

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.21
(515 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
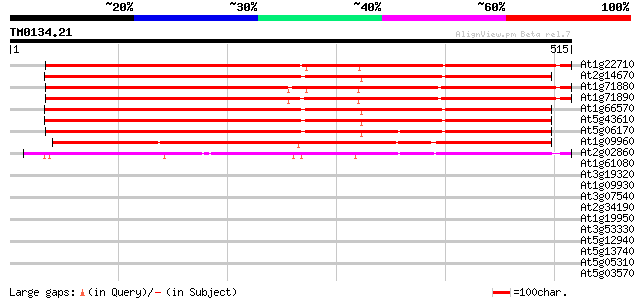
Score E
Sequences producing significant alignments: (bits) Value
At1g22710 putative sucrose transport protein, SUC2 649 0.0
At2g14670 putative sucrose-proton symporter 648 0.0
At1g71880 sucrose transport protein SUC1 645 0.0
At1g71890 putative sucrose transport protein 644 0.0
At1g66570 hypothetical protein 642 0.0
At5g43610 sucrose transporter protein 640 0.0
At5g06170 sucrose transporter protein 632 0.0
At1g09960 putative sucrose/H+ symporter 490 e-139
At2g02860 Sucrose transporter (suc3) 433 e-121
At1g61080 hypothetical protein 33 0.29
At3g19320 unknown protein 32 0.84
At1g09930 unknown protein 32 0.84
At3g07540 putative protein 31 1.9
At2g34190 putative membrane transporter 30 2.4
At1g19950 unknown protein 30 2.4
At3g53330 putative protein 30 3.2
At5g12940 unknown protein 30 4.2
At5g13740 transporter-like protein 29 5.5
At5g05310 unknown protein 29 7.1
At5g03570 transporter like protein 29 7.1
>At1g22710 putative sucrose transport protein, SUC2
Length = 512
Score = 649 bits (1673), Expect = 0.0
Identities = 325/491 (66%), Positives = 391/491 (79%), Gaps = 13/491 (2%)
Query: 34 PSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIV 93
P L K+I+V+SIAAGVQFGWALQLSLLTPYVQLLG+PHKW+S IWLCGPISG++VQPIV
Sbjct: 26 PERLRKIISVSSIAAGVQFGWALQLSLLTPYVQLLGIPHKWASLIWLCGPISGMLVQPIV 85
Query: 94 GYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVI 153
GY SDRCTSRFGRRRPFI AGA V +AVFLIG+AADIG+SMGD L K + RA+A F +
Sbjct: 86 GYHSDRCTSRFGRRRPFIVAGAGLVTVAVFLIGYAADIGHSMGDQLDKPPKTRAIAIFAL 145
Query: 154 GFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHK 213
GFWILDVANN LQGPCRAFL DLS G+ + RTAN FSFFM VGNVLGY AGSY L+K
Sbjct: 146 GFWILDVANNTLQGPCRAFLADLSAGNAKKTRTANAFFSFFMAVGNVLGYAAGSYRNLYK 205
Query: 214 IFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREEDDKAPKNV-- 271
+ PFT T++CD++CANLK+CFF SITLLL+++ +L YV + P D KA NV
Sbjct: 206 VVPFTMTESCDLYCANLKTCFFLSITLLLIVTFVSLCYVKEKPWTPEPTADGKA-SNVPF 264
Query: 272 FVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGK-------LGSKA 324
F E+FGAFKELK+PM ML++VT+LNWIAWFP++L+DTDWMG EVYGG K
Sbjct: 265 FGEIFGAFKELKRPMWMLLIVTALNWIAWFPFLLFDTDWMGREVYGGNSDATATAASKKL 324
Query: 325 YDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKV 384
Y+ GVRAGALGL+LN++VLG MSL VE +GR LGG KRLW IVN ILA+C+AMT+++TK
Sbjct: 325 YNDGVRAGALGLMLNAIVLGFMSLGVEWIGRKLGGAKRLWGIVNFILAICLAMTVVVTKQ 384
Query: 385 AEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLS 444
AE+ RR GGA G P V AGAL FA+LGIP AIT+S+PFALASI+S+++GAGQGLS
Sbjct: 385 AENHRRDHGGAKTG-PPGNVTAGALTLFAILGIPQAITFSIPFALASIFSTNSGAGQGLS 443
Query: 445 LGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSPKPEEM 504
LGVLN+AIV+PQM++S GP+D LFGGGN+PAF++GA+ AAVS VLA+ +LPSP P+
Sbjct: 444 LGVLNLAIVVPQMVISVGGGPFDELFGGGNIPAFVLGAIAAAVSGVLALTVLPSPPPD-- 501
Query: 505 AKASISASGFH 515
A A + GFH
Sbjct: 502 APAFKATMGFH 512
>At2g14670 putative sucrose-proton symporter
Length = 492
Score = 648 bits (1671), Expect = 0.0
Identities = 322/471 (68%), Positives = 382/471 (80%), Gaps = 10/471 (2%)
Query: 33 GPSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPI 92
GPSPL KMI+VASIAAG+QFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGP+SGL+VQP
Sbjct: 26 GPSPLRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPVSGLLVQPS 85
Query: 93 VGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFV 152
VGY SDRCTSRFGRRRPFI GA+ VA+AV LIG+AAD G+SMGD + K + RAV F
Sbjct: 86 VGYFSDRCTSRFGRRRPFIATGALLVAVAVVLIGYAADFGHSMGDKIDKPVKMRAVVIFA 145
Query: 153 IGFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLH 212
+GFWILDVANN LQGPCRAFLGDL+ G + RTAN FSFFM VGNVLGY AGSY L+
Sbjct: 146 LGFWILDVANNTLQGPCRAFLGDLAAGDAKKTRTANAFFSFFMAVGNVLGYAAGSYTNLY 205
Query: 213 KIFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREED-DKAPKNV 271
KIFPFT TKACD++CANLKSCFF SITLLLV++ AL+YV D + + D +K P
Sbjct: 206 KIFPFTMTKACDIYCANLKSCFFLSITLLLVVTIIALWYVEDKQWSPKADSDNEKTP--F 263
Query: 272 FVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLG-----SKAYD 326
F E+FGAFK +K+PM ML++VT+LNWIAWFP++LYDTDWMG EVYGG K Y+
Sbjct: 264 FGEIFGAFKVMKRPMWMLLIVTALNWIAWFPFLLYDTDWMGREVYGGDSKGDDKMKKLYN 323
Query: 327 AGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAE 386
G+ GALGL+LNS+VLG++SL +E + + +GG KRLW VNIILAVC+AMT+L+TK AE
Sbjct: 324 QGIHVGALGLMLNSIVLGIVSLGIEGISKKIGGAKRLWGAVNIILAVCLAMTVLVTKKAE 383
Query: 387 HDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLG 446
RR++G + PT G++AGAL FA+LGIPLAIT+S+PFALASI SSS+GAGQGLSLG
Sbjct: 384 EHRRIAGPMAL--PTDGIRAGALTLFALLGIPLAITFSIPFALASIISSSSGAGQGLSLG 441
Query: 447 VLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLP 497
VLN+AIVIPQMIVS GP D+LFGGGNLP F+VGA+ AA+S+V+A +LP
Sbjct: 442 VLNMAIVIPQMIVSFGVGPIDALFGGGNLPRFVVGAIAAAISSVVAFTVLP 492
>At1g71880 sucrose transport protein SUC1
Length = 513
Score = 645 bits (1663), Expect = 0.0
Identities = 323/493 (65%), Positives = 389/493 (78%), Gaps = 17/493 (3%)
Query: 34 PSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIV 93
PSPL K+I+VASIAAGVQFGWALQLSLLTPYVQLLG+PHKWSS IWLCGP+SG++VQPIV
Sbjct: 27 PSPLRKIISVASIAAGVQFGWALQLSLLTPYVQLLGIPHKWSSLIWLCGPVSGMIVQPIV 86
Query: 94 GYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVI 153
G+ SDRC S+FGRRRPFI GA VA+AVFLIG+AAD GY MGD L +K + RA+ F +
Sbjct: 87 GFHSDRCRSKFGRRRPFIATGAALVAVAVFLIGYAADFGYKMGDKLEEKVKVRAIGIFAL 146
Query: 154 GFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHK 213
GFWILDVANN LQGPCRAFL DL+ G R R AN FSFFM VGNVLGY AGSY LHK
Sbjct: 147 GFWILDVANNTLQGPCRAFLADLAAGDAKRTRVANAFFSFFMAVGNVLGYAAGSYTNLHK 206
Query: 214 IFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHD----PPIGSRREEDDKAPK 269
+FPFT TKACD++CANLK+CFF SITLLL+++ +L+YV+D PP R +DD+
Sbjct: 207 MFPFTMTKACDIYCANLKTCFFLSITLLLIVTVTSLWYVNDKQWSPP--PRNADDDEKTS 264
Query: 270 NV--FVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGG-----KLGS 322
+V F E+FGAFK +K+PM ML++VT+LNWIAWFP++L+DTDWMG EV+GG +
Sbjct: 265 SVPLFGEIFGAFKVMKRPMWMLLIVTALNWIAWFPFLLFDTDWMGREVFGGDSDGNERSK 324
Query: 323 KAYDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLIT 382
K Y GV++GA+GL+ NS+VLG MSL VE +GR LGG KRLW IVN ILA +AMT+L+T
Sbjct: 325 KLYSLGVQSGAMGLMFNSIVLGFMSLGVEWIGRKLGGAKRLWGIVNFILAAGLAMTVLVT 384
Query: 383 KVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQG 442
K AE R+ +G + P+ VKAGAL FAVLGIPLAIT+S PFALASI+SS +GAGQG
Sbjct: 385 KFAEDHRKTAG--DLAGPSASVKAGALSLFAVLGIPLAITFSTPFALASIFSSCSGAGQG 442
Query: 443 LSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSPKPE 502
LSLGVLN+AIVIPQMIVS GP+D+LFGGGNLPAF+V A+ AA+S VLA+ +LPSP P+
Sbjct: 443 LSLGVLNLAIVIPQMIVSLGGGPFDALFGGGNLPAFIVAAIAAAISGVLALTVLPSPPPD 502
Query: 503 EMAKASISASGFH 515
A + + GFH
Sbjct: 503 --APKATTMGGFH 513
>At1g71890 putative sucrose transport protein
Length = 512
Score = 644 bits (1662), Expect = 0.0
Identities = 317/491 (64%), Positives = 385/491 (77%), Gaps = 15/491 (3%)
Query: 34 PSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIV 93
PSPL K+I+VASIAAGVQFGWALQLSLLTPY+QLLG+PHKWSS++WLCGPISG++VQPIV
Sbjct: 28 PSPLRKIISVASIAAGVQFGWALQLSLLTPYIQLLGIPHKWSSYMWLCGPISGMIVQPIV 87
Query: 94 GYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVI 153
GY SDRC SRFGRRRPFI AG VA++VFLIGFAAD+G+S GD L K R RA+ F+
Sbjct: 88 GYHSDRCESRFGRRRPFIAAGVALVAVSVFLIGFAADMGHSFGDKLENKVRTRAIIIFLT 147
Query: 154 GFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHK 213
GFW LDVANN LQGPCRAFL DL+ G + R AN FSFFM VGNVLGY AGSY LHK
Sbjct: 148 GFWFLDVANNTLQGPCRAFLADLAAGDAKKTRVANACFSFFMAVGNVLGYAAGSYTNLHK 207
Query: 214 IFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHD----PPIGSRREEDDKAPK 269
+FPFT TKACD++CANLK+CFF SITLLL+++ +L+YV D PP G + E+
Sbjct: 208 MFPFTMTKACDIYCANLKTCFFLSITLLLIVTFSSLWYVKDKQWSPPQGDKEEKTSSL-- 265
Query: 270 NVFVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGG-----KLGSKA 324
F E+FGA + +K+PM+ML++VT +NWIAWFP++LYDTDWMG EVYGG + K
Sbjct: 266 FFFGEIFGAVRHMKRPMVMLLIVTVINWIAWFPFILYDTDWMGREVYGGNSDGDERSKKL 325
Query: 325 YDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKV 384
YD GV+AGALGL+ NS++LG +SL VE +GR +GG KRLW VN ILA+ +AMT+L+TK
Sbjct: 326 YDQGVQAGALGLMFNSILLGFVSLGVESIGRKMGGAKRLWGCVNFILAIGLAMTVLVTKS 385
Query: 385 AEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLS 444
AEH R ++G + P+ G+KAG F VLGIPLAITYS+PFALASI+S+++GAGQGLS
Sbjct: 386 AEHHREIAG--PLAGPSSGIKAGVFSLFTVLGIPLAITYSIPFALASIFSTNSGAGQGLS 443
Query: 445 LGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSPKPEEM 504
LGVLN+AI IPQMIVS +GP D+ FGGGNLP+F+VGA+ AAVS VLA+ +LPSP P+
Sbjct: 444 LGVLNIAICIPQMIVSFSSGPLDAQFGGGNLPSFVVGAIAAAVSGVLALTVLPSPPPD-- 501
Query: 505 AKASISASGFH 515
A A A GFH
Sbjct: 502 APAMSGAMGFH 512
>At1g66570 hypothetical protein
Length = 491
Score = 642 bits (1655), Expect = 0.0
Identities = 320/471 (67%), Positives = 379/471 (79%), Gaps = 10/471 (2%)
Query: 33 GPSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPI 92
GPSPL KMI+VASIAAG+QFGWALQLSLLTPYVQLLGVPHKW SFIWLCGP+SGL+VQP
Sbjct: 25 GPSPLRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWPSFIWLCGPVSGLLVQPS 84
Query: 93 VGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFV 152
VGY SDRCTSRFGRRRPFI GA+ VA++V LIG+AAD G+SMGD + K + RAV F
Sbjct: 85 VGYFSDRCTSRFGRRRPFIATGALLVAVSVVLIGYAADFGHSMGDKIDKPVKMRAVVIFA 144
Query: 153 IGFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLH 212
+GFWILDVANN LQGPCRAFLGDL+ G + RTAN FSFFM VGNVLGY AGSY L+
Sbjct: 145 LGFWILDVANNTLQGPCRAFLGDLAAGDAQKTRTANAFFSFFMAVGNVLGYAAGSYTNLY 204
Query: 213 KIFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREED-DKAPKNV 271
KIFPFT TKACD++CANLKSCFF SITLLLV++ AL+YV D + + D +K P
Sbjct: 205 KIFPFTMTKACDIYCANLKSCFFLSITLLLVVTIIALWYVEDKQWSPKADSDNEKTP--F 262
Query: 272 FVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLG-----SKAYD 326
F E+FGAFK +K+PM ML++VT+LNWIAWFP++LYDTDWMG EVYGG K Y+
Sbjct: 263 FGEIFGAFKVMKRPMWMLLIVTALNWIAWFPFLLYDTDWMGREVYGGDSKGDDKMKKLYN 322
Query: 327 AGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAE 386
G+ GALGL+LNS+VLG+MSL +E + R +GG KRLW VNIILAVC+AMT+L+TK AE
Sbjct: 323 QGIHVGALGLMLNSIVLGVMSLGIEGISRKMGGAKRLWGAVNIILAVCLAMTVLVTKKAE 382
Query: 387 HDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLG 446
RR++G + PT G++AGAL FA+LGIPLAIT+S+PFALASI SSS+GAGQ LSLG
Sbjct: 383 EHRRIAGPMAL--PTDGIRAGALTLFALLGIPLAITFSIPFALASIISSSSGAGQRLSLG 440
Query: 447 VLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLP 497
VLN+AIVIPQMIVS GP D+LFG GNLP F+VGA+ AAVS+++A +LP
Sbjct: 441 VLNMAIVIPQMIVSFGVGPIDALFGDGNLPGFVVGAIAAAVSSIVAFTVLP 491
>At5g43610 sucrose transporter protein
Length = 492
Score = 640 bits (1652), Expect = 0.0
Identities = 318/471 (67%), Positives = 378/471 (79%), Gaps = 10/471 (2%)
Query: 33 GPSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPI 92
GPSP+ KMI+VASIAAG+QFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGP+SGL+VQP
Sbjct: 26 GPSPMRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPVSGLLVQPS 85
Query: 93 VGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFV 152
VGY SDRC SRFGRRRPFI GA+ VA+AV LIG+AAD G+SMGD + + + RAV F
Sbjct: 86 VGYFSDRCKSRFGRRRPFIAMGALLVAVAVVLIGYAADFGHSMGDKVDEPVKMRAVVIFA 145
Query: 153 IGFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLH 212
+GFWILDVANN LQGPCRAFLGDL+ G + RTAN FSFFM VGNVLGY AGSY L+
Sbjct: 146 LGFWILDVANNTLQGPCRAFLGDLAAGDAKKTRTANAFFSFFMAVGNVLGYAAGSYTNLY 205
Query: 213 KIFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREED-DKAPKNV 271
KIFPFT TKACD++CANLKSCFF SITLLLV++ AL+YV D + + D +K P
Sbjct: 206 KIFPFTMTKACDIYCANLKSCFFLSITLLLVVTIIALWYVEDKQWSPKADSDNEKTP--F 263
Query: 272 FVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLG-----SKAYD 326
F E+FGAFK +K+PM ML++VT+LNWIAWFP++LYDTDWMG EVYGG K Y+
Sbjct: 264 FGEIFGAFKVMKRPMWMLLIVTALNWIAWFPFLLYDTDWMGREVYGGDSKGDDKMKKLYN 323
Query: 327 AGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAE 386
G+ G LGL+LNS+VLG MSL +E + R +GG KRLW VNIILAVC+AMT+L+TK AE
Sbjct: 324 QGIHVGGLGLMLNSIVLGFMSLGIEGISRKMGGAKRLWGAVNIILAVCLAMTVLVTKKAE 383
Query: 387 HDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLG 446
RR++G + PT G++AGAL FA+LGIPLAIT+S+PFALASI SSS+GAGQGLSLG
Sbjct: 384 EHRRIAGPMAL--PTDGIRAGALTLFALLGIPLAITFSIPFALASIISSSSGAGQGLSLG 441
Query: 447 VLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLP 497
VLN+ IVIPQM+VS GP D+LFGGGNLP F+VGA+ AA+S+V+A +LP
Sbjct: 442 VLNMTIVIPQMVVSFGVGPIDALFGGGNLPGFVVGAIAAAISSVVAFSVLP 492
>At5g06170 sucrose transporter protein
Length = 491
Score = 632 bits (1630), Expect = 0.0
Identities = 318/470 (67%), Positives = 377/470 (79%), Gaps = 11/470 (2%)
Query: 34 PSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIV 93
PSPL KMI+VASIAAG+QFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGL+VQP V
Sbjct: 27 PSPLRKMISVASIAAGIQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLLVQPTV 86
Query: 94 GYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVI 153
GY SDRC SRFGRRRPFI GA+ VA+AV LIGFAAD G++MGD L + + RAV FFV+
Sbjct: 87 GYFSDRCKSRFGRRRPFIATGALLVALAVILIGFAADFGHTMGDKLDEAVKIRAVGFFVV 146
Query: 154 GFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHK 213
GFWILDVANN LQGPCRAFLGDL+ G + RTAN IFSFFM VGNVLGY AGSY LHK
Sbjct: 147 GFWILDVANNTLQGPCRAFLGDLAAGDAKKTRTANAIFSFFMAVGNVLGYAAGSYTNLHK 206
Query: 214 IFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREED-DKAPKNVF 272
IFPFT TKACD++CANLKSCF SITLL+VL+ AL+YV D + D +K P F
Sbjct: 207 IFPFTVTKACDIYCANLKSCFIISITLLIVLTIIALWYVEDKQWSPNADSDNEKTP--FF 264
Query: 273 VELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLG-----SKAYDA 327
E+FGAFK +K+PM ML+ VT+LNWIAWFP++LYDTDWMG EVYGG K Y+
Sbjct: 265 GEIFGAFKVMKRPMWMLLAVTALNWIAWFPFLLYDTDWMGREVYGGDSAGDDKMKKLYNH 324
Query: 328 GVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEH 387
G++ G+LGL+LNS+VLG+MSL + + + + G KRLW VNIILAVC+AMT+L+TK AE
Sbjct: 325 GIQVGSLGLMLNSIVLGVMSLVIGVISKKI-GAKRLWGAVNIILAVCLAMTVLVTKKAEE 383
Query: 388 DRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGV 447
R+++G + PT ++ GAL FA+LGIPLAIT+S+PFALASI SSS+GAGQGLSLGV
Sbjct: 384 HRKIAGRMAL--PTNAIRDGALSLFAILGIPLAITFSIPFALASIISSSSGAGQGLSLGV 441
Query: 448 LNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLP 497
LN+AIVIPQMIVS GP D+LFGGGNLP F+VGA+ A +S+V+A+ +LP
Sbjct: 442 LNMAIVIPQMIVSFGVGPIDALFGGGNLPGFVVGAIAALISSVVALTVLP 491
>At1g09960 putative sucrose/H+ symporter
Length = 510
Score = 490 bits (1261), Expect = e-139
Identities = 248/463 (53%), Positives = 322/463 (68%), Gaps = 11/463 (2%)
Query: 40 MIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDR 99
++ VAS+A G+QFGWALQLSLLTPYVQ LG+PH W+S IWLCGP+SGL VQP+VG+SSDR
Sbjct: 43 LLRVASVACGIQFGWALQLSLLTPYVQELGIPHAWASVIWLCGPLSGLFVQPLVGHSSDR 102
Query: 100 CTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILD 159
CTS++GRRRPFI AGA+A++I+V +IG AADIG++ G D K +PRA+ FV+GFWILD
Sbjct: 103 CTSKYGRRRPFIVAGAVAISISVMVIGHAADIGWAFG-DREGKIKPRAIVAFVLGFWILD 161
Query: 160 VANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTE 219
VANNM QGPCRA L DL+ + R R AN FS FM VGNVLGY GSY G +KIF FT+
Sbjct: 162 VANNMTQGPCRALLADLTENDNRRTRVANGYFSLFMAVGNVLGYATGSYNGWYKIFTFTK 221
Query: 220 TKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREE---DDKAPKNVFV-EL 275
T AC+V CANLKS F+ + + + + ++ H+ P+ S E F+ E+
Sbjct: 222 TVACNVECANLKSAFYIDVVFIAITTILSVSAAHEVPLASLASEAHGQTSGTDEAFLSEI 281
Query: 276 FGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLG-SKAYDAGVRAGAL 334
FG F+ + +++LVT+L WI WFP++L+DTDWMG E+YGG+ +Y AGV GAL
Sbjct: 282 FGTFRYFPGNVWIILLVTALTWIGWFPFILFDTDWMGREIYGGEPNIGTSYSAGVSMGAL 341
Query: 335 GLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGG 394
GL+LNSV LG+ S+ +E L R G +W I NI++A+C ++ + VA H G
Sbjct: 342 GLMLNSVFLGITSVLMEKLCR-KWGAGFVWGISNILMAICFLGMIITSFVASH----LGY 396
Query: 395 ATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVI 454
+P + A++ F +LGIPLAITYSVP+AL SI S G GQGLSLGVLN+AIVI
Sbjct: 397 IGHEQPPASIVFAAVLIFTILGIPLAITYSVPYALISIRIESLGLGQGLSLGVLNLAIVI 456
Query: 455 PQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLP 497
PQ+IVS +GPWD LFGGGN PA VGA + ++A++ LP
Sbjct: 457 PQVIVSVGSGPWDQLFGGGNSPALAVGAATGFIGGIVAILALP 499
>At2g02860 Sucrose transporter (suc3)
Length = 594
Score = 433 bits (1114), Expect = e-121
Identities = 243/575 (42%), Positives = 339/575 (58%), Gaps = 82/575 (14%)
Query: 13 QNNNSIAPSSFQVEPAQH---AAGPS-----PLAKMIAVASIAAGVQFGWALQLSLLTPY 64
QN + + S P+ H A G S L ++ ++AAGVQFGWALQLSLLTPY
Sbjct: 28 QNESGSSSFSESASPSNHSDSADGESVSKNCSLVTLVLSCTVAAGVQFGWALQLSLLTPY 87
Query: 65 VQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFL 124
+Q LG+ H +SSFIWLCGPI+GLVVQP VG SD+CTS++GRRRPFI G+ ++IAV +
Sbjct: 88 IQTLGISHAFSSFIWLCGPITGLVVQPFVGIWSDKCTSKYGRRRPFILVGSFMISIAVII 147
Query: 125 IGFAADIGYSMGDDLS-----KKTRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTG 179
IGF+ADIGY +GD K TR RA F+IGFW+LD+ANN +QGP RA L DLS G
Sbjct: 148 IGFSADIGYLLGDSKEHCSTFKGTRTRAAVVFIIGFWLLDLANNTVQGPARALLADLS-G 206
Query: 180 HHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSIT 239
R TAN +F +M +GN+LG+ AG+ G + FPF ++AC C NLK+ F ++
Sbjct: 207 PDQR-NTANAVFCLWMAIGNILGFSAGASGKWQEWFPFLTSRACCAACGNLKAAFLLAVV 265
Query: 240 LLLVLSGFALFYVHDPPIGS---------------------------------------R 260
L + + +++ + P S
Sbjct: 266 FLTICTLVTIYFAKEIPFTSNKPTRIQDSAPLLDDLQSKGLEHSKLNNGTANGIKYERVE 325
Query: 261 REEDDK---------------APKNVFVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVL 305
R+ D++ P +V V L + + L M +++V +L W++WFP+ L
Sbjct: 326 RDTDEQFGNSENEHQDETYVDGPGSVLVNLLTSLRHLPPAMHSVLIVMALTWLSWFPFFL 385
Query: 306 YDTDWMGLEVY-----GGKLGSKAYDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGV 360
+DTDWMG EVY G L + YD GVR GALGL+LNSVVLG+ S +EP+ + + G
Sbjct: 386 FDTDWMGREVYHGDPTGDSLHMELYDQGVREGALGLLLNSVVLGISSFLIEPMCQRM-GA 444
Query: 361 KRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLA 420
+ +WA+ N + CMA T +I+ ++ D + +G I R + A++ FA+LG PLA
Sbjct: 445 RVVWALSNFTVFACMAGTAVISLMSLSDDK-NGIEYIMRGNETTRTAAVIVFALLGFPLA 503
Query: 421 ITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMV 480
ITYSVPF++ + ++ +G GQGL++GVLN+AIVIPQMIVS GPWD LFGGGNLPAF++
Sbjct: 504 ITYSVPFSVTAEVTADSGGGQGLAIGVLNLAIVIPQMIVSLGAGPWDQLFGGGNLPAFVL 563
Query: 481 GAVMAAVSAVLAMVLLPSPKPEEMAKASISASGFH 515
+V A + V+A+ LP+ +S ++GFH
Sbjct: 564 ASVAAFAAGVIALQRLPT------LSSSFKSTGFH 592
>At1g61080 hypothetical protein
Length = 907
Score = 33.5 bits (75), Expect = 0.29
Identities = 18/47 (38%), Positives = 23/47 (48%)
Query: 3 PPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAG 49
PPPPPP N + P + A AAGP P + +A+ AAG
Sbjct: 590 PPPPPPPMPLANGATPPPPPPPMAMANGAAGPPPPPPRMGMANGAAG 636
>At3g19320 unknown protein
Length = 493
Score = 32.0 bits (71), Expect = 0.84
Identities = 14/34 (41%), Positives = 16/34 (46%)
Query: 3 PPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSP 36
PPPPPP S + S P + P H PSP
Sbjct: 70 PPPPPPQSLPPPSPSPEPEHYPPPPYHHYITPSP 103
>At1g09930 unknown protein
Length = 734
Score = 32.0 bits (71), Expect = 0.84
Identities = 21/99 (21%), Positives = 42/99 (42%), Gaps = 25/99 (25%)
Query: 285 PMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGSKAYDAGVRAGALGLVLNSVVLG 344
P + + ++S++W+ W A+ + A LG ++ + +G
Sbjct: 226 PAYLFLTLSSISWVCW-----------------------AFPKSITAQQLGSGMSGLGIG 262
Query: 345 LMSLAVEPLGRYLGG--VKRLWAIVNIILAVCMAMTMLI 381
+L + YLG V +AIVN+++ + M M+I
Sbjct: 263 AFALDWSVIASYLGSPLVTPFFAIVNVLVGYVLVMYMVI 301
>At3g07540 putative protein
Length = 841
Score = 30.8 bits (68), Expect = 1.9
Identities = 13/23 (56%), Positives = 16/23 (69%)
Query: 3 PPPPPPNSTTQNNNSIAPSSFQV 25
P PPP N+ TQN S+APS+ V
Sbjct: 153 PCPPPRNNNTQNKLSVAPSTSDV 175
>At2g34190 putative membrane transporter
Length = 524
Score = 30.4 bits (67), Expect = 2.4
Identities = 55/217 (25%), Positives = 91/217 (41%), Gaps = 43/217 (19%)
Query: 288 MLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGSKAYDAGVRAGALGLVLNSVV----- 342
M L++S WI + Y W G+ ++DAG + VL S++
Sbjct: 254 MSNLISSAPWIK----IPYPLQW----------GAPSFDAGHAFAMMAAVLVSLIESTGA 299
Query: 343 ---LGLMSLAVEP----LGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGA 395
++ A P L R +G W + I+L T+ + V+ + + G
Sbjct: 300 FKAAARLASATPPPPHVLSRGIG-----WQGIGILLNGLFG-TLSGSSVSVENIGLLGST 353
Query: 396 TIG-RPTPGVKAGALMFFAVLGIPLAITYSVPFAL-ASIYSSSTG--AGQGLS-LGVLNV 450
+G R + AG ++FF++LG A+ S+PF + A++Y G A GLS L N+
Sbjct: 354 RVGSRRVIQISAGFMIFFSMLGKFGALFASIPFTIFAAVYCVLFGLVASVGLSFLQFTNM 413
Query: 451 AIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAV 487
+ IV SLF G ++P + M A+
Sbjct: 414 NSLRNLFIVGV------SLFLGLSIPEYFRDFSMKAL 444
>At1g19950 unknown protein
Length = 315
Score = 30.4 bits (67), Expect = 2.4
Identities = 18/48 (37%), Positives = 26/48 (53%), Gaps = 5/48 (10%)
Query: 3 PPPPPPNSTT---QNNNSIAPSSFQVEPAQH--AAGPSPLAKMIAVAS 45
PPPPPP+ TT +N + PS + E A AA P P +++ +S
Sbjct: 241 PPPPPPSPTTAAKRNADPAQPSPTEAEEASQTVAALPEPASEIQRASS 288
>At3g53330 putative protein
Length = 310
Score = 30.0 bits (66), Expect = 3.2
Identities = 18/58 (31%), Positives = 26/58 (44%), Gaps = 13/58 (22%)
Query: 3 PPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASI-AAGVQFGWALQLS 59
PPPPPP+ T + + I PS P P +K++ I G GW++ S
Sbjct: 156 PPPPPPSKTHEPSRRITPS------------PPPPSKILPFGKIYRVGDYGGWSVYYS 201
>At5g12940 unknown protein
Length = 371
Score = 29.6 bits (65), Expect = 4.2
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 6/33 (18%)
Query: 155 FWILDVANNMLQGPCRA------FLGDLSTGHH 181
F +LD+ANN LQGP A F+G L H+
Sbjct: 303 FTVLDLANNRLQGPIPASITAASFIGHLDVSHN 335
>At5g13740 transporter-like protein
Length = 486
Score = 29.3 bits (64), Expect = 5.5
Identities = 27/106 (25%), Positives = 49/106 (45%), Gaps = 21/106 (19%)
Query: 103 RFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVAN 162
R+GR+ P I G I++AI L G +++ ++G TR F++G +
Sbjct: 104 RYGRK-PIILLGTISIAIFNALFGLSSNFWMAIG------TR------FLLGSF------ 144
Query: 163 NMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSY 208
N L G +A+ ++ + TA + S G+G ++G G +
Sbjct: 145 NCLLGTMKAYASEIFRDEYQ--ATAMSAVSTAWGIGLIIGPALGGF 188
>At5g05310 unknown protein
Length = 496
Score = 28.9 bits (63), Expect = 7.1
Identities = 35/129 (27%), Positives = 53/129 (40%), Gaps = 22/129 (17%)
Query: 9 NSTTQNNNSIAP-----SSFQVEPAQHAAGPSPLAKMIAVASIAAGV---QFGWAL---- 56
N +Q++ ++P + Q E Q A P + +A QF WA+
Sbjct: 233 NDISQSSEELSPLRGTDNDHQRERKQEATSPKVGSPKVASPKSPISTTRPQF-WAILDGM 291
Query: 57 QLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAI 116
+L L +PY+ L+ S F+WL IS V + S GRRR F I
Sbjct: 292 RLILASPYLLLV------SLFLWLGAVISSFFYFQKVNIIATTIKSSIGRRRLF---AQI 342
Query: 117 AVAIAVFLI 125
+AVF++
Sbjct: 343 NSFVAVFIL 351
>At5g03570 transporter like protein
Length = 498
Score = 28.9 bits (63), Expect = 7.1
Identities = 26/89 (29%), Positives = 44/89 (49%), Gaps = 7/89 (7%)
Query: 402 PGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSA 461
PGV AL+FF VL +T ++ + I + G G+G+S GV A V+ ++ S
Sbjct: 313 PGVSL-ALLFFTVLSFGTLMTATLEW--KGIPTYIIGIGRGISAGVGLAATVLYPLMQSR 369
Query: 462 LN----GPWDSLFGGGNLPAFMVGAVMAA 486
++ G W + ++M+ A +AA
Sbjct: 370 ISPLRTGVWSFWSQKEKIASYMLMAGVAA 398
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,622,798
Number of Sequences: 26719
Number of extensions: 496086
Number of successful extensions: 3451
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 3291
Number of HSP's gapped (non-prelim): 102
length of query: 515
length of database: 11,318,596
effective HSP length: 104
effective length of query: 411
effective length of database: 8,539,820
effective search space: 3509866020
effective search space used: 3509866020
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0134.21


