

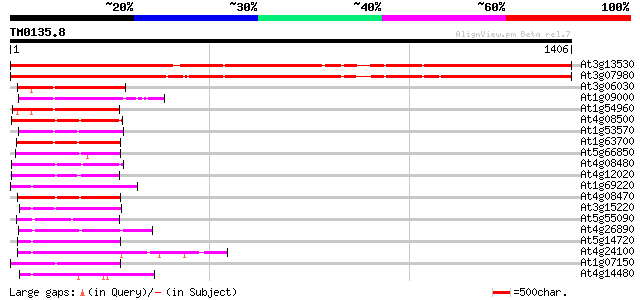
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.8
(1406 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g13530 MAP3K epsilon protein kinase 1899 0.0
At3g07980 putative MAP3K epsilon protein kinase 1809 0.0
At3g06030 NPK1-related protein kinase 3 227 3e-59
At1g09000 NPK1-related protein kinase 1S (ANP1) 225 2e-58
At1g54960 NPK1-related protein kinase 2 (ANP2) 223 8e-58
At4g08500 MEKK1/MAP kinase kinase kinase 208 2e-53
At1g53570 MEK kinase MAP3Ka, putative 203 5e-52
At1g63700 putative protein kinase 202 8e-52
At5g66850 MAP protein kinase like protein 197 4e-50
At4g08480 putative mitogen-activated protein kinase 196 8e-50
At4g12020 putative disease resistance protein 193 5e-49
At1g69220 serine/threonine kinase (SIK1) 193 5e-49
At4g08470 putative mitogen-activated protein kinase 192 1e-48
At3g15220 putative MAP kinase 191 3e-48
At5g55090 putative protein 152 1e-36
At4g26890 putative NPK1-related protein kinase 152 2e-36
At5g14720 protein kinase -like protein 151 3e-36
At4g24100 unknown protein 149 1e-35
At1g07150 hypothetical protein 148 2e-35
At4g14480 kinase like protein 145 2e-34
>At3g13530 MAP3K epsilon protein kinase
Length = 1368
Score = 1899 bits (4919), Expect = 0.0
Identities = 1022/1429 (71%), Positives = 1146/1429 (79%), Gaps = 84/1429 (5%)
Query: 1 MSRQSTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQED 60
M+RQ TSS F KSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENI QED
Sbjct: 1 MARQMTSSQFHKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIVQED 60
Query: 61 LNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESL 120
LN IM+ I++L+NLNHKNIVKYLGS KTK+HLHIILEYVENGSLANIIKPNKFGPFPESL
Sbjct: 61 LNTIMQEIDLLKNLNHKNIVKYLGSSKTKTHLHIILEYVENGSLANIIKPNKFGPFPESL 120
Query: 121 VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVV 180
VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKL EAD+NTHSVV
Sbjct: 121 VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLNEADVNTHSVV 180
Query: 181 GTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIP 240
GTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQD+ PPIP
Sbjct: 181 GTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDDNPPIP 240
Query: 241 DSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGTLRTIKDDGSA 300
DSLSP+ITDFL QCFKKD+RQRPDAKTLLSHPWI N RR LQSSLRHSGT++ +K+ ++
Sbjct: 241 DSLSPDITDFLRQCFKKDSRQRPDAKTLLSHPWIRNSRRALQSSLRHSGTIKYMKEATAS 300
Query: 301 VAEVSGGDHKS----TGEGSSVEKEDSAKEFSTGEANSRKSHEDNASDSNFSNERTEK-E 355
+ G + +GE + K DS + +S +S +D ++ S+ E T+ E
Sbjct: 301 SEKDDEGSQDAAESLSGENVGISKTDSKSKLPLVGVSSFRSEKDQSTPSDLGEEGTDNSE 360
Query: 356 DDIPSDQVLTLAIREKSFLRTGSGNLSSSIEVVSA-----EPTGTETSNAKDLHEVIMNG 410
DDI SDQV TL+I EKS G+ S S E TETS A+ I +
Sbjct: 361 DDIMSDQVPTLSIHEKSSDAKGTPQDVSDFHGKSERGETPENLVTETSEARKNTSAIKH- 419
Query: 411 EVESPQSRGKANMAVGKDSSINNRTKPFAFEPRGQDSGSLKAMKIPPPVEGNELSRFSDP 470
VGK+ SI +F +G++ G KA+K P V GNEL+RFSDP
Sbjct: 420 --------------VGKELSIPVDQTSHSFGRKGEERGIRKAVKTPSSVSGNELARFSDP 465
Query: 471 PGDAYLDDLFHPSDKQSGEVVAEASTSTSTSHIAKGNVSMNDGGKNDLAKELRATIARKQ 530
PGDA L DLFHP DK S EASTS TS++ +G+ + DGGKNDLA +LRATIA+KQ
Sbjct: 466 PGDASLHDLFHPLDKVSEGKPNEASTSMPTSNVNQGDSPVADGGKNDLATKLRATIAQKQ 525
Query: 531 WEKESEIGQENNGGNLLHRVMIGVLKDDVIDIDGLVFDDKLPGENLFPLQAVEFSKLVGS 590
E E+ G N+GG+L R+M+GVLKDDVIDIDGLVFD+K+P ENLFPLQAVEFS+LV S
Sbjct: 526 MEGET--GHSNDGGDLF-RLMMGVLKDDVIDIDGLVFDEKVPAENLFPLQAVEFSRLVSS 582
Query: 591 LRTEESEDVIVSACQKLIGIFHQRPEQKIVFVTQHGLLPLTDLLEVPKTRVICSVLQLIN 650
LR +ESED IVS+CQKL+ +F QRPEQK+VFVTQHG LPL DLL++PK+RVIC+VLQLIN
Sbjct: 583 LRPDESEDAIVSSCQKLVAMFRQRPEQKVVFVTQHGFLPLMDLLDIPKSRVICAVLQLIN 642
Query: 651 QIVKDNTDFQENACLVGLIPAVMSFA--VSDRPREIRMEAAYFLQQLCQSSSLTLQMFIA 708
+I+KDNTDFQENACLVGLIP VMSFA DR REIR EAAYFLQQLCQSS LTLQMFIA
Sbjct: 643 EIIKDNTDFQENACLVGLIPVVMSFAGPERDRSREIRKEAAYFLQQLCQSSPLTLQMFIA 702
Query: 709 CRGIPVLVGFLEADYAKYREMVHLAIDGMWQVFKLQQTTPRNDFCRIAAKNGILLRLINT 768
CRGIPVLVGFLEADYAKYREMVHLAIDGMWQVFKL+++TPRNDFCRIAAKNGILLRLINT
Sbjct: 703 CRGIPVLVGFLEADYAKYREMVHLAIDGMWQVFKLKRSTPRNDFCRIAAKNGILLRLINT 762
Query: 769 LYSLNESTRLASMSAGGGFLADGSAQRPRSGILDPAHPFMNQND-AQLSSADQQDLSKVR 827
LYSLNE+TRLAS+S G DG A R RSG LDP +P QN+ + LS DQ D+ K R
Sbjct: 763 LYSLNEATRLASISGG----LDGQAPRVRSGQLDPNNPIFGQNETSSLSMIDQPDVLKTR 818
Query: 828 RGVLDHHLEPMHASSSNPRRSDANYPT----DVDRPQSSNAAAEAVSLGKSLNLTSRESS 883
G + EP HAS+SN +RSD + P D D+P+ S+ A +A + G
Sbjct: 819 HGGGE---EPSHASTSNSQRSDVHQPDALHPDGDKPRVSSVAPDASTSG----------- 864
Query: 884 VVALKERENVDRWKTDPSRAEVEPRQQRSSISANRTSTDRPPKLAEPSSNGLSMTGATQQ 943
E +Q R S+SANRTSTD+ KLAE +SNG +T Q
Sbjct: 865 -------------------TEDVRQQHRISLSANRTSTDKLQKLAEGASNGFPVT---QT 902
Query: 944 EQVRPLLSLLEKEPPSGRFSGQLEYVRQFSGLERHESVLPLLHAT-EKKTNGELDFLMAE 1002
EQVRPLLSLL+KEPPS +SGQL+YV+ +G+ERHES LPLLH + EKK NG+LDFLMAE
Sbjct: 903 EQVRPLLSLLDKEPPSRHYSGQLDYVKHITGIERHESRLPLLHGSNEKKNNGDLDFLMAE 962
Query: 1003 FADVSQRGRENGNLDSSARVSHKVAPKKLGTFGSSEGAASTSGIVSQTASGVLSGSGVLN 1062
FA+VS RG+ENG+LD++ R K KK+ EG ASTSGI SQTASGVLSGSGVLN
Sbjct: 963 FAEVSGRGKENGSLDTTTRYPSKTMTKKVLAI---EGVASTSGIASQTASGVLSGSGVLN 1019
Query: 1063 ARPGSATSSGLLSHMVSSLNADVAREYLEKVADLLLEFAQADTTVKSYMCSQSLLSRLFQ 1122
ARPGSATSSGLL+HMVS+L+ADVAREYLEKVADLLLEFA+ADTTVKSYMCSQSLLSRLFQ
Sbjct: 1020 ARPGSATSSGLLAHMVSTLSADVAREYLEKVADLLLEFARADTTVKSYMCSQSLLSRLFQ 1079
Query: 1123 MFNRVEPPILLKILKCINHLSTDPNCLENLQRAEAIKHLIPNLELKEGTLVSEIHHEVLN 1182
MFNRVEPPILLKIL+C NHLSTDPNCLENLQRA+AIKHLIPNLELK+G LV +IHHEVL+
Sbjct: 1080 MFNRVEPPILLKILECTNHLSTDPNCLENLQRADAIKHLIPNLELKDGHLVYQIHHEVLS 1139
Query: 1183 ALFNLCKINKRRQEQAAENGIIPHLMQFITSNSPLKQYALPLLCDMAHASRNSREQLRAH 1242
ALFNLCKINKRRQEQAAENGIIPHLM FI S+SPLKQYALPLLCDMAHASRNSREQLRAH
Sbjct: 1140 ALFNLCKINKRRQEQAAENGIIPHLMLFIMSDSPLKQYALPLLCDMAHASRNSREQLRAH 1199
Query: 1243 GGLDVYLNLLEDELWSVTALDSIAVCLAHDNDNRKVEQSLLKKDAVQKLVKFFQSCPEQH 1302
GGLDVYL+LL+DE WSV ALDSIAVCLA DNDNRKVEQ+LLK+DA+QKLV FFQSCPE+H
Sbjct: 1200 GGLDVYLSLLDDEYWSVIALDSIAVCLAQDNDNRKVEQALLKQDAIQKLVDFFQSCPERH 1259
Query: 1303 FVHILEPFLKIITKSSRINTTLAVNGLTPLLIARLDHQDAIARLNLLRLIKAVYEHHPQP 1362
FVHILEPFLKIITKS RIN TLAVNGLTPLLI+RLDHQDAIARLNLL+LIKAVYEHHP+P
Sbjct: 1260 FVHILEPFLKIITKSYRINKTLAVNGLTPLLISRLDHQDAIARLNLLKLIKAVYEHHPRP 1319
Query: 1363 KKLIVENDLPEKLQNLIGERRD-----GQVLVKQMATSLLKALHINTVL 1406
K+LIVENDLP+KLQNLI ERRD GQVLVKQMATSLLKALHINT+L
Sbjct: 1320 KQLIVENDLPQKLQNLIEERRDGQRSGGQVLVKQMATSLLKALHINTIL 1368
>At3g07980 putative MAP3K epsilon protein kinase
Length = 1367
Score = 1809 bits (4686), Expect = 0.0
Identities = 981/1424 (68%), Positives = 1123/1424 (77%), Gaps = 75/1424 (5%)
Query: 1 MSRQSTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQED 60
M+RQ TSS F KSKTLDNKYMLGDEIGKGAYGRVY GLDLENGDFVAIKQVSLENI QED
Sbjct: 1 MARQMTSSQFHKSKTLDNKYMLGDEIGKGAYGRVYIGLDLENGDFVAIKQVSLENIGQED 60
Query: 61 LNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESL 120
LN IM+ I++L+NLNHKNIVKYLGSLKTK+HLHIILEYVENGSLANIIKPNKFGPFPESL
Sbjct: 61 LNTIMQEIDLLKNLNHKNIVKYLGSLKTKTHLHIILEYVENGSLANIIKPNKFGPFPESL 120
Query: 121 VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVV 180
V VYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKL EAD NTHSVV
Sbjct: 121 VTVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLNEADFNTHSVV 180
Query: 181 GTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIP 240
GTPYWMAPEVIE+SGVCAASDIWSVGCT+IELLTCVPPYYDLQPMPAL+RIVQD+ PPIP
Sbjct: 181 GTPYWMAPEVIELSGVCAASDIWSVGCTIIELLTCVPPYYDLQPMPALYRIVQDDTPPIP 240
Query: 241 DSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGTLRTIKD-DGS 299
DSLSP+ITDFL CFKKD+RQRPDAKTLLSHPWI N RR L+SSLRHSGT+R +K+ D S
Sbjct: 241 DSLSPDITDFLRLCFKKDSRQRPDAKTLLSHPWIRNSRRALRSSLRHSGTIRYMKETDSS 300
Query: 300 AVAEVSGGDH---KSTGEGSSVEKEDSAKEFSTGEANSRKSHEDNASDSNFSNERTEKED 356
+ + G + E V K +S + S +S +D +S S+ E T+ ED
Sbjct: 301 SEKDAEGSQEVVESVSAEKVEVTKTNSKSKLPVIGGASFRSEKDQSSPSDLGEEGTDSED 360
Query: 357 DIPSDQVLTLAIREKSFLRTGSGNLSSSIEVVSAEPTGTETSNAKDLHEVIMNGEVESPQ 416
DI SDQ TL++ +KS ++G+ ++SS + S + E D E+ N E E+ +
Sbjct: 361 DINSDQGPTLSMHDKSSRQSGTCSISSDAKGTSQDVL--ENHEKYDRDEIPGNLETEASE 418
Query: 417 SRGK--ANMAVGKDSSINNRTKPFAFEPRGQDSGSLKAMKIPPPVEGNELSRFSDPPGDA 474
R A VGK+ SI + +F +G+D G KA+K P GNEL+RFSDPPGDA
Sbjct: 419 GRRNTLATKLVGKEYSIQSS---HSFSQKGED-GLRKAVKTPSSFGGNELTRFSDPPGDA 474
Query: 475 YLDDLFHPSDKQSGEVVAEASTSTSTSHIAKGNVSMNDGGKNDLAKELRATIARKQWEKE 534
L DLFHP DK EASTST T+++ +G+ + DGGKNDLA +LRA IA+KQ E E
Sbjct: 475 SLHDLFHPLDKVPEGKTNEASTSTPTANVNQGDSPVADGGKNDLATKLRARIAQKQMEGE 534
Query: 535 SEIGQENNGGNLLHRVMIGVLKDDVIDIDGLVFDDKLPGENLFPLQAVEFSKLVGSLRTE 594
+ G +GG+L R+M+GVLKDDV++ID LVFD+K+P ENLFPLQAVEFS+LV SLR +
Sbjct: 535 T--GHSQDGGDLF-RLMMGVLKDDVLNIDDLVFDEKVPPENLFPLQAVEFSRLVSSLRPD 591
Query: 595 ESEDVIVSACQKLIGIFHQRPEQKIVFVTQHGLLPLTDLLEVPKTRVICSVLQLINQIVK 654
ESED IV++ KL+ +F QRP QK VFVTQ+G LPL DLL++PK+RVIC+VLQLIN+IVK
Sbjct: 592 ESEDAIVTSSLKLVAMFRQRPGQKAVFVTQNGFLPLMDLLDIPKSRVICAVLQLINEIVK 651
Query: 655 DNTDFQENACLVGLIPAVMSFA--VSDRPREIRMEAAYFLQQLCQSSSLTLQMFIACRGI 712
DNTDF ENACLVGLIP VMSFA DR REIR EAAYFLQQLCQSS LTLQMFI+CRGI
Sbjct: 652 DNTDFLENACLVGLIPLVMSFAGFERDRSREIRKEAAYFLQQLCQSSPLTLQMFISCRGI 711
Query: 713 PVLVGFLEADYAKYREMVHLAIDGMWQVFKLQQTTPRNDFCRIAAKNGILLRLINTLYSL 772
PVLVGFLEADYAK+REMVHLAIDGMWQVFKL+++T RNDFCRIAAKNGILLRL+NTLYSL
Sbjct: 712 PVLVGFLEADYAKHREMVHLAIDGMWQVFKLKKSTSRNDFCRIAAKNGILLRLVNTLYSL 771
Query: 773 NESTRLASMSAGGGFLADGSAQRPRSGILDPAHPFMNQNDAQLSSADQQDLSKVRRGVLD 832
+E+TRLAS+S G + DG R RSG LDP +P +Q + S D D K R G +
Sbjct: 772 SEATRLASIS-GDALILDGQTPRARSGQLDPNNPIFSQRETSPSVIDHPDGLKTRNGGGE 830
Query: 833 HHLEPMHASSSNPRRSDANYPT----DVDRPQSSNAAAEAVSLGKSLNLTSRESSVVALK 888
EP HA +SN + SD + P D DRP+ S+ A+A
Sbjct: 831 ---EPSHALTSNSQSSDVHQPDALHPDGDRPRLSSVVADAT------------------- 868
Query: 889 ERENVDRWKTDPSRAEVEPRQQRSSISANRTSTDRPPKLAEPSSNGLSMTGATQQEQVRP 948
E +Q R S+SANRTSTD+ KLAE +SNG +T Q +QVRP
Sbjct: 869 ---------------EDVIQQHRISLSANRTSTDKLQKLAEGASNGFPVT---QPDQVRP 910
Query: 949 LLSLLEKEPPSGRFSGQLEYVRQFSGLERHESVLPLLHAT-EKKTNGELDFLMAEFADVS 1007
LLSLLEKEPPS + SGQL+YV+ +G+ERHES LPLL+A+ EKKTNG+L+F+MAEFA+VS
Sbjct: 911 LLSLLEKEPPSRKISGQLDYVKHIAGIERHESRLPLLYASDEKKTNGDLEFIMAEFAEVS 970
Query: 1008 QRGRENGNLDSSARVSHKVAPKKLGTFGSSEGAASTSGIVSQTASGVLSGSGVLNARPGS 1067
RG+ENGNLD++ R S K KK+ E AST GI SQTASGVLSGSGVLNARPGS
Sbjct: 971 GRGKENGNLDTAPRYSSKTMTKKVMAI---ERVASTCGIASQTASGVLSGSGVLNARPGS 1027
Query: 1068 ATSSGLLSHMVSSLNADVAREYLEKVADLLLEFAQADTTVKSYMCSQSLLSRLFQMFNRV 1127
TSSGLL+H +L+ADV+ +YLEKVADLLLEFA+A+TTVKSYMCSQSLLSRLFQMFNRV
Sbjct: 1028 TTSSGLLAH---ALSADVSMDYLEKVADLLLEFARAETTVKSYMCSQSLLSRLFQMFNRV 1084
Query: 1128 EPPILLKILKCINHLSTDPNCLENLQRAEAIKHLIPNLELKEGTLVSEIHHEVLNALFNL 1187
EPPILLKIL+C NHLSTDPNCLENLQRA+AIK LIPNLELKEG LV +IHHEVL+ALFNL
Sbjct: 1085 EPPILLKILECTNHLSTDPNCLENLQRADAIKQLIPNLELKEGPLVYQIHHEVLSALFNL 1144
Query: 1188 CKINKRRQEQAAENGIIPHLMQFITSNSPLKQYALPLLCDMAHASRNSREQLRAHGGLDV 1247
CKINKRRQEQAAENGIIPHLM F+ S+SPLKQYALPLLCDMAHASRNSREQLRAHGGLDV
Sbjct: 1145 CKINKRRQEQAAENGIIPHLMLFVMSDSPLKQYALPLLCDMAHASRNSREQLRAHGGLDV 1204
Query: 1248 YLNLLEDELWSVTALDSIAVCLAHDNDNRKVEQSLLKKDAVQKLVKFFQSCPEQHFVHIL 1307
YL+LL+DE WSV ALDSIAVCLA D D +KVEQ+ LKKDA+QKLV FFQ+CPE+HFVHIL
Sbjct: 1205 YLSLLDDEYWSVIALDSIAVCLAQDVD-QKVEQAFLKKDAIQKLVNFFQNCPERHFVHIL 1263
Query: 1308 EPFLKIITKSSRINTTLAVNGLTPLLIARLDHQDAIARLNLLRLIKAVYEHHPQPKKLIV 1367
EPFLKIITKSS IN TLA+NGLTPLLIARLDHQDAIARLNLL+LIKAVYE HP+PK+LIV
Sbjct: 1264 EPFLKIITKSSSINKTLALNGLTPLLIARLDHQDAIARLNLLKLIKAVYEKHPKPKQLIV 1323
Query: 1368 ENDLPEKLQNLIGERRD-----GQVLVKQMATSLLKALHINTVL 1406
ENDLP+KLQNLI ERRD GQVLVKQMATSLLKALHINT+L
Sbjct: 1324 ENDLPQKLQNLIEERRDGQRSGGQVLVKQMATSLLKALHINTIL 1367
>At3g06030 NPK1-related protein kinase 3
Length = 651
Score = 227 bits (579), Expect = 3e-59
Identities = 122/282 (43%), Positives = 184/282 (64%), Gaps = 12/282 (4%)
Query: 19 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSL------ENIAQEDLNIIMEMINILQ 72
++ G+ IG GA+GRVY G++L++G+ +AIKQV + + Q + + E + +L+
Sbjct: 67 RWRKGELIGCGAFGRVYMGMNLDSGELLAIKQVLIAPSSASKEKTQGHIRELEEEVQLLK 126
Query: 73 NLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGL 132
NL+H NIV+YLG+++ L+I++E+V GS++++++ KFG FPE ++ +Y Q+L GL
Sbjct: 127 NLSHPNIVRYLGTVRESDSLNILMEFVPGGSISSLLE--KFGSFPEPVIIMYTKQLLLGL 184
Query: 133 VYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTE-ADIN-THSVVGTPYWMAPEV 190
YLH G++HRDIKGANIL +G ++LADFG + K+ E A +N S+ GTPYWMAPEV
Sbjct: 185 EYLHNNGIMHRDIKGANILVDNKGCIRLADFGASKKVVELATVNGAKSMKGTPYWMAPEV 244
Query: 191 IEMSGVCAASDIWSVGCTVIELLTCVPPYYD-LQPMPALFRIVQDE-QPPIPDSLSPNIT 248
I +G ++DIWSVGCTVIE+ T PP+ + Q A+ I + + PPIP+ LSP
Sbjct: 245 ILQTGHSFSADIWSVGCTVIEMATGKPPWSEQYQQFAAVLHIGRTKAHPPIPEDLSPEAK 304
Query: 249 DFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGT 290
DFL +C K+ R A LL HP++ R+ + R+S T
Sbjct: 305 DFLMKCLHKEPSLRLSATELLQHPFVTGKRQEPYPAYRNSLT 346
>At1g09000 NPK1-related protein kinase 1S (ANP1)
Length = 666
Score = 225 bits (573), Expect = 2e-58
Identities = 140/374 (37%), Positives = 222/374 (58%), Gaps = 25/374 (6%)
Query: 23 GDEIGKGAYGRVYKGLDLENGDFVAIKQVSLE-NIAQED-----LNIIMEMINILQNLNH 76
G IG+GA+G VY G++L++G+ +A+KQV + N A ++ + + E + +L+NL+H
Sbjct: 72 GQLIGRGAFGTVYMGMNLDSGELLAVKQVLIAANFASKEKTQAHIQELEEEVKLLKNLSH 131
Query: 77 KNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLH 136
NIV+YLG+++ L+I+LE+V GS++++++ KFGPFPES+V Y Q+L GL YLH
Sbjct: 132 PNIVRYLGTVREDDTLNILLEFVPGGSISSLLE--KFGPFPESVVRTYTRQLLLGLEYLH 189
Query: 137 EQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINT--HSVVGTPYWMAPEVIEMS 194
++HRDIKGANIL +G +KLADFG + ++ E T S+ GTPYWMAPEVI +
Sbjct: 190 NHAIMHRDIKGANILVDNKGCIKLADFGASKQVAELATMTGAKSMKGTPYWMAPEVILQT 249
Query: 195 GVCAASDIWSVGCTVIELLTCVPPY-YDLQPMPALFRI-VQDEQPPIPDSLSPNITDFLH 252
G ++DIWSVGCTVIE++T P+ + + A+F I PPIPD+LS + DFL
Sbjct: 250 GHSFSADIWSVGCTVIEMVTGKAPWSQQYKEVAAIFFIGTTKSHPPIPDTLSSDAKDFLL 309
Query: 253 QCFKKDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGTLRTIKDDGSAVAEVSGGDHKST 312
+C ++ RP A LL HP+++ + S+ L ++ ++ S + + KST
Sbjct: 310 KCLQEVPNLRPTASELLKHPFVMGKHKESAST-----DLGSVLNNLSTPLPLQINNTKST 364
Query: 313 GEGSSVEKEDSAKEFSTGEANSRKSHEDNASDSNFSNERTEKEDDIPSDQVLTLAIREKS 372
+ + +D + G N S D + N+ +++D D+ I +++
Sbjct: 365 PDSTC---DDVGDMCNFGSLN--YSLVDPV--KSIQNKNLWQQNDNGGDEDDMCLIDDEN 417
Query: 373 FLRTGSGNLSSSIE 386
FL T G +SS++E
Sbjct: 418 FL-TFDGEMSSTLE 430
>At1g54960 NPK1-related protein kinase 2 (ANP2)
Length = 651
Score = 223 bits (567), Expect = 8e-58
Identities = 121/288 (42%), Positives = 184/288 (63%), Gaps = 22/288 (7%)
Query: 7 SSAFTKSKTLDN----------KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSL--- 53
S F KS++ N ++ G IG+GA+G VY G++L++G+ +A+KQV +
Sbjct: 45 SMVFAKSQSPPNNSTVQIKPPIRWRKGQLIGRGAFGTVYMGMNLDSGELLAVKQVLITSN 104
Query: 54 ---ENIAQEDLNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKP 110
+ Q + + E + +L+NL+H NIV+YLG+++ L+I+LE+V GS++++++
Sbjct: 105 CASKEKTQAHIQELEEEVKLLKNLSHPNIVRYLGTVREDETLNILLEFVPGGSISSLLE- 163
Query: 111 NKFGPFPESLVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLT 170
KFG FPES+V Y Q+L GL YLH ++HRDIKGANIL +G +KLADFG + ++
Sbjct: 164 -KFGAFPESVVRTYTNQLLLGLEYLHNHAIMHRDIKGANILVDNQGCIKLADFGASKQVA 222
Query: 171 E-ADIN-THSVVGTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPY-YDLQPMPA 227
E A I+ S+ GTPYWMAPEVI +G ++DIWSVGCTVIE++T P+ + + A
Sbjct: 223 ELATISGAKSMKGTPYWMAPEVILQTGHSFSADIWSVGCTVIEMVTGKAPWSQQYKEIAA 282
Query: 228 LFRI-VQDEQPPIPDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWI 274
+F I PPIPD++S + DFL +C +++ RP A LL HP++
Sbjct: 283 IFHIGTTKSHPPIPDNISSDANDFLLKCLQQEPNLRPTASELLKHPFV 330
>At4g08500 MEKK1/MAP kinase kinase kinase
Length = 608
Score = 208 bits (529), Expect = 2e-53
Identities = 122/285 (42%), Positives = 176/285 (60%), Gaps = 15/285 (5%)
Query: 5 STSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSL---ENIAQEDL 61
+TS + + + G +G+G++G VY+G+ +GDF A+K+VSL + AQE +
Sbjct: 318 NTSPIYPDGGAIITSWQKGQLLGRGSFGSVYEGIS-GDGDFFAVKEVSLLDQGSQAQECI 376
Query: 62 NIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLV 121
+ I +L L H+NIV+Y G+ K S+L+I LE V GSL + + + +S+V
Sbjct: 377 QQLEGEIKLLSQLQHQNIVRYRGTAKDGSNLYIFLELVTQGSLLKLYQRYQLR---DSVV 433
Query: 122 AVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVG 181
++Y Q+L+GL YLH++G IHRDIK ANIL G VKLADFG+A DI S G
Sbjct: 434 SLYTRQILDGLKYLHDKGFIHRDIKCANILVDANGAVKLADFGLAKVSKFNDIK--SCKG 491
Query: 182 TPYWMAPEVI---EMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPP 238
TP+WMAPEVI + G + +DIWS+GCTV+E+ T PY DL+P+ ALFRI + P
Sbjct: 492 TPFWMAPEVINRKDSDGYGSPADIWSLGCTVLEMCTGQIPYSDLEPVQALFRIGRGTLPE 551
Query: 239 IPDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQS 283
+PD+LS + F+ +C K + +RP A LL+HP++ RR L S
Sbjct: 552 VPDTLSLDARLFILKCLKVNPEERPTAAELLNHPFV---RRPLPS 593
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 203 bits (517), Expect = 5e-52
Identities = 111/268 (41%), Positives = 161/268 (59%), Gaps = 8/268 (2%)
Query: 23 GDEIGKGAYGRVYKGLDLENGDFVAIKQVSL---ENIAQEDLNIIMEMINILQNLNHKNI 79
G +G G +G+VY G + E G AIK+V + + ++E L + + IN+L L H NI
Sbjct: 217 GKFLGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQLNQEINLLNQLCHPNI 276
Query: 80 VKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQG 139
V+Y GS ++ L + LEYV GS+ ++K +G F E ++ Y Q+L GL YLH +
Sbjct: 277 VQYYGSELSEETLSVYLEYVSGGSIHKLLKD--YGSFTEPVIQNYTRQILAGLAYLHGRN 334
Query: 140 VIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVI-EMSGVCA 198
+HRDIKGANIL G +KLADFG+A +T A S G+PYWMAPEV+ +G
Sbjct: 335 TVHRDIKGANILVDPNGEIKLADFGMAKHVT-AFSTMLSFKGSPYWMAPEVVMSQNGYTH 393
Query: 199 ASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQD-EQPPIPDSLSPNITDFLHQCFKK 257
A DIWS+GCT++E+ T PP+ + + A+F+I + P IPD LS + +F+ C ++
Sbjct: 394 AVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDHLSNDAKNFIRLCLQR 453
Query: 258 DARQRPDAKTLLSHPWILNCRRVLQSSL 285
+ RP A LL HP++ N RV +SL
Sbjct: 454 NPTVRPTASQLLEHPFLRNTTRVASTSL 481
>At1g63700 putative protein kinase
Length = 883
Score = 202 bits (515), Expect = 8e-52
Identities = 110/264 (41%), Positives = 163/264 (61%), Gaps = 8/264 (3%)
Query: 18 NKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSL---ENIAQEDLNIIMEMINILQNL 74
+++ G +G G++G VY G + E+G+ A+K+V+L + ++E + + I++L L
Sbjct: 398 SRWKKGRLLGMGSFGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRL 457
Query: 75 NHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVY 134
H+NIV+Y GS L+I LEYV GS+ +++ ++G F E+ + Y Q+L GL Y
Sbjct: 458 RHQNIVQYYGSETVDDKLYIYLEYVSGGSIYKLLQ--EYGQFGENAIRNYTQQILSGLAY 515
Query: 135 LHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIEMS 194
LH + +HRDIKGANIL G VK+ADFG+A +T A S G+PYWMAPEVI+ S
Sbjct: 516 LHAKNTVHRDIKGANILVDPHGRVKVADFGMAKHIT-AQSGPLSFKGSPYWMAPEVIKNS 574
Query: 195 -GVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQD-EQPPIPDSLSPNITDFLH 252
G A DIWS+GCTV+E+ T PP+ + +PA+F+I E P IPD LS DF+
Sbjct: 575 NGSNLAVDIWSLGCTVLEMATTKPPWSQYEGVPAMFKIGNSKELPDIPDHLSEEGKDFVR 634
Query: 253 QCFKKDARQRPDAKTLLSHPWILN 276
+C +++ RP A LL H ++ N
Sbjct: 635 KCLQRNPANRPTAAQLLDHAFVRN 658
>At5g66850 MAP protein kinase like protein
Length = 533
Score = 197 bits (501), Expect = 4e-50
Identities = 109/273 (39%), Positives = 164/273 (59%), Gaps = 17/273 (6%)
Query: 16 LDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSL---ENIAQEDLNIIMEMINILQ 72
+++++ G IG+G +G VY + E G A+K+V L + + E + + + I +L
Sbjct: 159 MNSQWKKGKLIGRGTFGSVYVASNSETGALCAMKEVELFPDDPKSAECIKQLEQEIKLLS 218
Query: 73 NLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGL 132
NL H NIV+Y GS + I LEYV GS+ I+ + G ES+V + +L GL
Sbjct: 219 NLQHPNIVQYFGSETVEDRFFIYLEYVHPGSINKYIRDH-CGTMTESVVRNFTRHILSGL 277
Query: 133 VYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLT--EADINTHSVVGTPYWMAPEV 190
YLH + +HRDIKGAN+L G+VKLADFG+A LT AD+ S+ G+PYWMAPE+
Sbjct: 278 AYLHNKKTVHRDIKGANLLVDASGVVKLADFGMAKHLTGQRADL---SLKGSPYWMAPEL 334
Query: 191 IEM-------SGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSL 243
++ + A DIWS+GCT+IE+ T PP+ + + A+F++++D PPIP+S+
Sbjct: 335 MQAVMQKDSNPDLAFAVDIWSLGCTIIEMFTGKPPWSEFEGAAAMFKVMRD-SPPIPESM 393
Query: 244 SPNITDFLHQCFKKDARQRPDAKTLLSHPWILN 276
SP DFL CF+++ +RP A LL H ++ N
Sbjct: 394 SPEGKDFLRLCFQRNPAERPTASMLLEHRFLKN 426
>At4g08480 putative mitogen-activated protein kinase
Length = 773
Score = 196 bits (498), Expect = 8e-50
Identities = 117/286 (40%), Positives = 173/286 (59%), Gaps = 15/286 (5%)
Query: 5 STSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSL---ENIAQEDL 61
+TS +++ + G + +G++G VY+ + E+GDF A+K+VSL + AQE +
Sbjct: 486 NTSPICVSGGSINTSWQKGQLLRQGSFGSVYEAIS-EDGDFFAVKEVSLLDQGSQAQECI 544
Query: 62 NIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLV 121
+ I +L L H+NI++Y G+ K S+L+I LE V GSL + + + +SL+
Sbjct: 545 QQLEGEIALLSQLEHQNILRYRGTDKDGSNLYIFLELVTQGSLLELYRRYQIR---DSLI 601
Query: 122 AVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVG 181
++Y Q+L+GL YLH +G IHRDIK A IL G VKLADFG+A DI +
Sbjct: 602 SLYTKQILDGLKYLHHKGFIHRDIKCATILVDANGTVKLADFGLAKVSKLNDIKSRKE-- 659
Query: 182 TPYWMAPEVI---EMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPP 238
T +WMAPEVI + G + +DIWS+GCTV+E+ T PY DL+P+ ALFRI + P
Sbjct: 660 TLFWMAPEVINRKDNDGYRSPADIWSLGCTVLEMCTGQIPYSDLEPVEALFRIRRGTLPE 719
Query: 239 IPDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSS 284
+PD+LS + F+ +C K + +RP A LL+HP++ RR L SS
Sbjct: 720 VPDTLSLDARHFILKCLKLNPEERPTATELLNHPFV---RRPLPSS 762
>At4g12020 putative disease resistance protein
Length = 1895
Score = 193 bits (491), Expect = 5e-49
Identities = 114/279 (40%), Positives = 167/279 (58%), Gaps = 21/279 (7%)
Query: 5 STSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNII 64
+TS + + + G +G+G+ G VY+G+ + GDF A K+VSL + + I
Sbjct: 1611 NTSPIYASEGSFITCWQKGQLLGRGSLGSVYEGISAD-GDFFAFKEVSLLDQGSQAHEWI 1669
Query: 65 MEM---INILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLV 121
++ I +L L H+NIV+Y G+ K +S+L+I LE V GSL + + N+ G +S+V
Sbjct: 1670 QQVEGGIALLSQLQHQNIVRYRGTTKDESNLYIFLELVTQGSLRKLYQRNQLG---DSVV 1726
Query: 122 AVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVG 181
++Y Q+L+GL YLH++G IHR+IK AN+L G VKLADFG+A + S+
Sbjct: 1727 SLYTRQILDGLKYLHDKGFIHRNIKCANVLVDANGTVKLADFGLAKVM--------SLWR 1778
Query: 182 TPYW--MAPEVI----EMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDE 235
TPYW MAPEVI + G +DIWS+GCTV+E+LT PY DL+ AL+ I +
Sbjct: 1779 TPYWNWMAPEVILNPKDYDGYGTPADIWSLGCTVLEMLTGQIPYSDLEIGTALYNIGTGK 1838
Query: 236 QPPIPDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWI 274
P IPD LS + DF+ C K + +RP A LL+HP++
Sbjct: 1839 LPKIPDILSLDARDFILTCLKVNPEERPTAAELLNHPFV 1877
>At1g69220 serine/threonine kinase (SIK1)
Length = 836
Score = 193 bits (491), Expect = 5e-49
Identities = 116/325 (35%), Positives = 176/325 (53%), Gaps = 9/325 (2%)
Query: 1 MSRQSTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQED 60
MS S + T+ KY +E+GKG+YG VYK DL+ + VA+K +SL +E
Sbjct: 231 MSTTSLPDSITREDPT-TKYEFLNELGKGSYGSVYKARDLKTSEIVAVKVISLTE-GEEG 288
Query: 61 LNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESL 120
I I +LQ NH N+V+YLGS + + +L I++EY GS+A+++ + E
Sbjct: 289 YEEIRGEIEMLQQCNHPNVVRYLGSYQGEDYLWIVMEYCGGGSVADLMNVTEEA-LEEYQ 347
Query: 121 VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVV 180
+A + L+GL YLH +HRDIKG NIL T++G VKL DFGVA +LT ++ +
Sbjct: 348 IAYICREALKGLAYLHSIYKVHRDIKGGNILLTEQGEVKLGDFGVAAQLTRTMSKRNTFI 407
Query: 181 GTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIP 240
GTP+WMAPEVI+ + D+W++G + IE+ +PP + PM LF I + P +
Sbjct: 408 GTPHWMAPEVIQENRYDGKVDVWALGVSAIEMAEGLPPRSSVHPMRVLFMISIEPAPMLE 467
Query: 241 DSLSPNIT--DFLHQCFKKDARQRPDAKTLLSHPWILNCR---RVLQSSLRHSGTLR-TI 294
D ++ DF+ +C K+ R RP A +L H ++ C+ + + S +R T+
Sbjct: 468 DKEKWSLVFHDFVAKCLTKEPRLRPTAAEMLKHKFVERCKTGASAMSPKIEKSRQIRATM 527
Query: 295 KDDGSAVAEVSGGDHKSTGEGSSVE 319
+V S D + G SS E
Sbjct: 528 ALQAQSVVAPSLEDTSTLGPKSSEE 552
>At4g08470 putative mitogen-activated protein kinase
Length = 560
Score = 192 bits (487), Expect = 1e-48
Identities = 110/263 (41%), Positives = 162/263 (60%), Gaps = 12/263 (4%)
Query: 20 YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENI---AQEDLNIIMEMINILQNLNH 76
++ G +G+G+Y VY+ + E+GDF A+K+VSL + AQE + + I +L L H
Sbjct: 303 WLKGQLLGRGSYASVYEAIS-EDGDFFAVKEVSLLDKGIQAQECIQQLEGEIALLSQLQH 361
Query: 77 KNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLH 136
+NIV+Y G+ K S L+I LE V GS+ + + + ++V++Y Q+L GL YLH
Sbjct: 362 QNIVRYRGTAKDVSKLYIFLELVTQGSVQKLYERYQLS---YTVVSLYTRQILAGLNYLH 418
Query: 137 EQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVI---EM 193
++G +HRDIK AN+L G VKLADFG+A DI S GT +WMAPEVI +
Sbjct: 419 DKGFVHRDIKCANMLVDANGTVKLADFGLAEASKFNDIM--SCKGTLFWMAPEVINRKDS 476
Query: 194 SGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNITDFLHQ 253
G + +DIWS+GCTV+E+ T PY DL+P+ A F+I + P +PD+LS + F+
Sbjct: 477 DGNGSPADIWSLGCTVLEMCTGQIPYSDLKPIQAAFKIGRGTLPDVPDTLSLDARHFILT 536
Query: 254 CFKKDARQRPDAKTLLSHPWILN 276
C K + +RP A LL HP+++N
Sbjct: 537 CLKVNPEERPTAAELLHHPFVIN 559
>At3g15220 putative MAP kinase
Length = 690
Score = 191 bits (485), Expect = 3e-48
Identities = 104/255 (40%), Positives = 154/255 (59%), Gaps = 4/255 (1%)
Query: 26 IGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLGS 85
IG+G++G VYK D + VAIK + LE +++++ I + I++L I +Y GS
Sbjct: 21 IGRGSFGDVYKAFDKDLNKEVAIKVIDLEE-SEDEIEDIQKEISVLSQCRCPYITEYYGS 79
Query: 86 LKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVIHRDI 145
++ L II+EY+ GS+A++++ N P E+ +A +L + YLH +G IHRDI
Sbjct: 80 YLHQTKLWIIMEYMAGGSVADLLQSNN--PLDETSIACITRDLLHAVEYLHNEGKIHRDI 137
Query: 146 KGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIEMS-GVCAASDIWS 204
K ANIL ++ G VK+ADFGV+ +LT + VGTP+WMAPEVI+ S G +DIWS
Sbjct: 138 KAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQNSEGYNEKADIWS 197
Query: 205 VGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNITDFLHQCFKKDARQRPD 264
+G TVIE+ PP DL PM LF I ++ P + + S + +F+ C KK +RP
Sbjct: 198 LGITVIEMAKGEPPLADLHPMRVLFIIPRETPPQLDEHFSRQVKEFVSLCLKKAPAERPS 257
Query: 265 AKTLLSHPWILNCRR 279
AK L+ H +I N R+
Sbjct: 258 AKELIKHRFIKNARK 272
>At5g55090 putative protein
Length = 448
Score = 152 bits (384), Expect = 1e-36
Identities = 95/264 (35%), Positives = 148/264 (55%), Gaps = 16/264 (6%)
Query: 17 DNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNH 76
+ ++ G IG+G+ V G+ +GDF A+K + A + +IL L+
Sbjct: 3 EQNWIRGPIIGRGSTATVSLGIT-NSGDFFAVKSAEFSSSA-----FLQREQSILSKLSS 56
Query: 77 KNIVKYLGSLKTKSH----LHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGL 132
IVKY+GS TK + ++++EYV GSL ++IK N G PE L+ Y Q+L+GL
Sbjct: 57 PYIVKYIGSNVTKENDKLMYNLLMEYVSGGSLHDLIK-NSGGKLPEPLIRSYTRQILKGL 115
Query: 133 VYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIE 192
+YLH+QG++H D+K N++ E + K+ D G A + E + S GTP +M+PEV
Sbjct: 116 MYLHDQGIVHCDVKSQNVMIGGE-IAKIVDLGCAKTVEENENLEFS--GTPAFMSPEVAR 172
Query: 193 MSGVCAASDIWSVGCTVIELLTCVPPYYDLQP-MPALFRI-VQDEQPPIPDSLSPNITDF 250
+D+W++GCTVIE+ T P+ +L + A+++I E P IP LS DF
Sbjct: 173 GEEQSFPADVWALGCTVIEMATGSSPWPELNDVVAAIYKIGFTGESPVIPVWLSEKGQDF 232
Query: 251 LHQCFKKDARQRPDAKTLLSHPWI 274
L +C +KD +QR + LL HP++
Sbjct: 233 LRKCLRKDPKQRWTVEELLQHPFL 256
>At4g26890 putative NPK1-related protein kinase
Length = 444
Score = 152 bits (383), Expect = 2e-36
Identities = 106/343 (30%), Positives = 175/343 (50%), Gaps = 25/343 (7%)
Query: 23 GDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKY 82
G IG+G+ V + +G+ A+K L + + ++ + +IL L+ ++VKY
Sbjct: 8 GPIIGRGSTATVSIAIS-SSGELFAVKSADLSSSS-----LLQKEQSILSTLSSPHMVKY 61
Query: 83 LGSLKTKSH----LHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQ 138
+G+ T+ +I++EYV G+L ++IK N G PE + Y Q+L GLVYLHE+
Sbjct: 62 IGTGLTRESNGLVYNILMEYVSGGNLHDLIK-NSGGKLPEPEIRSYTRQILNGLVYLHER 120
Query: 139 GVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVIEMSGVCA 198
G++H D+K N+L + G++K+AD G A + +++ + GTP +MAPEV
Sbjct: 121 GIVHCDLKSHNVLVEENGVLKIADMGCAKSVDKSEFS-----GTPAFMAPEVARGEEQRF 175
Query: 199 ASDIWSVGCTVIELLTCVPPYYDLQP-MPALFRI-VQDEQPPIPDSLSPNITDFLHQCFK 256
+D+W++GCT+IE++T P+ +L + A+++I E P IP +S DFL C K
Sbjct: 176 PADVWALGCTMIEMMTGSSPWPELNDVVAAMYKIGFSGESPAIPAWISDKAKDFLKNCLK 235
Query: 257 KDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGTLRTIKDDGSAVAEVSGGD--HKSTGE 314
+D +QR + LL HP++ + + S L+ S V + D S
Sbjct: 236 EDQKQRWTVEELLKHPFLDD-----DEESQTSDCLKNKTSSPSTVLDQRFWDSCESSKSH 290
Query: 315 GSSVEKEDSAKEFSTGEANSRKSHEDNASDSNFSNERTEKEDD 357
S++ ED E+S + E A D S + EDD
Sbjct: 291 LVSIDHEDPFAEYSESLDSPADRIEKLAGDEFSSLLDWDTEDD 333
>At5g14720 protein kinase -like protein
Length = 674
Score = 151 bits (381), Expect = 3e-36
Identities = 91/267 (34%), Positives = 139/267 (51%), Gaps = 9/267 (3%)
Query: 20 YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNI 79
Y L +EIG G V++ L + VAIK + LE DL+ I + + +NH N+
Sbjct: 16 YKLYEEIGDGVSATVHRALCIPLNVVVAIKVLDLEK-CNNDLDGIRREVQTMSLINHPNV 74
Query: 80 VKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQG 139
++ S T L +++ Y+ GS +IIK + F E ++A + + L+ LVYLH G
Sbjct: 75 LQAHCSFTTGHQLWVVMPYMAGGSCLHIIKSSYPDGFEEPVIATLLRETLKALVYLHAHG 134
Query: 140 VIHRDIKGANILTTKEGLVKLADFGVATKLTEA---DINTHSVVGTPYWMAPEVI-EMSG 195
IHRD+K NIL G VKLADFGV+ + + + ++ VGTP WMAPEV+ ++ G
Sbjct: 135 HIHRDVKAGNILLDSNGAVKLADFGVSACMFDTGDRQRSRNTFVGTPCWMAPEVMQQLHG 194
Query: 196 VCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPI----PDSLSPNITDFL 251
+D+WS G T +EL P+ PM L +Q+ P + S + +
Sbjct: 195 YDFKADVWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYERDKRFSKAFKEMV 254
Query: 252 HQCFKKDARQRPDAKTLLSHPWILNCR 278
C KD ++RP ++ LL HP+ + R
Sbjct: 255 GTCLVKDPKKRPTSEKLLKHPFFKHAR 281
>At4g24100 unknown protein
Length = 709
Score = 149 bits (376), Expect = 1e-35
Identities = 139/562 (24%), Positives = 250/562 (43%), Gaps = 51/562 (9%)
Query: 20 YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNI 79
Y L +EIG GA VY+ + L + VAIK + L+ +L+ I + ++H N+
Sbjct: 33 YKLMEEIGHGASAVVYRAIYLPTNEVVAIKCLDLDR-CNSNLDDIRRESQTMSLIDHPNV 91
Query: 80 VKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQG 139
+K S L +++ ++ GS +++K F ES + + + L+ L YLH QG
Sbjct: 92 IKSFCSFSVDHSLWVVMPFMAQGSCLHLMKTAYSDGFEESAICCVLKETLKALDYLHRQG 151
Query: 140 VIHRDIKGANILTTKEGLVKLADFGVATKL---TEADINTHSVVGTPYWMAPEVIEM-SG 195
IHRD+K NIL G +KL DFGV+ L + ++ VGTP WMAPEV++ +G
Sbjct: 152 HIHRDVKAGNILLDDNGEIKLGDFGVSACLFDNGDRQRARNTFVGTPCWMAPEVLQPGNG 211
Query: 196 VCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPI----PDSLSPNITDFL 251
+ +DIWS G T +EL P+ PM L +Q+ P + S + + +
Sbjct: 212 YNSKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGLDYDRDKKFSKSFKEMV 271
Query: 252 HQCFKKDARQRPDAKTLLSHPWILNCR------RVLQSSLRHSGT-LRTIKDDGS---AV 301
C KD +RP A+ LL H + + ++L S L T +++++D + A+
Sbjct: 272 AMCLVKDQTKRPTAEKLLKHSCFKHTKPPEQTVKILFSDLPPLWTRVKSLQDKDAQQLAL 331
Query: 302 AEVSGGDHKSTGEGSSVEKEDSAKEFSTGEANSRKSHEDNASDSNFSNERTEKEDDIPSD 361
++ D ++ + S ++ SA F + ++ S + D E ++++++I
Sbjct: 332 KRMATADEEAISQ-SEYQRGVSAWNFDVRDLKTQASLLIDDDDL----EESKEDEEILCA 386
Query: 362 QVLTLAIREKSF-------LRTGSGNLSSS-IEVVSAEPTGTETSNAKDLHEVIMNGEVE 413
Q + RE+ F G +S++ +E + + T + L + N E +
Sbjct: 387 QFNKVNDREQVFDSLQLYENMNGKEKVSNTEVEEPTCKEKFTFVTTTSSLERMSPNSEHD 446
Query: 414 SPQSRGKANMAVGKDSSINNRT--------KPFAFEPRGQDSGSLKAMKIPPPVEG--NE 463
P+++ K + + +RT K FE + + ++ P G N
Sbjct: 447 IPEAKVKPLRRQSQSGPLTSRTVLSHSASEKSHIFERSESEPQTAPTVRRAPSFSGPLNL 506
Query: 464 LSRFSDPPGDAYLDDLFHPSDKQSGEVVAEASTSTSTSHIAKGNVSMNDGGKNDLAKELR 523
+R S A + K SG + + + KG S+ G DLAK++
Sbjct: 507 STRASSNSLSAPI--------KYSGGFRDSLDDKSKANLVQKGRFSVTSGNV-DLAKDVP 557
Query: 524 ATIARKQWEKESEIGQENNGGN 545
+I ++ + + + + + GN
Sbjct: 558 LSIVPRRSPQATPLRKSASVGN 579
>At1g07150 hypothetical protein
Length = 499
Score = 148 bits (374), Expect = 2e-35
Identities = 100/287 (34%), Positives = 150/287 (51%), Gaps = 15/287 (5%)
Query: 1 MSRQSTSSAFTKSKTLDNK---YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIA 57
M +QS + + S L + ++ G IG+G +G V + NG+ A+K V L
Sbjct: 1 MEKQSIRNTCSSSLMLSSPSSFWVRGACIGRGCFGAVSTAISKTNGEVFAVKSVDLATSL 60
Query: 58 QEDLNIIMEMINILQNLN-HKNIVKYLGSLKTK----SHLHIILEYVENGSLANIIKPNK 112
+ I++ ++L H IVK+LG +K + ++ LEY+ NG +A+ K
Sbjct: 61 PTQSESLENEISVFRSLKPHPYIVKFLGDGVSKEGTTTFRNLYLEYLPNGDVASHRAGGK 120
Query: 113 FGPFPESLVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKL-TE 171
E+L+ Y A ++ L ++H QG +H D+K NIL ++ +VKLADFG A ++ T
Sbjct: 121 IED--ETLLQRYTACLVSALRHVHSQGFVHCDVKARNILVSQSSMVKLADFGSAFRIHTP 178
Query: 172 ADINTHSVVGTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRI 231
+ T G+P WMAPEVI SD+WS+GCT+IE+ T P + D + +L RI
Sbjct: 179 RALITPR--GSPLWMAPEVIRREYQGPESDVWSLGCTIIEMFTGKPAWED-HGIDSLSRI 235
Query: 232 -VQDEQPPIPDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWILNC 277
DE P P LS DFL +C K+D QR LL HP++ C
Sbjct: 236 SFSDELPVFPSKLSEIGRDFLEKCLKRDPNQRWSCDQLLQHPFLSQC 282
>At4g14480 kinase like protein
Length = 487
Score = 145 bits (366), Expect = 2e-34
Identities = 103/367 (28%), Positives = 175/367 (47%), Gaps = 31/367 (8%)
Query: 25 EIGKGAYGRVYKGLDLE-NGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYL 83
+IG G VYK + + N VAIK + L+ ++ D + + + L+H NI+
Sbjct: 20 KIGVGVSASVYKAICIPMNSMVVAIKAIDLDQ-SRADFDSLRRETKTMSLLSHPNILNAY 78
Query: 84 GSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVIHR 143
S L +++ ++ GSL +I+ + PE+ ++V++ + L + YLH+QG +HR
Sbjct: 79 CSFTVDRCLWVVMPFMSCGSLHSIVSSSFPSGLPENCISVFLKETLNAISYLHDQGHLHR 138
Query: 144 DIKGANILTTKEGLVKLADFGVATKL----------TEADINTHSVVGTPYWMAPEVIEM 193
DIK NIL +G VKLADFGV+ + T + + + GTPYWMAPEV+
Sbjct: 139 DIKAGNILVDSDGSVKLADFGVSASIYEPVTSSSGTTSSSLRLTDIAGTPYWMAPEVVHS 198
Query: 194 -SGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIV------QDEQPPIPDS---- 242
+G +DIWS G T +EL PP L P+ +L + D + S
Sbjct: 199 HTGYGFKADIWSFGITALELAHGRPPLSHLPPLKSLLMKITKRFHFSDYEINTSGSSKKG 258
Query: 243 ---LSPNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRV--LQSSLRH--SGTLRTIK 295
S + + C ++D +RP A+ LL HP+ NC+ + + ++ H S +
Sbjct: 259 NKKFSKAFREMVGLCLEQDPTKRPSAEKLLKHPFFKNCKGLDFVVKNVLHSLSNAEQMFM 318
Query: 296 DDGSAVAEVSGGDHKSTGEGSSVEKEDSAKEFSTGEANSRKSHEDNASDSNFSNERTEKE 355
+ + V D + E + K ++ E + + S A++S+ S+E + +E
Sbjct: 319 ESQILIKSVGDDDEEEEEEDEEIVKNRRISGWNFREDDLQLSPVFPATESD-SSESSPRE 377
Query: 356 DDIPSDQ 362
+D D+
Sbjct: 378 EDQSKDK 384
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,027,073
Number of Sequences: 26719
Number of extensions: 1377406
Number of successful extensions: 8535
Number of sequences better than 10.0: 1058
Number of HSP's better than 10.0 without gapping: 849
Number of HSP's successfully gapped in prelim test: 211
Number of HSP's that attempted gapping in prelim test: 4922
Number of HSP's gapped (non-prelim): 1448
length of query: 1406
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1294
effective length of database: 8,326,068
effective search space: 10773931992
effective search space used: 10773931992
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0135.8


