjct: 242 FT--GLRRPARVTSLLGLLLFGPPGNGKTMLAKAVASESQATFFNVSASSLTSKW----- 294
Query: 432 KLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQR 491
+DS++ R + E+EA+RR+++EF+ +DG+ S +
Sbjct: 295 ----------------------IDSIMSTRSTS-ENEASRRLKSEFLIQFDGVTSNPDDL 331
Query: 492 ILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILK---HENLDPDFQFDKL 548
++I+GATN+P +LDDAV+RRL +RIYV LPD++ RK + + LK H D D DK+
Sbjct: 332 VIIIGATNKPQELDDAVLRRLVKRIYVPLPDSNVRKLLFKTKLKCQPHSLSDGD--IDKI 389
Query: 549 ANLTDG--------------------YSGSDLKNLCITAAYRPVQELLEEEKAGDNNITN 588
T+G YSGSDL+ LC AA P++EL G N +T
Sbjct: 390 VKETEGKLYRLCIKKHRFISQVTDKRYSGSDLQALCEEAAMMPIREL------GANILTI 443
Query: 589 VALRPLN 595
A + LN
Sbjct: 444 QANKVLN 450
>At1g03000 putative peroxisome assembly factor-2
Length = 941
Score = 161 bits (407), Expect = 1e-39
Identities = 105/306 (34%), Positives = 175/306 (56%), Gaps = 21/306 (6%)
Query: 328 AKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVK 387
A D + SA+ P VK+DD+G LEDVK ++ + V LP+ +LF+ G LR
Sbjct: 635 ALDRSKKRNASALGAPKVPNVKWDDVGGLEDVKTSILDTVQLPLLHKDLFSSG--LRKRS 692
Query: 388 GILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPV 447
G+LL+GPPGTGKTL+AKA+ATE NF+SV G L + + G++EK + +F A P
Sbjct: 693 GVLLYGPPGTGKTLLAKAVATECSLNFLSVKGPELINMYIGESEKNVRDIFEKARSARPC 752
Query: 448 IIFVDEVDSLLGARGGAFEHEATR-RMRNEFMAAWDGLRSKENQRILILGATNRPFDLDD 506
+IF DE+DSL ARG + + R+ ++ +A DGL S +Q + I+GA+NRP +D
Sbjct: 753 VIFFDELDSLAPARGASGDSGGVMDRVVSQMLAEIDGL-SDSSQDLFIIGASNRPDLIDP 811
Query: 507 AVIR--RLPRRIYVDL-PDADNRKKILRIILKHENLDPDFQFDKLA-NLTDGYSGSDLKN 562
A++R R + +YV + DA R+++L+ + + L D +A ++G+D+
Sbjct: 812 ALLRPGRFDKLLYVGVNADASYRERVLKALTRKFKLSEDVSLYSVAKKCPSTFTGADMYA 871
Query: 563 LCITAAYRPVQELLEEEKAGD-----NNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSV 617
LC A ++ + + + +GD ++ +V + + DF+++ ++ PS +S+
Sbjct: 872 LCADAWFQAAKRKVSKSDSGDMPTEEDDPDSVVVEYV---DFIKAMDQLSPS-----LSI 923
Query: 618 NELRKW 623
EL+K+
Sbjct: 924 TELKKY 929
Score = 31.6 bits (70), Expect = 1.4
Identities = 20/78 (25%), Positives = 35/78 (44%), Gaps = 2/78 (2%)
Query: 389 ILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVI 448
+LL G PG GK + K +A G + + S +L + F+ A + +P I
Sbjct: 380 VLLHGIPGCGKRTVVKYVARRLGLHVVEFSCHSLLASSERKTSTALAQTFNMARRYSPTI 439
Query: 449 IFVDEVDSL--LGARGGA 464
+ + D LG++ G+
Sbjct: 440 LLLRHFDVFKNLGSQDGS 457
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,968,656
Number of Sequences: 26719
Number of extensions: 666820
Number of successful extensions: 2407
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 124
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 2043
Number of HSP's gapped (non-prelim): 224
length of query: 645
length of database: 11,318,596
effective HSP length: 106
effective length of query: 539
effective length of database: 8,486,382
effective search space: 4574159898
effective search space used: 4574159898
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0135.9


