

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.3
(361 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
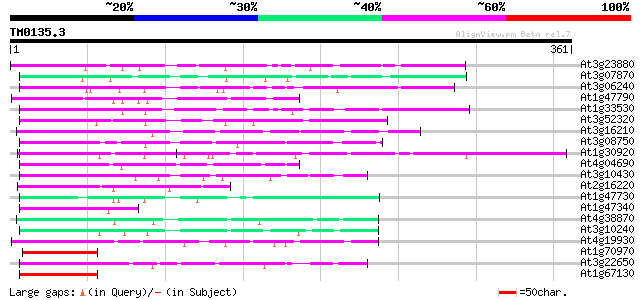
Score E
Sequences producing significant alignments: (bits) Value
At3g23880 unknown protein 124 8e-29
At3g07870 unknown protein 74 1e-13
At3g06240 unknown protein 73 3e-13
At1g47790 hypothetical protein 72 6e-13
At1g33530 hypothetical protein 72 6e-13
At3g52320 putative protein 68 7e-12
At3g16210 hypothetical protein 66 3e-11
At3g08750 hypothetical protein 64 1e-10
At1g30920 F17F8.21 64 2e-10
At4g04690 62 5e-10
At3g10430 hypothetical protein 62 5e-10
At2g16220 hypothetical protein 61 8e-10
At1g47730 hypothetical protein 60 2e-09
At1g47340 unknown protein (At1g47340) 58 7e-09
At4g38870 putative protein 57 2e-08
At3g10240 hypothetical protein 56 3e-08
At4g19930 putative protein 55 5e-08
At1g70970 55 5e-08
At3g22650 hypothetical protein 55 6e-08
At1g67130 hypothetical protein 55 6e-08
>At3g23880 unknown protein
Length = 364
Score = 124 bits (311), Expect = 8e-29
Identities = 109/319 (34%), Positives = 147/319 (45%), Gaps = 49/319 (15%)
Query: 1 MEPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLH--LHRSSSTTRNT 58
M P LP E+ EIL LPVKSL RF+CV SW+S+IS++ F H + +S T +T
Sbjct: 8 MFSPHNLPLEMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTST 67
Query: 59 DFAYLQSLITSPR---------KSRSASTIAIP---------DDYDFCGTCNGLVCLHSS 100
Y +IT+ R +AST+ + D Y GTC+GLVC H
Sbjct: 68 KSPY--GVITTSRYHLKSCCIHSLYNASTVYVSEHDGELLGRDYYQVVGTCHGLVCFHVD 125
Query: 101 KYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDS---YFGFDYDSSTDTYKVVAVAV 157
Y + LWNP ++ + QR L D +GF YD S D YKVVA+
Sbjct: 126 -------YDKSLYLWNPTIK-LQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVALLQ 177
Query: 158 ALSWDTLETMVNVYNIGDDTCWRTIPVSHLPSMYL---KDDDAVYVNNTLNRLAILPNVE 214
+ET +Y+ WR+ + PS + K +Y+N TLN A
Sbjct: 178 QRHQVKIET--KIYST-RQKLWRS--NTSFPSGVVVADKSRSGIYINGTLNWAA----TS 228
Query: 215 DRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVW 274
TI+S+D ++ +LP P C R F TLG LR CL + K + VW
Sbjct: 229 SSSSWTIISYDMSRDEFKELPGPVC-CGRGCFTM---TLGDLRGCLSMVCYCKGANADVW 284
Query: 275 QMKEFGLYKSWTLLFNIAG 293
MKEFG SW+ L +I G
Sbjct: 285 VMKEFGEVYSWSKLLSIPG 303
>At3g07870 unknown protein
Length = 417
Score = 73.9 bits (180), Expect = 1e-13
Identities = 81/327 (24%), Positives = 128/327 (38%), Gaps = 62/327 (18%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIK----------LHLHRSSSTTR 56
LPE++ +I S LP+ S+ R V +SW+S+++ + L LH S
Sbjct: 28 LPEDIIADIFSRLPISSIARLMFVCRSWRSVLTQHGRLSSSSSSPTKPCLLLHCDSPIRN 87
Query: 57 NTDFAYL----QSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHV 112
F L + + T R AS++ ++D G+CNGL+CL S Y+ +
Sbjct: 88 GLHFLDLSEEEKRIKTKKFTLRFASSM---PEFDVVGSCNGLLCLSDSLYN------DSL 138
Query: 113 RLWNPAMRSMSQRSPPLYLPIVYDSY------FGFDYDSSTDTYKVVAVAVALSWDT--- 163
L+NP + L LP + Y FGF + T YKV+ + +
Sbjct: 139 YLYNPFTTN------SLELPECSNKYHDQELVFGFGFHEMTKEYKVLKIVYFRGSSSNNN 192
Query: 164 -----------LETMVNVYNIGDDT-----CWRTIPVSHLPSMYLKDDDAVYVNNTLNRL 207
++ V + + T WR++ P ++K VN L+
Sbjct: 193 GIYRGRGRIQYKQSEVQILTLSSKTTDQSLSWRSL--GKAPYKFVKRSSEALVNGRLH-F 249
Query: 208 AILPNVEDRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDK 267
P DR +SFD E ++P P C RT L L+ CLC
Sbjct: 250 VTRPRRHVPDR-KFVSFDLEDEEFKEIPKPDCG----GLNRTNHRLVNLKGCLCAVVYGN 304
Query: 268 NMHFVVWQMKEFGLYKSWTLLFNIAGY 294
+W MK +G+ +SW ++I Y
Sbjct: 305 YGKLDIWVMKTYGVKESWGKEYSIGTY 331
>At3g06240 unknown protein
Length = 427
Score = 72.8 bits (177), Expect = 3e-13
Identities = 88/337 (26%), Positives = 141/337 (41%), Gaps = 76/337 (22%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL------------HR---- 50
LP E+ EIL LP KS+ RFRCVSK + ++ SD F K+HL HR
Sbjct: 36 LPPEIITEILLRLPAKSIGRFRCVSKLFCTLSSDPGFAKIHLDLILRNESVRSLHRKLIV 95
Query: 51 SSSTTRNTDFAYLQSLITS--------PRKSRSASTIAIPDDY----------------- 85
SS + DF + I P K + + +Y
Sbjct: 96 SSHNLYSLDFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRRVMLKLNA 155
Query: 86 --------DFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLP--IVY 135
+ G+ NGLVC+ + V L+NP S+R P + P + Y
Sbjct: 156 KSYRRNWVEIVGSSNGLVCISPGE--------GAVFLYNPTTGD-SKRLPENFRPKSVEY 206
Query: 136 D----SYFGFDYDSSTDTYKVVAVAVALSWDTLETMVNVYNIGDDTCWRTIPVSHLPSMY 191
+ +GF +D TD YK+V + VA S D L+ +VY++ D+ WR I ++
Sbjct: 207 ERDNFQTYGFGFDGLTDDYKLVKL-VATSEDILDA--SVYSLKADS-WRRIC-----NLN 257
Query: 192 LKDDDAVYVNNTLNRLAI--LPNVEDRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRT 249
+ +D Y + AI + ++ +++FD E ++P+P + +
Sbjct: 258 YEHNDGSYTSGVHFNGAIHWVFTESRHNQRVVVAFDIQTEEFREMPVPDEAEDCSHRFSN 317
Query: 250 WPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWT 286
+ +G L LC+ ++H +W M E+G KSW+
Sbjct: 318 F-VVGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWS 353
>At1g47790 hypothetical protein
Length = 389
Score = 71.6 bits (174), Expect = 6e-13
Identities = 64/203 (31%), Positives = 92/203 (44%), Gaps = 34/203 (16%)
Query: 2 EPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFA 61
+P P +L EIL LPVKS+VRFRCVSK W SII+D FIK + SST ++ F
Sbjct: 20 KPTSSFPLDLASEILLRLPVKSVVRFRCVSKLWSSIITDPYFIKTY-ETQSSTRQSLLFC 78
Query: 62 YLQS----LITSPR---KSRSASTIAI-------PDDYDF---CGTCNGLVCLHSSKYDR 104
+ QS + + P+ S S+S AI P ++ + + +GL+C H
Sbjct: 79 FKQSDKLFVFSIPKHHYDSNSSSQAAIDRFQVKLPQEFSYPSPTESVHGLICFH------ 132
Query: 105 KDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTL 164
+ V +WNP+MR P + G YD +KVV + + D
Sbjct: 133 ---VLATVIVWNPSMRQFLTLPKPRKSWKELTVFLG--YDPIEGKHKVVCLPRNRTCDEC 187
Query: 165 ETMVNVYNIGD-DTCWRTIPVSH 186
+ V +G WRT+ H
Sbjct: 188 Q----VLTLGSAQKSWRTVKTKH 206
>At1g33530 hypothetical protein
Length = 441
Score = 71.6 bits (174), Expect = 6e-13
Identities = 84/307 (27%), Positives = 126/307 (40%), Gaps = 44/307 (14%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQSL 66
LP+ L EIL LPVK LVR + +SK WKS+I + HL +
Sbjct: 97 LPDVLVEEILQRLPVKYLVRLKSISKGWKSLIESDHLAEKHLRLLEKKYGLKEIKITVER 156
Query: 67 ITSPR-----KSRSASTIAIPDDYD----FCGTCNGLVCLHSSKYDRKDKYTSHVRLWNP 117
TS SR + AI D D G+CNGLVC++ + + ++ L NP
Sbjct: 157 STSKSICIKFFSRRSGMNAINSDSDDLLRVPGSCNGLVCVY-------ELDSVYIYLLNP 209
Query: 118 AMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTLETMVNVYNIGDDT 177
+PP + GF D T TYKV+ V +D + T+ V+++ D
Sbjct: 210 MTGVTRTLTPPRGTKL----SVGFGIDVVTGTYKVM---VLYGFDRVGTV--VFDL-DTN 259
Query: 178 CWR-------TIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFDFGKES 230
WR +P+S +P+ + + V+VN +L L D IL D E
Sbjct: 260 KWRQRYKTAGPMPLSCIPT---PERNPVFVNGSLFWLLA------SDFSEILVMDLHTEK 310
Query: 231 CAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSW-TLLF 289
L P D + + L D LC+ + +H VW + + L + W F
Sbjct: 311 FRTLSQPN-DMDDVDVSSGYIYMWSLEDRLCVSNVRQGLHSYVWVLVQDELSEKWERTRF 369
Query: 290 NIAGYAY 296
N+ G+ +
Sbjct: 370 NLLGHVF 376
>At3g52320 putative protein
Length = 390
Score = 68.2 bits (165), Expect = 7e-12
Identities = 64/259 (24%), Positives = 108/259 (40%), Gaps = 41/259 (15%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSST---------TRN 57
+PEE+ ++IL LP KSL+RF+CVSK W S+I+ F SS +
Sbjct: 27 IPEEMLIDILIRLPAKSLMRFKCVSKLWLSLITSRYFTNRFFKPSSPSCLFAYLVDRENQ 86
Query: 58 TDFAYLQSLITSPRKSRSASTIAIPDDYD--------FCGTCNGLVCLHSSKYDRKDKYT 109
+ + LQS +S R S +++++ D + GL+C + +
Sbjct: 87 SKYLLLQS-SSSSRHDHSDTSVSVIDQHSTIPIMGGYLVNAARGLLCYRTGR-------- 137
Query: 110 SHVRLWNPAMRSMSQRSPPLYLPIVYDS---YFGFDYDSSTDTYKVVAV--AVALSWDTL 164
V++ NP+ R + + LPI+ + F +D D YKV+++ V +
Sbjct: 138 -RVKVCNPSTRQIVE------LPIMRSKTNVWNWFGHDPFHDEYKVLSLFWEVTKEQTVV 190
Query: 165 ETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSF 224
+ V +G WR H P + T++ + D +RC ++SF
Sbjct: 191 RSEHQVLVLGVGASWRNTKSHHTPH---RPFHPYSRGMTIDGVLYYSARTDANRCVLMSF 247
Query: 225 DFGKESCAQLPLPYCPQSR 243
D E + LP+ SR
Sbjct: 248 DLSSEEFNLIELPFENWSR 266
>At3g16210 hypothetical protein
Length = 360
Score = 66.2 bits (160), Expect = 3e-11
Identities = 66/267 (24%), Positives = 109/267 (40%), Gaps = 32/267 (11%)
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQ 64
+FLPEEL +EIL L +K L RFRCV K+W+ +I+D F + + S + +
Sbjct: 3 KFLPEELAIEILVRLSMKDLARFRCVCKTWRDLINDPGFTETYRDMSPAKFVSFYDKNFY 62
Query: 65 SLITSPRKSRSASTIAIPDDYDFCGT------CNGLVCLHSSKYDRKDKYTSHVRLWNPA 118
L + + + P D C+G +C+ + + +WNP
Sbjct: 63 MLDVEGKHPVITNKLDFPLDQSMIDESTCVLHCDGTLCVTLKNHT--------LMVWNPF 114
Query: 119 MRSMS-QRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTLETMVNVYNIGDDT 177
+ +P +Y + GF YD D YKVV L T +V+ +
Sbjct: 115 SKQFKIVPNPGIYQD---SNILGFGYDPVHDDYKVVTFIDRLDVST----AHVFEFRTGS 167
Query: 178 CWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFDFGKESCAQLPLP 237
++ +S+ P + +D +++ L +A + + IL F+ +LPLP
Sbjct: 168 WGESLRISY-PDWHYRDRRGTFLDQYLYWIAYRSSADR----FILCFNLSTHEYRKLPLP 222
Query: 238 YCPQSRYDFYRTWPTLGVLRDCLCICQ 264
Y+ T LGV LCI +
Sbjct: 223 V-----YNQGVTSSWLGVTSQKLCITE 244
>At3g08750 hypothetical protein
Length = 369
Score = 64.3 bits (155), Expect = 1e-10
Identities = 63/244 (25%), Positives = 111/244 (44%), Gaps = 31/244 (12%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL-HRSSSTTRNTDFAYLQS 65
LP EL EIL +P +SL+RF+ K W ++I++ +F+ HL H S T Y Q
Sbjct: 12 LPFELIEEILYKIPAESLIRFKSTCKKWYNLITEKRFMYNHLDHYSPERFIRT---YDQQ 68
Query: 66 LITSPRKSRSASTIAIPDDYD------FCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAM 119
+I + S IPD++ C+GL+ K+D + + +WNP +
Sbjct: 69 IIDP--VTEILSDALIPDEFRDLYPIYSMVHCDGLMLCTCRKWD------NSLAVWNPVL 120
Query: 120 RSMSQRSPPLYLPIVYDSYFGFDYDS--STDTYKVVAVAVAL-SWDTLETMVNVYNIGDD 176
R + P + ++ Y G YD S D YK++ + L D + +Y D
Sbjct: 121 REIKWIKPSVC--YLHTDYVGIGYDDNVSRDNYKILKLLGRLPKDDDSDPNCEIYEFKSD 178
Query: 177 TCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFDFGKESCAQLPL 236
+ W+T+ + ++ ++ V V + +A + ++ TI+ FDF E+ ++ +
Sbjct: 179 S-WKTLVAKFDWDIDIRCNNGVSVKGKMYWIA-----KKKEDFTIIRFDFSTETFKEICV 232
Query: 237 PYCP 240
CP
Sbjct: 233 --CP 234
>At1g30920 F17F8.21
Length = 1201
Score = 63.5 bits (153), Expect = 2e-10
Identities = 89/386 (23%), Positives = 158/386 (40%), Gaps = 56/386 (14%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQSL 66
+P +L L+ILS LP KS+ R RCVSK W+S+I S F +L L RSSS L
Sbjct: 811 IPIDLILDILSRLPSKSIARCRCVSKLWESMIRQSYFTELFLTRSSSRPH---------L 861
Query: 67 ITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSH--------VRLWNPA 118
+ + + ++P ++ G + +V + +DK H + N
Sbjct: 862 LIAVEQEGEWKFFSLPQPKNYLGKSSLVVAANLHLKFFEDKRPQHGCSYASSLIYFPNMT 921
Query: 119 MRSMSQRS-----------------PPL--YLPIVYDSYFGFDYDSSTDTYKVVAVAVAL 159
+R PPL + + Y + GFD D V + +
Sbjct: 922 IRKKGDDHLGVICNPSTGQYGYVILPPLLDFKSVPYGKFLGFD---PIDKQFKVLIPI-F 977
Query: 160 SWDTLETMVNVYNIGDDTC-WRTI--PVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDR 216
+D +T ++ +G +T WR I P+ +LP + + +N L LA + D+
Sbjct: 978 DFDKHQTDHHILTLGAETVGWRKIQSPLRYLP----HSNGTICINGILYYLAKINYAMDK 1033
Query: 217 DRCTILSFDFGKESCAQLPL-PYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQ 275
+ ++ FD E+ L L YC ++ Y+ LG++ + + VW
Sbjct: 1034 N--VLVCFDVRSENFVFLRLNTYCSSTKLVNYK--GKLGMINQ-EYVDDGGFPLKLSVWV 1088
Query: 276 MKEFGLYKSWTLLFNIA--GYAYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQKD 333
+++ G + T ++ + + + ++ +G+ +L+ K +P VLY D
Sbjct: 1089 LEDVGKEEWSTYVYTLRDDNKVDQVKYNLSVVGVTASGEIVLVKKTQTLKPFYVLYFNPD 1148
Query: 334 -NKLEVTDVANYIFNCYAMNYIESLV 358
N L +V YA++ I + V
Sbjct: 1149 KNTLLTVEVKGLHRGLYAVHRIYAFV 1174
Score = 47.0 bits (110), Expect = 2e-05
Identities = 36/120 (30%), Positives = 55/120 (45%), Gaps = 20/120 (16%)
Query: 6 FLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTR--------N 57
F ++L EIL LP KS+ R CVSK +SI+S F +L L +SS+ R N
Sbjct: 4 FPNDDLVYEILLRLPAKSVARCSCVSKLRRSILSRQDFTELFLTKSSARPRLLFGVKRAN 63
Query: 58 TDFAYLQSLITSPRKSRSASTIAIPDDY----------DFCGTCNGLVCLHSSKYDRKDK 107
+ + S T PR S S++ + D+ + C +GL+ L + D+
Sbjct: 64 GEGLFFSS--TQPRNSYEKSSLVVAADFHTKFSEVISREICSYASGLIYLSDMRISVNDE 121
>At4g04690
Length = 378
Score = 62.0 bits (149), Expect = 5e-10
Identities = 57/190 (30%), Positives = 89/190 (46%), Gaps = 20/190 (10%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL-HRSSSTTRNTDFAYLQS 65
LP EL EIL P +SL RF+ K W II+ +F+ HL H R D +Q
Sbjct: 11 LPFELVEEILKKTPAESLNRFKSTCKQWYGIITSKRFMYNHLDHSPERFIRIDDHKTVQ- 69
Query: 66 LITSP-----RKSRSASTIAIPDDYDFCGTCNGL-VCLHS-SKYDRKDKYTSHVRLWNPA 118
I P S P + C+GL +C+ S S Y+R + +++ +WNP
Sbjct: 70 -IMDPMTGIFSDSPVPDVFRSPHSFASMVHCDGLMLCICSDSSYERTRE--ANLAVWNPV 126
Query: 119 MRSMSQRSPPLYLPIVYDS-YFGFDYDSS-TDTYKVVAVAVALSWDTLETMVNVYNIGDD 176
+ + P L Y++ YFG YD++ + YK+V + +S+D +T +Y D
Sbjct: 127 TKKIKWIEP---LDSYYETDYFGIGYDNTCRENYKIVRFSGPMSFD--DTECEIYEFKSD 181
Query: 177 TCWRTIPVSH 186
+ WRT+ +
Sbjct: 182 S-WRTLDTKY 190
>At3g10430 hypothetical protein
Length = 370
Score = 62.0 bits (149), Expect = 5e-10
Identities = 67/239 (28%), Positives = 106/239 (44%), Gaps = 33/239 (13%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIIS-DSQFIKLHLHRSSSTTRNTDFAYLQS 65
LP +L LEIL P +SL+RF+ K W +IS D +F+ HL +S+ +
Sbjct: 5 LPFDLILEILQRTPAESLLRFKSTCKKWYELISNDKRFMYKHLDKSTKRFLRIENRERVQ 64
Query: 66 LITSPRKSRSASTI--AIPDDYDFCGTCNGL---VCLHSSKYDRKDKYTSHVRLWNPAMR 120
++ + + STI + Y C+GL +C D ++ +WNP MR
Sbjct: 65 ILDPVTEILAVSTIPNELRHKYFTLIHCDGLMLGMCYEELGSD------PNLAVWNPVMR 118
Query: 121 SMS--QRSPPLYLPIVY--DSYFGFDYDSS-TDTYKVVAVAVALSWDTLET----MVNVY 171
+ + SPPL + Y Y GF YD + D YK++ D E+ + +
Sbjct: 119 KIKWIKPSPPL---VCYWGSDYLGFGYDKTFRDNYKILRFTYLGDDDDDESYPKCQIYEF 175
Query: 172 NIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFDFGKES 230
N G WR+I + + + D V VN ++ + + + ILSFDF KE+
Sbjct: 176 NSGS---WRSIEAKFDGEIDV-EVDGVSVNGSMYWIEL-----QEKKNFILSFDFSKET 225
>At2g16220 hypothetical protein
Length = 407
Score = 61.2 bits (147), Expect = 8e-10
Identities = 50/154 (32%), Positives = 72/154 (46%), Gaps = 19/154 (12%)
Query: 6 FLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQS 65
FL +L LE+LS LP+KS+ RF CVSK W S+ F +L L RSS+ R FA +Q+
Sbjct: 5 FLTNDLILEVLSRLPLKSVARFHCVSKRWASMFGSPYFKELFLTRSSAKPRLL-FAIVQN 63
Query: 66 ----LITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSK-------------YDRKDKY 108
+SPR +S+ST+ ++ + N L H + Y D+Y
Sbjct: 64 GVWRFFSSPRLEKSSSTLVATAEFHMKLSPNNLRIYHDNTPRYFSIGYASGLIYLYGDRY 123
Query: 109 TSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFD 142
+ + NP + P Y S+FGFD
Sbjct: 124 EATPLICNPNTGRYT-ILPKCYTYRKAFSFFGFD 156
>At1g47730 hypothetical protein
Length = 391
Score = 60.1 bits (144), Expect = 2e-09
Identities = 67/259 (25%), Positives = 98/259 (36%), Gaps = 53/259 (20%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQS- 65
LP +L EI S LP KS+VRFRCVSK W SI + F S T N F + +
Sbjct: 25 LPLDLTSEIFSRLPAKSVVRFRCVSKLWSSITTAPYFT-----NSFETRPNLMFFFKEGD 79
Query: 66 ---LIT------SPRKSRSASTIAIPDDY-----------DFCGTCNGLVCLHSSKYDRK 105
+ T +P +S S S+ I D Y + +GL+C +
Sbjct: 80 KFFVFTIPQHNQNPNESYSYSSSEIIDSYHTTYPKRCCVTTLTESVHGLICFRKA----- 134
Query: 106 DKYTSHVRLWNPAM------RSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVAL 159
+ +WNP M R + +R + + + YD G +KV V +
Sbjct: 135 ----ATPIIWNPTMRKFKPLRKLDERWKNIKVSLGYDPVDG--------KHKV----VCM 178
Query: 160 SWDTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRC 219
+ V +G D WRT+ +H S + Y + I +
Sbjct: 179 PYGNAFYECRVLTLGSDQEWRTVKTNHKNSPFTFHGGVCYRQSRCINGVIYYRADTNSGR 238
Query: 220 TILSFDFGKESCAQLPLPY 238
ILSFD E + LP+
Sbjct: 239 VILSFDVRSERFDVIELPW 257
>At1g47340 unknown protein (At1g47340)
Length = 428
Score = 58.2 bits (139), Expect = 7e-09
Identities = 37/87 (42%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAY---- 62
LP+EL LEIL LP KS+ RF CVSK W S++S F +L L RSSS FA
Sbjct: 6 LPKELILEILKRLPAKSVKRFHCVSKQWASMLSCPHFRELFLTRSSSAQPRLLFAIEKHN 65
Query: 63 ------LQSLITSPRKSRSASTIAIPD 83
L +T KS S+S + P+
Sbjct: 66 QWSLFSLPQRLTPYEKSSSSSVVVTPE 92
>At4g38870 putative protein
Length = 426
Score = 56.6 bits (135), Expect = 2e-08
Identities = 67/259 (25%), Positives = 105/259 (39%), Gaps = 41/259 (15%)
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAY-L 63
+ LP +L +EIL L +K L+RF CVSK W SII D F+KL L+ S ++ F +
Sbjct: 51 ELLPVDLIMEILKKLSLKPLIRFLCVSKLWASIIRDPYFMKLFLNESLKRPKSLVFVFRA 110
Query: 64 QSL--------ITSPRKSRSASTIAIPDDYDFCGTC------------NGLVCLHSSKYD 103
QSL + S R+ S+S+ + + TC +GL+C
Sbjct: 111 QSLGSIFSSVHLKSTREISSSSSSSSASSITYHVTCYTQQRMTISPSVHGLICYGP---- 166
Query: 104 RKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVAL---- 159
S + ++NP R + Y G YD YKVV + +
Sbjct: 167 -----PSSLVIYNPCTRRSITLPKIKAGRRAINQYIG--YDPLDGNYKVVCITRGMPMLR 219
Query: 160 SWDTLETMVNVYNIGD-DTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDR 218
+ L + V +G D+ WR I P + ++ + +N L A + +
Sbjct: 220 NRRGLAEEIQVLTLGTRDSSWRMIHDIIPPHSPVSEE--LCINGVLYYRAFIGT--KLNE 275
Query: 219 CTILSFDFGKESCAQLPLP 237
I+SFD E + +P
Sbjct: 276 SAIMSFDVRSEKFDLIKVP 294
>At3g10240 hypothetical protein
Length = 389
Score = 56.2 bits (134), Expect = 3e-08
Identities = 66/252 (26%), Positives = 100/252 (39%), Gaps = 42/252 (16%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTR--NTDFAYLQ 64
+P +L EIL LP KS+ RFRCVSK W SI ++ FI L RS + +
Sbjct: 27 IPLDLVSEILLRLPEKSVARFRCVSKPWSSITTEPYFINLLTTRSPRLLLCFKANEKFFV 86
Query: 65 SLITSPRK-----SRSASTIAIPDDYDF-----------CGTCNGLVCLHSSKYDRKDKY 108
S I R+ ++S S + D Y + NGL+C S
Sbjct: 87 SSIPQHRQTFETWNKSHSYSQLIDRYHMEFSEEMNYFPPTESVNGLICFQES-------- 138
Query: 109 TSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTLETMV 168
+ + +WNP+ R + P + G YD +KV+ + + ++DT
Sbjct: 139 -ARLIVWNPSTRQLLILPKPNGNSNDLTIFLG--YDPVEGKHKVMCMEFSATYDT----C 191
Query: 169 NVYNIGD-DTCWRTIPV--SHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFD 225
V +G WRT+ H Y D +N + +A V+D ++SFD
Sbjct: 192 RVLTLGSAQKLWRTVKTHNKHRSDYY---DSGRCINGVVYHIAY---VKDMCVWVLMSFD 245
Query: 226 FGKESCAQLPLP 237
E + LP
Sbjct: 246 VRSEIFDMIELP 257
>At4g19930 putative protein
Length = 431
Score = 55.5 bits (132), Expect = 5e-08
Identities = 70/270 (25%), Positives = 112/270 (40%), Gaps = 51/270 (18%)
Query: 2 EPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTR-NTDF 60
EP ++P +L +EIL+ LP KSL+RF+ VSK W S+I F L S R
Sbjct: 38 EPMPYIPFDLVIEILTRLPAKSLMRFKSVSKLWSSLICSRTFTNRLLRVPSFIQRLYVTL 97
Query: 61 AYLQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSH--------- 111
+L + + RKS+ S+ + P D + +V + K Y SH
Sbjct: 98 TFLDNSL--QRKSKLLSSSSSPGS-DISTMSSFVVDRDLTTPSMKGYYLSHVLRGLMCFV 154
Query: 112 ----VRLWNPAMRSMSQRSPPLYLPIVYDS------------YFGFDYDSSTDTYKVVAV 155
V+++N R + + LP + +S + +D D YKVV +
Sbjct: 155 KEPSVKIYNTTTRQL------VVLPDIEESNIIAEDHKNKKIMYRIGHDPVGDQYKVVCI 208
Query: 156 AVALSWDTLETMVN------VYNIGDD--TCWRTIPVSHLPSMYLKDDDAVYVNNTLNRL 207
VA D + V+ +G D + WR IP PS +L + +N ++ L
Sbjct: 209 -VARPNDEFGELRRYLSEHWVFILGGDKSSGWRKIP---CPSPHLPITQILSINGRMHYL 264
Query: 208 AILPNVEDRDRCTILSFDFGKESCAQLPLP 237
A + + +++FDF E + L P
Sbjct: 265 AWVQKFDP----MLVTFDFSSEEISILQAP 290
>At1g70970
Length = 402
Score = 55.5 bits (132), Expect = 5e-08
Identities = 27/48 (56%), Positives = 34/48 (70%)
Query: 9 EELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTR 56
E+L+ EI+SWLP+KSL+RF VSK W SII QF L+L RS + R
Sbjct: 12 EDLQTEIMSWLPLKSLLRFVIVSKKWASIIRGEQFKALYLRRSMTRPR 59
>At3g22650 hypothetical protein
Length = 372
Score = 55.1 bits (131), Expect = 6e-08
Identities = 58/237 (24%), Positives = 102/237 (42%), Gaps = 35/237 (14%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQSL 66
LP ++ EI +PV+ L +F+ K W +++ D +FI +L R + +
Sbjct: 9 LPIDIIEEICCRIPVEYLTQFKLTCKQWFALLKDKRFIYKYLDLFQEQERFIRIDRIVQI 68
Query: 67 ITSPRKSRSASTIAIPDDYDFCGT------CNGLVCLHSSKYDRKDKYTSHVRLWNPAMR 120
I + +RS+S IP ++D C+GL+ L K +R Y + +WNP +
Sbjct: 69 IDPVKGARSSS--PIPQEFDNVAQISTMVHCDGLL-LCRCKNERSRSY--KLAVWNPFLS 123
Query: 121 SMSQRSPPLYLPIVYDSYFGFDYDS-STDTYKVVAVAVALSWD------TLETMVNVYNI 173
+ P + + ++GF YD+ D YK++ + D + E + +Y+
Sbjct: 124 RVKWIEPMDFYS--SNDFYGFGYDNVCRDEYKLLRIFDGEIEDESEIAGSYEPKIQIYDF 181
Query: 174 GDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFDFGKES 230
D+ WR + DD V V + +A + +R I SFDF E+
Sbjct: 182 KSDS-WRIV-----------DDTRVSVKGNMYWIA---HWNNRPEIFIQSFDFSTET 223
>At1g67130 hypothetical protein
Length = 391
Score = 55.1 bits (131), Expect = 6e-08
Identities = 25/50 (50%), Positives = 35/50 (70%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTR 56
+P +L L+ILS LP KS+ RF CVSK W S+++ F +L ++RSSS R
Sbjct: 10 IPTDLILDILSRLPTKSIARFHCVSKLWSSMLASQDFTRLFVNRSSSNPR 59
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,724,809
Number of Sequences: 26719
Number of extensions: 369520
Number of successful extensions: 1261
Number of sequences better than 10.0: 319
Number of HSP's better than 10.0 without gapping: 284
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 896
Number of HSP's gapped (non-prelim): 363
length of query: 361
length of database: 11,318,596
effective HSP length: 101
effective length of query: 260
effective length of database: 8,619,977
effective search space: 2241194020
effective search space used: 2241194020
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0135.3


