

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.5
(510 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
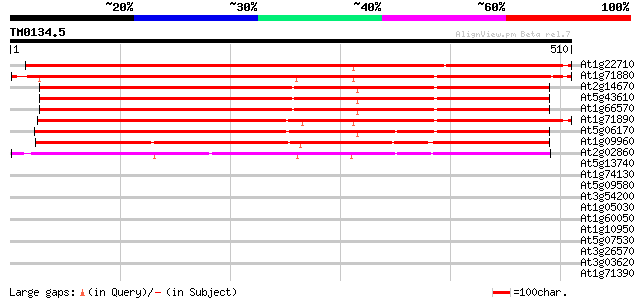
Score E
Sequences producing significant alignments: (bits) Value
At1g22710 putative sucrose transport protein, SUC2 670 0.0
At1g71880 sucrose transport protein SUC1 649 0.0
At2g14670 putative sucrose-proton symporter 644 0.0
At5g43610 sucrose transporter protein 640 0.0
At1g66570 hypothetical protein 640 0.0
At1g71890 putative sucrose transport protein 637 0.0
At5g06170 sucrose transporter protein 628 e-180
At1g09960 putative sucrose/H+ symporter 487 e-138
At2g02860 Sucrose transporter (suc3) 418 e-117
At5g13740 transporter-like protein 33 0.49
At1g74130 unknown protein 32 0.83
At5g09580 putative protein 30 2.4
At3g54200 unknown protein 30 3.2
At1g05030 sugar transporter like protein 30 4.1
At1g60050 hypothetical protein 29 7.0
At1g10950 putative endomembrane protein EMP70 precusor isolog (A... 29 7.0
At5g07530 glycine-rich protein atGRP-7 28 9.2
At3g26570 phosphate transporter like protein 28 9.2
At3g03620 unknown protein 28 9.2
At1g71390 putative disease resistance protein 28 9.2
>At1g22710 putative sucrose transport protein, SUC2
Length = 512
Score = 670 bits (1728), Expect = 0.0
Identities = 331/504 (65%), Positives = 403/504 (79%), Gaps = 13/504 (2%)
Query: 15 SSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLC 74
S+L+ + Q LRK+I+V+SIAAG+QFGWALQLSLLTPYVQ LG+PH WAS IWLC
Sbjct: 14 SALETQTGELDQPERLRKIISVSSIAAGVQFGWALQLSLLTPYVQLLGIPHKWASLIWLC 73
Query: 75 GPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAK 134
GP+SG+LVQPIVGY+SDR T RFGRRRPFI+AGA V ++VFLIGYAADIGHSMGD L K
Sbjct: 74 GPISGMLVQPIVGYHSDRCTSRFGRRRPFIVAGAGLVTVAVFLIGYAADIGHSMGDQLDK 133
Query: 135 KTRPRAVAIFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVL 194
+ RA+AIF GFWILDVANN LQGPCRAFL DL+AG+ KKTRTA FFSFFMAVGNVL
Sbjct: 134 PPKTRAIAIFALGFWILDVANNTLQGPCRAFLADLSAGNAKKTRTANAFFSFFMAVGNVL 193
Query: 195 GYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKP 254
GYAAG+Y L+K+ PFT+T++C +CANLK+CFF SI LLLI++ +L YV++ P T +P
Sbjct: 194 GYAAGSYRNLYKVVPFTMTESCDLYCANLKTCFFLSITLLLIVTFVSLCYVKEKPWTPEP 253
Query: 255 AAD-VDSPVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGG-- 311
AD S V FG++FGAFKELK+PMW+L++VTA+NW+AWFPF LFDTDWMGREVYGG
Sbjct: 254 TADGKASNVPFFGEIFGAFKELKRPMWMLLIVTALNWIAWFPFLLFDTDWMGREVYGGNS 313
Query: 312 -----VVGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAI 366
+K Y+DGVRAG+LGLM+NA+VL FMSL VE +GR LGG K LWGIVNFILAI
Sbjct: 314 DATATAASKKLYNDGVRAGALGLMLNAIVLGFMSLGVEWIGRKLGGAKRLWGIVNFILAI 373
Query: 367 CMAMTVLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIY 426
C+AMTV++TK AE+ R GGA G P + V AGA+ F ++GIP AI FS+PFALASI+
Sbjct: 374 CLAMTVVVTKQAENHRRDHGGAKTGPPGN-VTAGALTLFAILGIPQAITFSIPFALASIF 432
Query: 427 SSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAI 486
S+ +GAGQGLSLGVLNLAIVVPQM++S GP+D LFGGGN+PAFV+GA+ AAVS ++A+
Sbjct: 433 STNSGAGQGLSLGVLNLAIVVPQMVISVGGGPFDELFGGGNIPAFVLGAIAAAVSGVLAL 492
Query: 487 VLLPTPKPADVAKASSLPPGGGFH 510
+LP+P P A +++ GFH
Sbjct: 493 TVLPSPPPDAPAFKATM----GFH 512
>At1g71880 sucrose transport protein SUC1
Length = 513
Score = 649 bits (1675), Expect = 0.0
Identities = 329/521 (63%), Positives = 402/521 (77%), Gaps = 26/521 (4%)
Query: 2 ESPTKSNHLDTKGSSLQIEAHIPQ---QSSPLRKMIAVASIAAGIQFGWALQLSLLTPYV 58
E PTK + +E P+ Q SPLRK+I+VASIAAG+QFGWALQLSLLTPYV
Sbjct: 7 EKPTKD--------AAALETQSPEDFDQPSPLRKIISVASIAAGVQFGWALQLSLLTPYV 58
Query: 59 QTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLI 118
Q LG+PH W+S IWLCGPVSG++VQPIVG++SDR +FGRRRPFI GA VA++VFLI
Sbjct: 59 QLLGIPHKWSSLIWLCGPVSGMIVQPIVGFHSDRCRSKFGRRRPFIATGAALVAVAVFLI 118
Query: 119 GYAADIGHSMGDDLAKKTRPRAVAIFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTR 178
GYAAD G+ MGD L +K + RA+ IF GFWILDVANN LQGPCRAFL DLAAGD K+TR
Sbjct: 119 GYAADFGYKMGDKLEEKVKVRAIGIFALGFWILDVANNTLQGPCRAFLADLAAGDAKRTR 178
Query: 179 TAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILS 238
A FFSFFMAVGNVLGYAAG+Y+ LHK+FPFT+T AC +CANLK+CFF SI LLLI++
Sbjct: 179 VANAFFSFFMAVGNVLGYAAGSYTNLHKMFPFTMTKACDIYCANLKTCFFLSITLLLIVT 238
Query: 239 IAALIYVEDTPLTKKPA-ADVD---SPVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWF 294
+ +L YV D + P AD D S V FG++FGAFK +K+PMW+L++VTA+NW+AWF
Sbjct: 239 VTSLWYVNDKQWSPPPRNADDDEKTSSVPLFGEIFGAFKVMKRPMWMLLIVTALNWIAWF 298
Query: 295 PFFLFDTDWMGREVYGG-----VVGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRI 349
PF LFDTDWMGREV+GG +K Y GV++G++GLM N++VL FMSL VE +GR
Sbjct: 299 PFLLFDTDWMGREVFGGDSDGNERSKKLYSLGVQSGAMGLMFNSIVLGFMSLGVEWIGRK 358
Query: 350 LGGVKNLWGIVNFILAICMAMTVLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMG 409
LGG K LWGIVNFILA +AMTVL+TK AE R +G + PS+ VKAGA++ F V+G
Sbjct: 359 LGGAKRLWGIVNFILAAGLAMTVLVTKFAEDHRKTAG--DLAGPSASVKAGALSLFAVLG 416
Query: 410 IPLAINFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLP 469
IPLAI FS PFALASI+SS +GAGQGLSLGVLNLAIV+PQM+VS GP+DALFGGGNLP
Sbjct: 417 IPLAITFSTPFALASIFSSCSGAGQGLSLGVLNLAIVIPQMIVSLGGGPFDALFGGGNLP 476
Query: 470 AFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGFH 510
AF+V A+ AA+S ++A+ +LP+P P D KA+++ GGFH
Sbjct: 477 AFIVAAIAAAISGVLALTVLPSP-PPDAPKATTM---GGFH 513
>At2g14670 putative sucrose-proton symporter
Length = 492
Score = 644 bits (1660), Expect = 0.0
Identities = 315/468 (67%), Positives = 379/468 (80%), Gaps = 8/468 (1%)
Query: 28 SPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVG 87
SPLRKMI+VASIAAGIQFGWALQLSLLTPYVQ LGVPH W+SFIWLCGPVSGLLVQP VG
Sbjct: 28 SPLRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPVSGLLVQPSVG 87
Query: 88 YYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFG 147
Y+SDR T RFGRRRPFI GA+ VA++V LIGYAAD GHSMGD + K + RAV IF G
Sbjct: 88 YFSDRCTSRFGRRRPFIATGALLVAVAVVLIGYAADFGHSMGDKIDKPVKMRAVVIFALG 147
Query: 148 FWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKI 207
FWILDVANN LQGPCRAFLGDLAAGD KKTRTA FFSFFMAVGNVLGYAAG+Y+ L+KI
Sbjct: 148 FWILDVANNTLQGPCRAFLGDLAAGDAKKTRTANAFFSFFMAVGNVLGYAAGSYTNLYKI 207
Query: 208 FPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSCFGD 267
FPFT+T AC +CANLKSCFF SI LLL+++I AL YVED + K +D + FG+
Sbjct: 208 FPFTMTKACDIYCANLKSCFFLSITLLLVVTIIALWYVEDKQWSPKADSD-NEKTPFFGE 266
Query: 268 LFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVG-----EKAYDDGV 322
+FGAFK +K+PMW+L++VTA+NW+AWFPF L+DTDWMGREVYGG +K Y+ G+
Sbjct: 267 IFGAFKVMKRPMWMLLIVTALNWIAWFPFLLYDTDWMGREVYGGDSKGDDKMKKLYNQGI 326
Query: 323 RAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKTAEHDR 382
G+LGLM+N++VL +SL +E + + +GG K LWG VN ILA+C+AMTVL+TK AE R
Sbjct: 327 HVGALGLMLNSIVLGIVSLGIEGISKKIGGAKRLWGAVNIILAVCLAMTVLVTKKAEEHR 386
Query: 383 LISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLN 442
I+G + P+ G++AGA+ F ++GIPLAI FS+PFALASI SS++GAGQGLSLGVLN
Sbjct: 387 RIAG--PMALPTDGIRAGALTLFALLGIPLAITFSIPFALASIISSSSGAGQGLSLGVLN 444
Query: 443 LAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP 490
+AIV+PQM+VS GP DALFGGGNLP FVVGA+ AA+S+++A +LP
Sbjct: 445 MAIVIPQMIVSFGVGPIDALFGGGNLPRFVVGAIAAAISSVVAFTVLP 492
>At5g43610 sucrose transporter protein
Length = 492
Score = 640 bits (1652), Expect = 0.0
Identities = 314/468 (67%), Positives = 377/468 (80%), Gaps = 8/468 (1%)
Query: 28 SPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVG 87
SP+RKMI+VASIAAGIQFGWALQLSLLTPYVQ LGVPH W+SFIWLCGPVSGLLVQP VG
Sbjct: 28 SPMRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPVSGLLVQPSVG 87
Query: 88 YYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFG 147
Y+SDR RFGRRRPFI GA+ VA++V LIGYAAD GHSMGD + + + RAV IF G
Sbjct: 88 YFSDRCKSRFGRRRPFIAMGALLVAVAVVLIGYAADFGHSMGDKVDEPVKMRAVVIFALG 147
Query: 148 FWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKI 207
FWILDVANN LQGPCRAFLGDLAAGD KKTRTA FFSFFMAVGNVLGYAAG+Y+ L+KI
Sbjct: 148 FWILDVANNTLQGPCRAFLGDLAAGDAKKTRTANAFFSFFMAVGNVLGYAAGSYTNLYKI 207
Query: 208 FPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSCFGD 267
FPFT+T AC +CANLKSCFF SI LLL+++I AL YVED + K +D + FG+
Sbjct: 208 FPFTMTKACDIYCANLKSCFFLSITLLLVVTIIALWYVEDKQWSPKADSD-NEKTPFFGE 266
Query: 268 LFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVG-----EKAYDDGV 322
+FGAFK +K+PMW+L++VTA+NW+AWFPF L+DTDWMGREVYGG +K Y+ G+
Sbjct: 267 IFGAFKVMKRPMWMLLIVTALNWIAWFPFLLYDTDWMGREVYGGDSKGDDKMKKLYNQGI 326
Query: 323 RAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKTAEHDR 382
G LGLM+N++VL FMSL +E + R +GG K LWG VN ILA+C+AMTVL+TK AE R
Sbjct: 327 HVGGLGLMLNSIVLGFMSLGIEGISRKMGGAKRLWGAVNIILAVCLAMTVLVTKKAEEHR 386
Query: 383 LISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLN 442
I+G + P+ G++AGA+ F ++GIPLAI FS+PFALASI SS++GAGQGLSLGVLN
Sbjct: 387 RIAG--PMALPTDGIRAGALTLFALLGIPLAITFSIPFALASIISSSSGAGQGLSLGVLN 444
Query: 443 LAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP 490
+ IV+PQM+VS GP DALFGGGNLP FVVGA+ AA+S+++A +LP
Sbjct: 445 MTIVIPQMVVSFGVGPIDALFGGGNLPGFVVGAIAAAISSVVAFSVLP 492
>At1g66570 hypothetical protein
Length = 491
Score = 640 bits (1651), Expect = 0.0
Identities = 317/468 (67%), Positives = 376/468 (79%), Gaps = 8/468 (1%)
Query: 28 SPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVG 87
SPLRKMI+VASIAAGIQFGWALQLSLLTPYVQ LGVPH W SFIWLCGPVSGLLVQP VG
Sbjct: 27 SPLRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWPSFIWLCGPVSGLLVQPSVG 86
Query: 88 YYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFG 147
Y+SDR T RFGRRRPFI GA+ VA+SV LIGYAAD GHSMGD + K + RAV IF G
Sbjct: 87 YFSDRCTSRFGRRRPFIATGALLVAVSVVLIGYAADFGHSMGDKIDKPVKMRAVVIFALG 146
Query: 148 FWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKI 207
FWILDVANN LQGPCRAFLGDLAAGD +KTRTA FFSFFMAVGNVLGYAAG+Y+ L+KI
Sbjct: 147 FWILDVANNTLQGPCRAFLGDLAAGDAQKTRTANAFFSFFMAVGNVLGYAAGSYTNLYKI 206
Query: 208 FPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSCFGD 267
FPFT+T AC +CANLKSCFF SI LLL+++I AL YVED + K +D + FG+
Sbjct: 207 FPFTMTKACDIYCANLKSCFFLSITLLLVVTIIALWYVEDKQWSPKADSD-NEKTPFFGE 265
Query: 268 LFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVG-----EKAYDDGV 322
+FGAFK +K+PMW+L++VTA+NW+AWFPF L+DTDWMGREVYGG +K Y+ G+
Sbjct: 266 IFGAFKVMKRPMWMLLIVTALNWIAWFPFLLYDTDWMGREVYGGDSKGDDKMKKLYNQGI 325
Query: 323 RAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKTAEHDR 382
G+LGLM+N++VL MSL +E + R +GG K LWG VN ILA+C+AMTVL+TK AE R
Sbjct: 326 HVGALGLMLNSIVLGVMSLGIEGISRKMGGAKRLWGAVNIILAVCLAMTVLVTKKAEEHR 385
Query: 383 LISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLN 442
I+G + P+ G++AGA+ F ++GIPLAI FS+PFALASI SS++GAGQ LSLGVLN
Sbjct: 386 RIAG--PMALPTDGIRAGALTLFALLGIPLAITFSIPFALASIISSSSGAGQRLSLGVLN 443
Query: 443 LAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP 490
+AIV+PQM+VS GP DALFG GNLP FVVGA+ AAVS+I+A +LP
Sbjct: 444 MAIVIPQMIVSFGVGPIDALFGDGNLPGFVVGAIAAAVSSIVAFTVLP 491
>At1g71890 putative sucrose transport protein
Length = 512
Score = 637 bits (1644), Expect = 0.0
Identities = 313/493 (63%), Positives = 389/493 (78%), Gaps = 15/493 (3%)
Query: 26 QSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPI 85
Q SPLRK+I+VASIAAG+QFGWALQLSLLTPY+Q LG+PH W+S++WLCGP+SG++VQPI
Sbjct: 27 QPSPLRKIISVASIAAGVQFGWALQLSLLTPYIQLLGIPHKWSSYMWLCGPISGMIVQPI 86
Query: 86 VGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFV 145
VGY+SDR RFGRRRPFI AG VA+SVFLIG+AAD+GHS GD L K R RA+ IF+
Sbjct: 87 VGYHSDRCESRFGRRRPFIAAGVALVAVSVFLIGFAADMGHSFGDKLENKVRTRAIIIFL 146
Query: 146 FGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLH 205
GFW LDVANN LQGPCRAFL DLAAGD KKTR A FSFFMAVGNVLGYAAG+Y+ LH
Sbjct: 147 TGFWFLDVANNTLQGPCRAFLADLAAGDAKKTRVANACFSFFMAVGNVLGYAAGSYTNLH 206
Query: 206 KIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSC- 264
K+FPFT+T AC +CANLK+CFF SI LLLI++ ++L YV+D + P D + S
Sbjct: 207 KMFPFTMTKACDIYCANLKTCFFLSITLLLIVTFSSLWYVKDKQWS-PPQGDKEEKTSSL 265
Query: 265 --FGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGG-----VVGEKA 317
FG++FGA + +K+PM +L++VT +NW+AWFPF L+DTDWMGREVYGG +K
Sbjct: 266 FFFGEIFGAVRHMKRPMVMLLIVTVINWIAWFPFILYDTDWMGREVYGGNSDGDERSKKL 325
Query: 318 YDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKT 377
YD GV+AG+LGLM N+++L F+SL VE +GR +GG K LWG VNFILAI +AMTVL+TK+
Sbjct: 326 YDQGVQAGALGLMFNSILLGFVSLGVESIGRKMGGAKRLWGCVNFILAIGLAMTVLVTKS 385
Query: 378 AEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGLS 437
AEH R I+G + PSSG+KAG + F V+GIPLAI +S+PFALASI+S+ +GAGQGLS
Sbjct: 386 AEHHREIAG--PLAGPSSGIKAGVFSLFTVLGIPLAITYSIPFALASIFSTNSGAGQGLS 443
Query: 438 LGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLPTPKPADV 497
LGVLN+AI +PQM+VS SGP DA FGGGNLP+FVVGA+ AAVS ++A+ +LP+P P
Sbjct: 444 LGVLNIAICIPQMIVSFSSGPLDAQFGGGNLPSFVVGAIAAAVSGVLALTVLPSPPPDAP 503
Query: 498 AKASSLPPGGGFH 510
A + ++ GFH
Sbjct: 504 AMSGAM----GFH 512
>At5g06170 sucrose transporter protein
Length = 491
Score = 628 bits (1620), Expect = e-180
Identities = 311/474 (65%), Positives = 378/474 (79%), Gaps = 11/474 (2%)
Query: 23 IPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLV 82
+P + SPLRKMI+VASIAAGIQFGWALQLSLLTPYVQ LGVPH W+SFIWLCGP+SGLLV
Sbjct: 23 VPDEPSPLRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLLV 82
Query: 83 QPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVA 142
QP VGY+SDR RFGRRRPFI GA+ VA++V LIG+AAD GH+MGD L + + RAV
Sbjct: 83 QPTVGYFSDRCKSRFGRRRPFIATGALLVALAVILIGFAADFGHTMGDKLDEAVKIRAVG 142
Query: 143 IFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYS 202
FV GFWILDVANN LQGPCRAFLGDLAAGD KKTRTA FSFFMAVGNVLGYAAG+Y+
Sbjct: 143 FFVVGFWILDVANNTLQGPCRAFLGDLAAGDAKKTRTANAIFSFFMAVGNVLGYAAGSYT 202
Query: 203 GLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDS-P 261
LHKIFPFTVT AC +CANLKSCF SI LL++L+I AL YVED + P AD D+
Sbjct: 203 NLHKIFPFTVTKACDIYCANLKSCFIISITLLIVLTIIALWYVEDKQWS--PNADSDNEK 260
Query: 262 VSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVG-----EK 316
FG++FGAFK +K+PMW+L+ VTA+NW+AWFPF L+DTDWMGREVYGG +K
Sbjct: 261 TPFFGEIFGAFKVMKRPMWMLLAVTALNWIAWFPFLLYDTDWMGREVYGGDSAGDDKMKK 320
Query: 317 AYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITK 376
Y+ G++ GSLGLM+N++VL MSL + + + + G K LWG VN ILA+C+AMTVL+TK
Sbjct: 321 LYNHGIQVGSLGLMLNSIVLGVMSLVIGVISKKI-GAKRLWGAVNIILAVCLAMTVLVTK 379
Query: 377 TAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGL 436
AE R I+G + P++ ++ GA++ F ++GIPLAI FS+PFALASI SS++GAGQGL
Sbjct: 380 KAEEHRKIAG--RMALPTNAIRDGALSLFAILGIPLAITFSIPFALASIISSSSGAGQGL 437
Query: 437 SLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP 490
SLGVLN+AIV+PQM+VS GP DALFGGGNLP FVVGA+ A +S+++A+ +LP
Sbjct: 438 SLGVLNMAIVIPQMIVSFGVGPIDALFGGGNLPGFVVGAIAALISSVVALTVLP 491
>At1g09960 putative sucrose/H+ symporter
Length = 510
Score = 487 bits (1253), Expect = e-138
Identities = 250/473 (52%), Positives = 319/473 (66%), Gaps = 13/473 (2%)
Query: 24 PQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQ 83
P+ R ++ VAS+A GIQFGWALQLSLLTPYVQ LG+PH WAS IWLCGP+SGL VQ
Sbjct: 34 PRSKVSKRVLLRVASVACGIQFGWALQLSLLTPYVQELGIPHAWASVIWLCGPLSGLFVQ 93
Query: 84 PIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAI 143
P+VG+ SDR T ++GRRRPFI+AGAVA++ISV +IG+AADIG + G D K +PRA+
Sbjct: 94 PLVGHSSDRCTSKYGRRRPFIVAGAVAISISVMVIGHAADIGWAFG-DREGKIKPRAIVA 152
Query: 144 FVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSG 203
FV GFWILDVANNM QGPCRA L DL D ++TR A G+FS FMAVGNVLGYA G+Y+G
Sbjct: 153 FVLGFWILDVANNMTQGPCRALLADLTENDNRRTRVANGYFSLFMAVGNVLGYATGSYNG 212
Query: 204 LHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVS 263
+KIF FT T AC CANLKS F+ ++ + I +I ++ + PL A++ S
Sbjct: 213 WYKIFTFTKTVACNVECANLKSAFYIDVVFIAITTILSVSAAHEVPLASL-ASEAHGQTS 271
Query: 264 -----CFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVG-EKA 317
++FG F+ +WI++LVTA+ W+ WFPF LFDTDWMGRE+YGG +
Sbjct: 272 GTDEAFLSEIFGTFRYFPGNVWIILLVTALTWIGWFPFILFDTDWMGREIYGGEPNIGTS 331
Query: 318 YDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKT 377
Y GV G+LGLM+N+V L S+ +E L R G +WGI N ++AIC ++ +
Sbjct: 332 YSAGVSMGALGLMLNSVFLGITSVLMEKLCR-KWGAGFVWGISNILMAICFLGMIITSFV 390
Query: 378 AEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGLS 437
A H G P + + A+ F ++GIPLAI +SVP+AL SI + G GQGLS
Sbjct: 391 ASH----LGYIGHEQPPASIVFAAVLIFTILGIPLAITYSVPYALISIRIESLGLGQGLS 446
Query: 438 LGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP 490
LGVLNLAIV+PQ++VS SGPWD LFGGGN PA VGA + I+AI+ LP
Sbjct: 447 LGVLNLAIVIPQVIVSVGSGPWDQLFGGGNSPALAVGAATGFIGGIVAILALP 499
>At2g02860 Sucrose transporter (suc3)
Length = 594
Score = 418 bits (1074), Expect = e-117
Identities = 223/554 (40%), Positives = 330/554 (59%), Gaps = 74/554 (13%)
Query: 2 ESPTKSNHLDTKGSSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTL 61
ES + SNH D+ + ++ L ++ ++AAG+QFGWALQLSLLTPY+QTL
Sbjct: 38 ESASPSNHSDSA------DGESVSKNCSLVTLVLSCTVAAGVQFGWALQLSLLTPYIQTL 91
Query: 62 GVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYA 121
G+ H ++SFIWLCGP++GL+VQP VG +SD+ T ++GRRRPFIL G+ ++I+V +IG++
Sbjct: 92 GISHAFSSFIWLCGPITGLVVQPFVGIWSDKCTSKYGRRRPFILVGSFMISIAVIIIGFS 151
Query: 122 ADIGHSMGD-----DLAKKTRPRAVAIFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKK 176
ADIG+ +GD K TR RA +F+ GFW+LD+ANN +QGP RA L DL+ DQ+
Sbjct: 152 ADIGYLLGDSKEHCSTFKGTRTRAAVVFIIGFWLLDLANNTVQGPARALLADLSGPDQRN 211
Query: 177 TRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLI 236
T A+ F +MA+GN+LG++AGA + FPF + AC + C NLK+ F +++ L I
Sbjct: 212 TANAV--FCLWMAIGNILGFSAGASGKWQEWFPFLTSRACCAACGNLKAAFLLAVVFLTI 269
Query: 237 LSIAALIYVEDTPLT-KKPAADVDS----------------------------------- 260
++ + + ++ P T KP DS
Sbjct: 270 CTLVTIYFAKEIPFTSNKPTRIQDSAPLLDDLQSKGLEHSKLNNGTANGIKYERVERDTD 329
Query: 261 ------------------PVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTD 302
P S +L + + L M +++V A+ W++WFPFFLFDTD
Sbjct: 330 EQFGNSENEHQDETYVDGPGSVLVNLLTSLRHLPPAMHSVLIVMALTWLSWFPFFLFDTD 389
Query: 303 WMGREVY-----GGVVGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLW 357
WMGREVY G + + YD GVR G+LGL++N+VVL S +EP+ + + G + +W
Sbjct: 390 WMGREVYHGDPTGDSLHMELYDQGVREGALGLLLNSVVLGISSFLIEPMCQRM-GARVVW 448
Query: 358 GIVNFILAICMAMTVLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFS 417
+ NF + CMA T +I+ + D +G + + + A+ F ++G PLAI +S
Sbjct: 449 ALSNFTVFACMAGTAVISLMSLSDDK-NGIEYIMRGNETTRTAAVIVFALLGFPLAITYS 507
Query: 418 VPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVM 477
VPF++ + ++ +G GQGL++GVLNLAIV+PQM+VS +GPWD LFGGGNLPAFV+ +V
Sbjct: 508 VPFSVTAEVTADSGGGQGLAIGVLNLAIVIPQMIVSLGAGPWDQLFGGGNLPAFVLASVA 567
Query: 478 AAVSAIMAIVLLPT 491
A + ++A+ LPT
Sbjct: 568 AFAAGVIALQRLPT 581
>At5g13740 transporter-like protein
Length = 486
Score = 32.7 bits (73), Expect = 0.49
Identities = 40/176 (22%), Positives = 70/176 (39%), Gaps = 56/176 (31%)
Query: 62 GVPHIWASFIWLCG-----PVSGLL--------------VQPIVGYYSDRTTCRF--GR- 99
G P++ SF+W+ P+S L + +G+Y+ C F GR
Sbjct: 33 GYPYLELSFVWIIVLSTSLPISSLYPFLYYMIEDFGVAKTEKDIGFYAGFVGCSFMLGRA 92
Query: 100 --------------RRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFV 145
R+P IL G +++AI L G +++ ++G TR F+
Sbjct: 93 LTSVFWGIVADRYGRKPIILLGTISIAIFNALFGLSSNFWMAIG------TR------FL 140
Query: 146 FGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAY 201
G + N L G +A+ ++ + + TAM S +G ++G A G +
Sbjct: 141 LGSF------NCLLGTMKAYASEIFRDEYQ--ATAMSAVSTAWGIGLIIGPALGGF 188
>At1g74130 unknown protein
Length = 322
Score = 32.0 bits (71), Expect = 0.83
Identities = 32/118 (27%), Positives = 53/118 (44%), Gaps = 16/118 (13%)
Query: 314 GEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVL 373
G K++ +G GL+I + AV + R+LG N+W + NF+L+ MT
Sbjct: 106 GWKSWINGANGVVFGLVI-------ANAAVFTMWRVLGK-DNMWMVKNFMLSRYSFMTGR 157
Query: 374 ITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAG 431
I H + SG + VGA + + +FG +A +F + L ++ A G
Sbjct: 158 I-----HTLITSGFSHVGATHIILNMMGLCYFGAR---IARSFGPRYLLKLYFAGALG 207
>At5g09580 putative protein
Length = 393
Score = 30.4 bits (67), Expect = 2.4
Identities = 23/91 (25%), Positives = 42/91 (45%), Gaps = 10/91 (10%)
Query: 304 MGREVYGGVVGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFI 363
+GR +YG +G +A + + LG +++ L + + P+G + +N
Sbjct: 298 LGRGLYGECLGMRADGNHQLSDELGKLLS---LQSSAAGLRPIGAVTFVQRNN------- 347
Query: 364 LAICMAMTVLITKTAEHDRLISGGATVGAPS 394
L +C+ T IT T+E + GG T + S
Sbjct: 348 LKMCLRSTDAITNTSEVAKAYGGGGTSSSSS 378
>At3g54200 unknown protein
Length = 235
Score = 30.0 bits (66), Expect = 3.2
Identities = 14/39 (35%), Positives = 22/39 (55%)
Query: 222 NLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDS 260
N K C F+ILL+L+++I +I K+P +DS
Sbjct: 50 NCKICICFTILLILLIAIVIVILAFTLFKPKRPTTTIDS 88
>At1g05030 sugar transporter like protein
Length = 524
Score = 29.6 bits (65), Expect = 4.1
Identities = 14/51 (27%), Positives = 29/51 (56%), Gaps = 2/51 (3%)
Query: 100 RRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWI 150
R+ ++ + +A+S+FLI YA +G + +DL++ +++F F I
Sbjct: 382 RKKLLIGSYLGMAVSMFLIVYA--VGFPLDEDLSQSLSILGTLMYIFSFAI 430
>At1g60050 hypothetical protein
Length = 374
Score = 28.9 bits (63), Expect = 7.0
Identities = 26/98 (26%), Positives = 36/98 (36%), Gaps = 17/98 (17%)
Query: 94 TCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWILDV 153
T G PF+ S+ L+ Y+ DD T+P V IF+ GF + +
Sbjct: 33 TALTGGMSPFVFIVYTNALGSLLLLPYSFYFHRDESDDEPFLTKPSLVRIFLLGFTGVFL 92
Query: 154 ANNM-----------------LQGPCRAFLGDLAAGDQ 174
NM LQ P +FL LA G +
Sbjct: 93 FQNMAFLGLSYSSPIVVCAMGLQSPAFSFLLSLALGKE 130
>At1g10950 putative endomembrane protein EMP70 precusor isolog
(At1g10950)
Length = 589
Score = 28.9 bits (63), Expect = 7.0
Identities = 13/40 (32%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query: 327 LGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAI 366
+G ++N + + + SLA P G ++ V +WG ++F LA+
Sbjct: 373 IGFLLNTIAIFYGSLAAIPFGTMV-VVFVIWGFISFPLAL 411
>At5g07530 glycine-rich protein atGRP-7
Length = 543
Score = 28.5 bits (62), Expect = 9.2
Identities = 22/91 (24%), Positives = 37/91 (40%), Gaps = 8/91 (8%)
Query: 418 VPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVM 477
V FA ++ SAA + ++ ++VP + + + GG+ A +G +M
Sbjct: 79 VGFACVTLAGSAAALVVSTPVFIIFSPVLVPATIATVVLAT--GFTAGGSFGATALGLIM 136
Query: 478 AAVSAIMAIVLLPTPKPADVAKASSLPPGGG 508
V M + KP D + LPP G
Sbjct: 137 WLVKRRMGV------KPKDNPPPAGLPPNSG 161
>At3g26570 phosphate transporter like protein
Length = 587
Score = 28.5 bits (62), Expect = 9.2
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 9/69 (13%)
Query: 98 GRRRPFILAGAVAVAISVFLIGYAADIG---HSMGDDLAKKTRPRAVAIFVFGFWIL--- 151
G + F ++ A AIS+ + A + S+G LA KT+ + A +FGF++
Sbjct: 113 GMAQAFHISSTTARAISIVIAFSALTLPIFMKSLGQGLALKTKLLSYATLLFGFYMAWNI 172
Query: 152 ---DVANNM 157
DVAN M
Sbjct: 173 GANDVANAM 181
>At3g03620 unknown protein
Length = 466
Score = 28.5 bits (62), Expect = 9.2
Identities = 17/60 (28%), Positives = 33/60 (54%), Gaps = 8/60 (13%)
Query: 30 LRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYY 89
++ ++AV ++A+ G + L L+ YV LGV +W+ G ++G+ +Q I+ Y
Sbjct: 402 MQSIVAVVNLASYYAIG--IPLGLILTYVFHLGVKGLWS------GMLAGIAIQTIILCY 453
>At1g71390 putative disease resistance protein
Length = 784
Score = 28.5 bits (62), Expect = 9.2
Identities = 18/67 (26%), Positives = 35/67 (51%), Gaps = 5/67 (7%)
Query: 105 LAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWILDVANNMLQGPCRA 164
L G++ +IS FL D+ H+ ++ + V++ +FGF +NN L+G +
Sbjct: 306 LDGSIPESISKFLNLVLLDVAHNNISGPVPRSMSKLVSLRIFGF-----SNNKLEGEVPS 360
Query: 165 FLGDLAA 171
+L L++
Sbjct: 361 WLWRLSS 367
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,998,883
Number of Sequences: 26719
Number of extensions: 455655
Number of successful extensions: 1577
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1531
Number of HSP's gapped (non-prelim): 22
length of query: 510
length of database: 11,318,596
effective HSP length: 104
effective length of query: 406
effective length of database: 8,539,820
effective search space: 3467166920
effective search space used: 3467166920
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0134.5


