

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.17
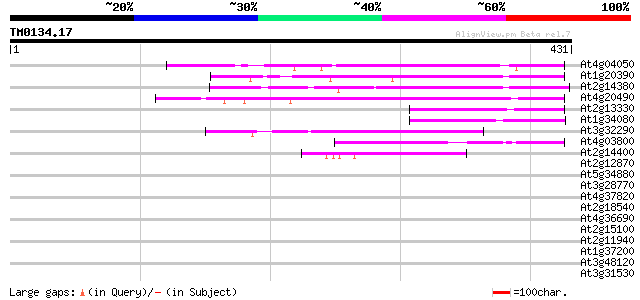
(431 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g04050 putative transposon protein 90 2e-18
At1g20390 hypothetical protein 84 2e-16
At2g14380 putative retroelement pol polyprotein 74 2e-13
At4g20490 putative protein 70 2e-12
At2g13330 F14O4.9 65 9e-11
At1g34080 putative protein 64 1e-10
At3g32290 hypothetical protein 64 2e-10
At4g03800 49 5e-06
At2g14400 putative retroelement pol polyprotein 42 7e-04
At2g12870 putative retroelement pol polyprotein 41 0.001
At5g34880 putative protein 37 0.016
At3g28770 hypothetical protein 34 0.14
At4g37820 unknown protein 34 0.18
At2g18540 putative vicilin storage protein (globulin-like) 33 0.30
At4g36690 splicing factor like protein 33 0.40
At2g15100 putative retroelement pol polyprotein 33 0.40
At2g11940 putative retroelement gag/pol polyprotein 32 0.52
At1g37200 hypothetical protein 32 0.52
At3g48120 putative protein 32 0.68
At3g31530 hypothetical protein 32 0.68
>At4g04050 putative transposon protein
Length = 681
Score = 90.1 bits (222), Expect = 2e-18
Identities = 77/324 (23%), Positives = 142/324 (43%), Gaps = 42/324 (12%)
Query: 121 AHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLI 180
AHL + ++ + + + +D K+C+YH+ GY T +C +K +LI
Sbjct: 351 AHLRSFRFAITRAYLTDEENEAAENESPELDLGKYCKYHKKRGYSTEECRAVK----KLI 406
Query: 181 RAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRRRDD-------RRRTPTSDKKETLATR 233
AG G ++G+ K + D + ++ R RTP A
Sbjct: 407 AAG------------GKTKKGSNPKVETPPPDEQEEEQTPKQKKRERTPEGITTPPPAPH 454
Query: 234 KKGSE---------ETFNKDLEPPVGTINTIAGGFGGGGDIASTRRRNVRAVTSVREYEA 284
KK + + L P I+ I GG D ++ + + R S + ++
Sbjct: 455 KKNMRFPLRLLKFCRSATEHLSPK--RIDFIMGGSQLCNDSINSIKTHQRKADSYTKGKS 512
Query: 285 PFGFHHPDIVISSADF*RIKTHKDDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQL 344
P + I ++ + DD +V+ I + ++ + ++++ GSS D+++ DAF ++
Sbjct: 513 PMMGPNHQITFWESETTDLNKPHDDALVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRM 572
Query: 345 GLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHTVFGVDENAKLLR--VRYLVLQVVASYNV 402
G D +L L GF+G+ + G + L T+ A+ +R +LV+ A +N
Sbjct: 573 GHLDSELQGRKTPLTGFAGDTTFSLGTIQLPTI------ARGVRRLTSFLVVNKKAPFNA 626
Query: 403 IIGRNTLNRLCAVISTDHLAVKYP 426
I+GR L+ + AV ST H +K+P
Sbjct: 627 ILGRPWLHAMKAVPSTYHQCIKFP 650
>At1g20390 hypothetical protein
Length = 1791
Score = 84.0 bits (206), Expect = 2e-16
Identities = 74/287 (25%), Positives = 128/287 (43%), Gaps = 29/287 (10%)
Query: 155 WCEYHRSAGYDTGDCFTLKNEIERLIRAG-------RSQLNDRGDRWRG*RQQGNRYKGD 207
+CEYH+S + T +C L+ + ++G R +N++ R +Y D
Sbjct: 410 YCEYHKSRAHSTENCRFLQGLLMAKYKSGGITIECDRPPINNKNQRRN--ETTARQYLND 467
Query: 208 R*QDDRRRDDRRRTPTSDKKETLATRKKGSEETFNKDL--EPPV-GTINTIAGGFGGGGD 264
+ + TP T A + K + EP V ++ I GG D
Sbjct: 468 Q--------TKPPTPAEQGIITSADDPAAKRQRNGKAIAAEPVVVRQVHVIMGGLQNCSD 519
Query: 265 -IASTRRRNVRAVTSVREYEAPFGFHHPD----IVISSADF*RIKTHKDDPVVVMIRINS 319
+ S ++ +A V + +P+ I + D + T +DP+VV + I+
Sbjct: 520 SVRSIKQYRKKAEMVVAWPSSTSTTRNPNQSAPISFTDVDLEGLDTPHNDPLVVELIISD 579
Query: 320 FNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHTVFG 379
V RVL++ GSS D+I+ D + +TD+ + P + L GF G+ V G + L G
Sbjct: 580 SRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTIKLPIFVG 639
Query: 380 VDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
+ V+++V+ A YNVI+G ++++ A+ ST H VK+P
Sbjct: 640 ----GLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFP 682
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 73.6 bits (179), Expect = 2e-13
Identities = 59/293 (20%), Positives = 121/293 (41%), Gaps = 30/293 (10%)
Query: 154 KWCEYHRSAGYDTGDCFTLKNEIERLIRAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDR 213
++C++H+ +G+ T C L++ + + G ++ R + N Y +D
Sbjct: 185 QYCDFHKRSGHSTAACRHLQSILLNKYKKGDIEVQHRQYK-----SHNNTYAARGGRDGN 239
Query: 214 RRDDRRRTPTSDKKETLATRKKGSEETFNKDLEPPVGT----------------INTIAG 257
R T ++E +EE + D+EPP +N I G
Sbjct: 240 NVFHRLGPHTGRQQEA----PPANEEERHPDMEPPKKNRENDQQHNDAPVPRRRVNMIMG 295
Query: 258 GFGGGGDIASTRRRNVRAVTSVREYEAPFGFHHPDIVISSADF*RIKTHKDDPVVVMIRI 317
G D + + +++ + + +P + +S D + +DP+V+ + I
Sbjct: 296 GLTACRDSFRSIKEYIKSGAATL-WSSPATKEMTPLTFTSEDLFGVDLPHNDPLVIELHI 354
Query: 318 NSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHTV 377
V R+L++ GSS ++++ D ++ + D+ + P L GF G + G + L
Sbjct: 355 GESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTNGTIKLPIY 414
Query: 378 FGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYPLICG 430
G +++V+ YN+I+G ++ + A+ S+ H +K P G
Sbjct: 415 LG----GAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG 463
>At4g20490 putative protein
Length = 853
Score = 70.5 bits (171), Expect = 2e-12
Identities = 71/324 (21%), Positives = 138/324 (41%), Gaps = 18/324 (5%)
Query: 113 EESGKSLNAHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAG--YDTGDCF 170
+E+G++LN +L + + H + K+ R ++++ +AG + T +C
Sbjct: 277 QENGETLNDYLGRFKEILSSYHHGRSRGKQARNDR---RRSRFGRPTSTAGGGHSTEECK 333
Query: 171 TLKNEIERL----IRAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRR---RDDRRRTPT 223
L N + + + A N + R G +Q + D R DDR
Sbjct: 334 HLLNILGKYKSGEVEAVYHPKNGKNKRKSGSKQSQAGPTHEEQHVDHRLLLEDDRVEEAR 393
Query: 224 SDKKETLATRKKGSEETFNKDLEPPVGTINTIAGGFGGGGDIASTRRRNVRAVTSVREYE 283
+ E K+ + + + P G I+ I G D S ++ R V SVR
Sbjct: 394 VPENELPGPPKRWRGQQPQRAPDHPRGRISMIMAGLSDDEDSVSALKKRARQVCSVRVAP 453
Query: 284 APFGFHHPDIVISSADF*RIKTHKDDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQ 343
I+ +S D I+ +D +V+ + + F+V R+L++ GSS ++++ +
Sbjct: 454 EAEPLSQEPIIFTSEDASGIQHPHNDALVIKLVMEDFDVERILIDTGSSVNLMFLKTLFK 513
Query: 344 LGLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHT-VFGVDENAKLLRVRYLVLQVVASYNV 402
+G+++ +MP + G+ GE G + L V G+ + K ++V+ YN
Sbjct: 514 MGISESKIMPKIRPMTGYDGEAKMSIGEIKLQVQVGGITQKTK-----FVVIDSEPIYNA 568
Query: 403 IIGRNTLNRLCAVISTDHLAVKYP 426
I+G + + A+ ST+ ++ P
Sbjct: 569 ILGSPWIYSMKAIPSTNLHTIRQP 592
>At2g13330 F14O4.9
Length = 889
Score = 64.7 bits (156), Expect = 9e-11
Identities = 38/120 (31%), Positives = 66/120 (54%), Gaps = 6/120 (5%)
Query: 308 DDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVW 367
DD +V+ I + ++ + ++++ GSS D+++ DAF + G D L L GF+G+ +
Sbjct: 62 DDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTGFAGDTTF 121
Query: 368 VRGYLDLHTVF-GVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G + L T+ GV + +LV+ A +N I+GR L+ + AV ST H +K+P
Sbjct: 122 SIGTIQLPTIARGVRQ-----LTNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQCIKFP 176
>At1g34080 putative protein
Length = 811
Score = 64.3 bits (155), Expect = 1e-10
Identities = 38/120 (31%), Positives = 64/120 (52%), Gaps = 4/120 (3%)
Query: 308 DDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVW 367
DD +V+ + + + V+R+L++ GSS D+I+ D ++G++ KD+ LV F+ E
Sbjct: 474 DDALVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKDIKGAPSPLVSFTIETSM 533
Query: 368 VRGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYPL 427
G + L V + + + V A+YN+I+G L + AV ST H VK+PL
Sbjct: 534 SLGTITL----PVTAQGVVKMIEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQCVKFPL 589
>At3g32290 hypothetical protein
Length = 612
Score = 63.5 bits (153), Expect = 2e-10
Identities = 51/218 (23%), Positives = 95/218 (43%), Gaps = 17/218 (7%)
Query: 151 DKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAGRS----QLNDRGDRWRG*RQQGNRYKG 206
++ ++CE H+S G+ T C +L ++ AG + D KG
Sbjct: 406 NEQEYCELHKSYGHHTSRCRSLGAKLVAKFLAGEIGGGLTIEDL-----------EAEKG 454
Query: 207 DR*QDDRRRDDRRRTPTSDKKETLATRKKGSEETFNKDLEPPVGTINTIAGGFGGGGDIA 266
Q + + + P ++ + R +G+ E + + E G I TI G D A
Sbjct: 455 KTEQVNAVANPEQAAPAANPERP--KRGRGNREADDDEPEAARGRIFTILGDSAFCQDTA 512
Query: 267 STRRRNVRAVTSVREYEAPFGFHHPDIVISSADF*RIKTHKDDPVVVMIRINSFNVRRVL 326
+ + R + R + PF + ++ D + +DP+V+ + I FNV RVL
Sbjct: 513 PSIKAYQRKADANRNWARPFNGPNDEVTFHERDTNGLDRPHNDPLVITLTIGDFNVERVL 572
Query: 327 LNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGE 364
++ GS+ DII+ ++ + ++P ++GFSG+
Sbjct: 573 VDTGSTLDIIFLTTLREMKIDMTQIVPTPRPVLGFSGK 610
>At4g03800
Length = 637
Score = 48.9 bits (115), Expect = 5e-06
Identities = 41/177 (23%), Positives = 78/177 (43%), Gaps = 18/177 (10%)
Query: 250 GTINTIAGGFGGGGDIASTRRRNVRAVTSVREYEAPFGFHHPDIVISSADF*RIKTHKDD 309
G IN I GG + S +++ V + + + I+ DD
Sbjct: 142 GKINVILGGSKLCHNSVSAIKKHRPNVLLKSSLSEEVNYQCSSVSFDEEETRHIERPHDD 201
Query: 310 PVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWVR 369
+++ + + +F + R+L++ GSS D+I+ + LV F+ E
Sbjct: 202 ALIITLDVANFKISRILVDTGSSVDLIF--------------LGPPSPLVAFTSESAMSL 247
Query: 370 GYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G + L + G + +K+ V ++V A+YN+I+G + ++ AV ST H +K+P
Sbjct: 248 GTIKLPVLAG--KMSKI--VDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWLKFP 300
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 42.0 bits (97), Expect = 7e-04
Identities = 35/147 (23%), Positives = 64/147 (42%), Gaps = 20/147 (13%)
Query: 225 DKKETLATRKKGSEETFN---KDLEP-------PVGTI---NTIAGGFGGGG-------D 264
D+KE+ A R K ++ K EP G + N + F G G D
Sbjct: 321 DEKESFAKRDKQAKPAVTFPPKKFEPRENQGPRKFGLLPFNNNVGKQFQGKGRSNTWIRD 380
Query: 265 IASTRRRNVRAVTSVREYEAPFGFHHPDIVISSADF*RIKTHKDDPVVVMIRINSFNVRR 324
+S +++ R V F I+ + ++ DD +VV + + +F V R
Sbjct: 381 ESSVIKKHSRNVHLKLSLSEEVDFQSTSILFDEKETQHLERSHDDALVVTLDVANFEVSR 440
Query: 325 VLLNQGSSADIIYGDAFDQLGLTDKDL 351
+L++ GSS D+I+ +++G++ D+
Sbjct: 441 ILIDTGSSVDLIFLSTLERMGISRADV 467
>At2g12870 putative retroelement pol polyprotein
Length = 411
Score = 40.8 bits (94), Expect = 0.001
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 4/103 (3%)
Query: 233 RKKGSEETFNKDLEPPVGTINTIAGGFGGGGDIASTRRRNVRAVTSVREYEAPFGFHHPD 292
R +G+ E ++ + E G I TI G D ++ ++A S R PF
Sbjct: 305 RGRGNREAYDDESEAARGRIFTILGDSVFCHDTTTS----IKAYHSNRNLTRPFNGPDDK 360
Query: 293 IVISSADF*RIKTHKDDPVVVMIRINSFNVRRVLLNQGSSADI 335
+ ++ + +DP+V+ + I +FNV RVL++ GS +
Sbjct: 361 VTFHESNINCLDQPHNDPLVITLTIGAFNVERVLVDTGSDISL 403
>At5g34880 putative protein
Length = 420
Score = 37.4 bits (85), Expect = 0.016
Identities = 17/67 (25%), Positives = 38/67 (56%)
Query: 308 DDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVW 367
DD +V+ + + +F V R+L++ SS D+I+ +++ ++ D++ +LV F+ +
Sbjct: 338 DDALVLTLDVANFEVSRILIDTRSSVDLIFLSTLERMRISRADIIGPPSSLVAFTSDTAM 397
Query: 368 VRGYLDL 374
G + L
Sbjct: 398 SLGTIKL 404
>At3g28770 hypothetical protein
Length = 2081
Score = 34.3 bits (77), Expect = 0.14
Identities = 44/177 (24%), Positives = 76/177 (42%), Gaps = 17/177 (9%)
Query: 70 RETIQRRDKKIKSGERTAKEPLYPRKESFERRRPWHQTDSRRQEESGKSLNAHLTELLRK 129
RE + +KK K+ E KE +K+S +++R + DS +E K +L K
Sbjct: 1010 REKKEYEEKKSKTKEEAKKE----KKKSQDKKR--EEKDS--EERKSKKEKEESRDLKAK 1061
Query: 130 VKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAGRSQLND 189
K E E E + DK K E ++S + K+E + R + D
Sbjct: 1062 KKEEETKEKKESENHKSKKKEDK-KEHEDNKSMKKEEDKKEKKKHEESK----SRKKEED 1116
Query: 190 RGDRWRG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDKKETLATRKKGSEETFNKDLE 246
+ D + Q N+ K D+ + + + + SDKKE +K+ E++ K++E
Sbjct: 1117 KKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKE----KKENEEKSETKEIE 1169
Score = 28.5 bits (62), Expect = 7.5
Identities = 40/199 (20%), Positives = 80/199 (40%), Gaps = 21/199 (10%)
Query: 57 QRAPVGQGKSKAIRETIQRRDKKIKSGERTAKEPLYPRKESFERRRPWHQTDSRRQEESG 116
Q+ V + + K+ ++ ++++K++K E +K +R++ ++++Q+E+
Sbjct: 1174 QKNEVDKKEKKSSKDQQKKKEKEMKESEEKKL-----KKNEEDRKKQTSVEENKKQKETK 1228
Query: 117 KSLNAHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDT-GDCFTLKNE 175
K N + K + G+KE+ + ++K E + + T D KNE
Sbjct: 1229 KEKNKPKDDKKNTTKQS----GGKKES-----MESESKEAENQQKSQATTQADSDESKNE 1279
Query: 176 IERLIRAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDKKETLATRKK 235
I + SQ + D + N Q D + +R KK+T K
Sbjct: 1280 I---LMQADSQADSHSDSQADSDESKNEILM---QADSQATTQRNNEEDRKKQTSVAENK 1333
Query: 236 GSEETFNKDLEPPVGTINT 254
+ET + +P NT
Sbjct: 1334 KQKETKEEKNKPKDDKKNT 1352
>At4g37820 unknown protein
Length = 532
Score = 33.9 bits (76), Expect = 0.18
Identities = 38/185 (20%), Positives = 75/185 (40%), Gaps = 15/185 (8%)
Query: 59 APVGQGKSKAIRETIQRRDKKIKSGERTAKEPLYPRKESFERRRPWHQTDSRRQEESGKS 118
A Q +SK + +++++ GE +EP E E+ Q +S+ +E K
Sbjct: 322 ASSSQDESKEEKPERKKKEESSSQGEGKEEEP-----EKREKEDSSSQEESKEEEPENKE 376
Query: 119 LNAHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIER 178
A ++ ++K T + E E + + K +S+ + + +IE+
Sbjct: 377 KEASSSQEENEIKETEIKEKEESSSQEGNENKETEK-----KSSESQRKENTNSEKKIEQ 431
Query: 179 LIRAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDKKETLATRKKGSE 238
+ + S +GD + + R G+ + DD +T + K+E R +E
Sbjct: 432 -VESTDSSNTQKGDEQK--TDESKRESGNDTSNKETEDDSSKTESEKKEEN--NRNGETE 486
Query: 239 ETFNK 243
ET N+
Sbjct: 487 ETQNE 491
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 33.1 bits (74), Expect = 0.30
Identities = 51/235 (21%), Positives = 98/235 (41%), Gaps = 19/235 (8%)
Query: 17 DGVVHDLASRIYHELPRFLVKVPCSVFREQEQASCDRGLIQRAPVGQGKSKAIRETIQRR 76
+ V+ + AS EL + + ++ RE+E+ R + A + + E +RR
Sbjct: 403 ESVIFECASCAEGELSKLMREIEERKRREEEEIERRRKEEEEARKREEAKRREEEEAKRR 462
Query: 77 DKKIKSGERTAKEPLYPRKESFERRRPWHQTDSRRQEESGK-----SLNAHLTELLRKVK 131
++ + ER +E RK ER+R + +++R+EE K + A E R+ +
Sbjct: 463 EE--EETERKKREEEEARKREEERKR--EEEEAKRREEERKKREEEAEQARKREEEREKE 518
Query: 132 ATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAGRSQLNDRG 191
+ E+ R V++ + E R + K E E R ++ R
Sbjct: 519 EEMAKKREEERQRKEREEVERKRREEQERKRREEE----ARKREEE---RKREEEMAKRR 571
Query: 192 DRWRG*RQQGNRYKGDR*QDDRRRDD---RRRTPTSDKKETLATRKKGSEETFNK 243
++ R +++ + R + +R+R++ +RR KKE +K EE K
Sbjct: 572 EQERQRKEREEVERKIREEQERKREEEMAKRREQERQKKEREEMERKKREEEARK 626
Score = 33.1 bits (74), Expect = 0.30
Identities = 42/200 (21%), Positives = 79/200 (39%), Gaps = 26/200 (13%)
Query: 47 EQASCDRGLIQRAPVGQGKSKAIRETIQRRDKKIKSGERTAKEPLYPRKESFERRRPWHQ 106
E ASC G + SK +RE +R+ ++ + ER KE RK +RR +
Sbjct: 408 ECASCAEGEL---------SKLMREIEERKRREEEEIERRRKEEEEARKREEAKRREEEE 458
Query: 107 TDSRRQEESGKSLNAHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDT 166
R +EE+ + K EA ++E + R + + E + +
Sbjct: 459 AKRREEEETER-------------KKREEEEARKREEERKREEEEAKRREEERKKREEEA 505
Query: 167 GDCFTLKNEIERLIRAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDK 226
+ E E+ + + +R R R++ R + + Q+ +RR++ R ++
Sbjct: 506 EQARKREEEREKEEEMAKKR---EEERQRKEREEVERKRREE-QERKRREEEARKREEER 561
Query: 227 KETLATRKKGSEETFNKDLE 246
K K+ +E K+ E
Sbjct: 562 KREEEMAKRREQERQRKERE 581
Score = 32.0 bits (71), Expect = 0.68
Identities = 42/183 (22%), Positives = 73/183 (38%), Gaps = 10/183 (5%)
Query: 65 KSKAIRETIQRRDKKIKSGERTAKEPLYPRKESFERRRPWHQTDSRRQEESGKSLN---- 120
+++ R+ + R+K+ + ++ +E +E ER+R Q RR+EE+ K
Sbjct: 504 EAEQARKREEEREKEEEMAKKREEERQRKEREEVERKRREEQERKRREEEARKREEERKR 563
Query: 121 ----AHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEI 176
A E R+ K VE +E + + K E R + + K E
Sbjct: 564 EEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKK-EREEMERKKREE 622
Query: 177 ERLIRAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDKKETLATRKKG 236
E R +R R R+ R + + + RR ++R+R + K+ RKK
Sbjct: 623 EARKREEEMAKIREEERQRKEREDVERKRREE-EAMRREEERKREEEAAKRAEEERRKKE 681
Query: 237 SEE 239
EE
Sbjct: 682 EEE 684
>At4g36690 splicing factor like protein
Length = 573
Score = 32.7 bits (73), Expect = 0.40
Identities = 43/202 (21%), Positives = 76/202 (37%), Gaps = 25/202 (12%)
Query: 69 IRETIQRRDKKIKSGERTAKEPLYPRKESFERRRPWHQTDSRRQEESGKSLNAHLTELLR 128
+ + I + + GE + P++ES + R ++ R +E+ +E+ R
Sbjct: 13 VADAIYDEENGGRDGEIEDQLDSKPKRESRDHERETSRSKDREREKGRDKDRERDSEVSR 72
Query: 129 KVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAGRSQLN 188
+ + ++ E+ D R ++ HR + G ER R GR +
Sbjct: 73 RSRDRDGEKSKERSRDKDRDHRERHHRSSRHRDHSRERG---------ERRERGGRDDDD 123
Query: 189 DRGDRWRG*RQQGNRYKGDR*QDDR--RRDDRRRTPTSDKKETLATRK---KGSEETFNK 243
R R R DR +DDR RR R R+ + D+ E + K +
Sbjct: 124 YRRSRDR---------DHDRRRDDRGGRRSRRSRSRSKDRSERRTRSRSPSKSKQRVSGF 174
Query: 244 DLEPPVGTINTIAGGFGGGGDI 265
D+ PP + +A G G +
Sbjct: 175 DMAPPASAM--LAAGAAVTGQV 194
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 32.7 bits (73), Expect = 0.40
Identities = 18/43 (41%), Positives = 23/43 (52%)
Query: 389 VRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYPLICGK 431
V + V A+YNVI+G L + V ST H VK+P GK
Sbjct: 369 VEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGK 411
>At2g11940 putative retroelement gag/pol polyprotein
Length = 1212
Score = 32.3 bits (72), Expect = 0.52
Identities = 16/38 (42%), Positives = 23/38 (60%)
Query: 308 DDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLG 345
DD V+ + I + V+RVL+ GSS D+I+ D LG
Sbjct: 320 DDAFVITLDIANCKVQRVLIVTGSSVDLIFLDTLANLG 357
>At1g37200 hypothetical protein
Length = 1564
Score = 32.3 bits (72), Expect = 0.52
Identities = 23/100 (23%), Positives = 45/100 (45%), Gaps = 18/100 (18%)
Query: 129 KVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAGRSQLN 188
+ + H + A E+ + +D K+C+YH G+ T +C +++LI AG
Sbjct: 297 QTQGQHTLFAIEEAAEDESPELDLGKYCKYHNKRGHSTEEC----RAVKKLIAAG----- 347
Query: 189 DRGDRWRG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDKKE 228
G ++G+ K + D + ++ +TP K+E
Sbjct: 348 -------GKTKKGSNPKVETPPPDEQEEE--QTPKQKKRE 378
Score = 28.9 bits (63), Expect = 5.7
Identities = 13/29 (44%), Positives = 19/29 (64%)
Query: 398 ASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
A +N I+GR L+ + AV ST H +K+P
Sbjct: 530 APFNAILGRPWLHAMKAVPSTYHQCIKFP 558
Score = 28.1 bits (61), Expect = 9.8
Identities = 9/27 (33%), Positives = 19/27 (70%)
Query: 308 DDPVVVMIRINSFNVRRVLLNQGSSAD 334
DD +V+ I + ++ + R++++ GSS D
Sbjct: 502 DDAIVIRIDVGNYKLSRIMIDTGSSVD 528
>At3g48120 putative protein
Length = 347
Score = 32.0 bits (71), Expect = 0.68
Identities = 26/123 (21%), Positives = 54/123 (43%), Gaps = 5/123 (4%)
Query: 43 FREQEQASCDRGLIQRAPVGQGKSKAIRETIQRRDKKIKSGERTAKEPLYPRKESFERRR 102
+R +E+ D+ +R+ + +S + +RR + +S ER + Y +ES R
Sbjct: 100 YRSRERDERDKSHRRRSRSSERRSSYVDR--ERRRSRSRSAERRNR---YGDRESRRRSN 154
Query: 103 PWHQTDSRRQEESGKSLNAHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSA 162
RR+ S + + + R +K + A EK ++ +D+T + + A
Sbjct: 155 RSRSLSPRRERRSREDVKEKKPDYSRLIKGYDEMSAAEKVKAKMKLQLDETAEKDTSKGA 214
Query: 163 GYD 165
G++
Sbjct: 215 GWE 217
>At3g31530 hypothetical protein
Length = 831
Score = 32.0 bits (71), Expect = 0.68
Identities = 15/27 (55%), Positives = 18/27 (66%)
Query: 400 YNVIIGRNTLNRLCAVISTDHLAVKYP 426
YN IIG LN+ AV ST HL +K+P
Sbjct: 30 YNAIIGTPWLNQFRAVASTYHLCLKFP 56
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.143 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,672,102
Number of Sequences: 26719
Number of extensions: 426708
Number of successful extensions: 1701
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 1621
Number of HSP's gapped (non-prelim): 82
length of query: 431
length of database: 11,318,596
effective HSP length: 102
effective length of query: 329
effective length of database: 8,593,258
effective search space: 2827181882
effective search space used: 2827181882
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0134.17


