

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.10
(1564 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
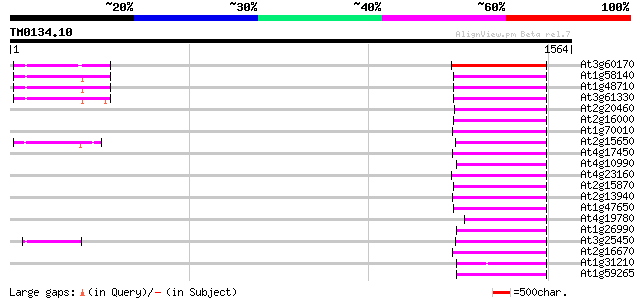
Score E
Sequences producing significant alignments: (bits) Value
At3g60170 putative protein 226 6e-59
At1g58140 hypothetical protein 210 5e-54
At1g48710 hypothetical protein 210 5e-54
At3g61330 copia-type polyprotein 209 1e-53
At2g20460 putative retroelement pol polyprotein 193 6e-49
At2g16000 putative retroelement pol polyprotein 192 1e-48
At1g70010 hypothetical protein 189 8e-48
At2g15650 putative retroelement pol polyprotein 186 1e-46
At4g17450 retrotransposon like protein 182 1e-45
At4g10990 putative retrotransposon polyprotein 177 4e-44
At4g23160 putative protein 176 1e-43
At2g15870 putative retroelement pol polyprotein 174 4e-43
At2g13940 putative retroelement pol polyprotein 170 7e-42
At1g47650 hypothetical protein 170 7e-42
At4g19780 putative LTR retrotransposon (fragment) 168 3e-41
At1g26990 polyprotein, putative 166 1e-40
At3g25450 hypothetical protein 164 5e-40
At2g16670 putative retroelement pol polyprotein 164 5e-40
At1g31210 putative reverse transcriptase 164 5e-40
At1g59265 polyprotein, putative 162 1e-39
>At3g60170 putative protein
Length = 1339
Score = 226 bits (577), Expect = 6e-59
Identities = 116/268 (43%), Positives = 171/268 (63%), Gaps = 3/268 (1%)
Query: 1232 SRYNSLLKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQ 1291
S + K +ILIV +YVDD+IF +++++C EF + M EFEMS +G++K+FLGI+
Sbjct: 1001 SEHTLFTKTRVGNILIVSLYVDDLIFTGSDKAMCDEFKKSMMLEFEMSDLGKMKHFLGIE 1060
Query: 1292 VDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIG 1351
V Q+ G +I Q +Y +E+L +F M ES K P+ P L K++ KV + ++ ++G
Sbjct: 1061 VKQSDGGIFICQRRYAREVLARFGMDESNAVKNPIVPGTKLTKDENGEKVDETMFKQLVG 1120
Query: 1352 TLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMY--KKT*EYK 1409
+L+YLT +RPD+++ V L +RF S+PR +H A KRILRYLKGT LG+ Y +K K
Sbjct: 1121 SLMYLTVTRPDLMYGVCLISRFMSNPRMSHWLAAKRILRYLKGTVELGIFYRRRKNRSLK 1180
Query: 1410 LSGYCDADYAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQM 1469
L + D+DYAGD +R+STS + S + WASK Q +ALST EAEYI A C+ Q
Sbjct: 1181 LMAFTDSDYAGDLNDRRSTSGFVFLMASGAICWASKKQPVVALSTTEAEYIAAAFCACQC 1240
Query: 1470 LWMKHQLEDYQILESNIS-IYCDNTAAI 1496
+W++ LE E + + I CDN++ I
Sbjct: 1241 VWLRKVLEKLGAEEKSATVINCDNSSTI 1268
Score = 89.4 bits (220), Expect = 2e-17
Identities = 63/274 (22%), Positives = 130/274 (46%), Gaps = 13/274 (4%)
Query: 11 DLWDIIVDGYERPVDEEGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYEKITDRE 70
+LW ++ +G V ++ KL + K + L AI E E I D+
Sbjct: 34 ELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKL--KDLKVKNFLFQAIDREILETILDKS 91
Query: 71 YAKGIFESLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
+K I+ES+K ++G+ KVK ++ +L +++E M+ E I+ R +V ++
Sbjct: 92 TSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAMKEGEKIDTFLGRTLTVVNKMKTNG 151
Query: 131 KSYTTKYHVIRVIRSLPESWMPLVTSIELTRDVERMSLEELISILKCHELKHSEMLDSDE 190
+ V +++RSL + +V SIE + D+ +S++EL L HE + + + ++
Sbjct: 152 EVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTLSIDELHGSLLVHEQRLNGHVQEEQ 211
Query: 191 DELTLISKRLNRIWKHKQSKYRGSG---KAKGKSESSGQKKSSIKEVTCFECKESGHYKS 247
L + + S+ RG G ++G+ G+ ++ V C++C GH++
Sbjct: 212 --------ALKVTHEERPSQGRGRGVFRGSRGRGRGRGRSGTNRAIVECYKCHNLGHFQY 263
Query: 248 DCPKLKKDKRPKKHFKTKKSLMVTFDESESEDVD 281
+CP+ +K+ + + ++ L++ + E + D
Sbjct: 264 ECPEWEKNANYAELEEEEELLLMAYVEQNQANRD 297
>At1g58140 hypothetical protein
Length = 1320
Score = 210 bits (535), Expect = 5e-54
Identities = 110/260 (42%), Positives = 156/260 (59%), Gaps = 1/260 (0%)
Query: 1238 LKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPE 1297
+K + DILI +YVDD+IF N S+ +EF + M EFEM+ +G + Y+LGI+V Q
Sbjct: 986 IKIQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDN 1045
Query: 1298 GTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLT 1357
G +I Q Y KE+LKKF M +S TPM L K++ V + ++G+L YLT
Sbjct: 1046 GIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLT 1105
Query: 1358 ASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDAD 1417
+RPDIL++V + +R+ P TH A KRILRY+KGT N GL Y T +YKL GY D+D
Sbjct: 1106 CTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSD 1165
Query: 1418 YAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLE 1477
+ GD +RKSTS ++G +W SK Q + LST EAEY+ C +W+++ L+
Sbjct: 1166 WGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLK 1225
Query: 1478 DYQI-LESNISIYCDNTAAI 1496
+ + E I+ DN +AI
Sbjct: 1226 ELSLPQEEPTKIFVDNKSAI 1245
Score = 97.8 bits (242), Expect = 4e-20
Identities = 71/302 (23%), Positives = 139/302 (45%), Gaps = 36/302 (11%)
Query: 11 DLWDIIVDGYERPVDEEGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYEKITDRE 70
D+W+I+ G+ P +E + + D +K + KA ++ + + +EK+ +
Sbjct: 33 DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---RDKKALCLIYQGLDEDTFEKVVEAT 89
Query: 71 YAKGIFESLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
AK +E L+ S++G +VK+ + +L ++E+ M+ E + + FSR + ++
Sbjct: 90 SAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNG 149
Query: 131 KSYTTKYHVIRVIRSLPESWMPLVTSIELTRDVERMSLEELISILKCHELKHSEMLDSDE 190
+ + +V+RSL + +VT IE T+D+E M++E+L+ L+ +E K + D E
Sbjct: 150 EKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIVE 209
Query: 191 DELTL-ISKRLN-----------------------RIWK--HKQSKYRGSGKAKGKSESS 224
L + I+K N R W+ + RG ++G+ +
Sbjct: 210 QVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGH 269
Query: 225 GQKKSSIKEVTCFECKESGHYKSDC--PKLKKDKRPKKHFKTK-----KSLMVTFDESES 277
+ + V C+ C + GHY S+C P KK + + + K LM ++ + E
Sbjct: 270 PKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKANYVEEKIQEEDMLLMASYKKDEQ 329
Query: 278 ED 279
E+
Sbjct: 330 EE 331
>At1g48710 hypothetical protein
Length = 1352
Score = 210 bits (535), Expect = 5e-54
Identities = 110/260 (42%), Positives = 156/260 (59%), Gaps = 1/260 (0%)
Query: 1238 LKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPE 1297
+K + DILI +YVDD+IF N S+ +EF + M EFEM+ +G + Y+LGI+V Q
Sbjct: 1018 IKIQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDN 1077
Query: 1298 GTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLT 1357
G +I Q Y KE+LKKF M +S TPM L K++ V + ++G+L YLT
Sbjct: 1078 GIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLT 1137
Query: 1358 ASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDAD 1417
+RPDIL++V + +R+ P TH A KRILRY+KGT N GL Y T +YKL GY D+D
Sbjct: 1138 CTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSD 1197
Query: 1418 YAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLE 1477
+ GD +RKSTS ++G +W SK Q + LST EAEY+ C +W+++ L+
Sbjct: 1198 WGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVVLSTCEAEYVAATSCVCHAIWLRNLLK 1257
Query: 1478 DYQI-LESNISIYCDNTAAI 1496
+ + E I+ DN +AI
Sbjct: 1258 ELSLPQEEPTKIFVDNKSAI 1277
Score = 97.8 bits (242), Expect = 4e-20
Identities = 71/302 (23%), Positives = 139/302 (45%), Gaps = 36/302 (11%)
Query: 11 DLWDIIVDGYERPVDEEGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYEKITDRE 70
D+W+I+ G+ P +E + + D +K + KA ++ + + +EK+ +
Sbjct: 33 DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---RDKKALCLIYQGLDEDTFEKVVEAT 89
Query: 71 YAKGIFESLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
AK +E L+ S++G +VK+ + +L ++E+ M+ E + + FSR + ++
Sbjct: 90 SAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNG 149
Query: 131 KSYTTKYHVIRVIRSLPESWMPLVTSIELTRDVERMSLEELISILKCHELKHSEMLDSDE 190
+ + +V+RSL + +VT IE T+D+E M++E+L+ L+ +E K + D E
Sbjct: 150 EKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIIE 209
Query: 191 DELTL-ISKRLN-----------------------RIWK--HKQSKYRGSGKAKGKSESS 224
L + I+K N R W+ + RG ++G+ +
Sbjct: 210 QVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGH 269
Query: 225 GQKKSSIKEVTCFECKESGHYKSDC--PKLKKDKRPKKHFKTK-----KSLMVTFDESES 277
+ + V C+ C + GHY S+C P KK + + + K LM ++ + E
Sbjct: 270 PKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKANYVEEKIQEEDMLLMASYKKDEQ 329
Query: 278 ED 279
E+
Sbjct: 330 EE 331
>At3g61330 copia-type polyprotein
Length = 1352
Score = 209 bits (531), Expect = 1e-53
Identities = 109/260 (41%), Positives = 156/260 (59%), Gaps = 1/260 (0%)
Query: 1238 LKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPE 1297
+K + DILI +YVDD+IF N S+ +EF + M EFEM+ +G + Y+LGI+V Q
Sbjct: 1018 IKIQKEDILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDN 1077
Query: 1298 GTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLT 1357
G +I Q Y KE+LKKF + +S TPM L K++ V + ++G+L YLT
Sbjct: 1078 GIFITQEGYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLT 1137
Query: 1358 ASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDAD 1417
+RPDIL++V + +R+ P TH A KRILRY+KGT N GL Y T +YKL GY D+D
Sbjct: 1138 CTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSD 1197
Query: 1418 YAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLE 1477
+ GD +RKSTS ++G +W SK Q + LST EAEY+ C +W+++ L+
Sbjct: 1198 WGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLK 1257
Query: 1478 DYQI-LESNISIYCDNTAAI 1496
+ + E I+ DN +AI
Sbjct: 1258 ELSLPQEEPTKIFVDNKSAI 1277
Score = 97.4 bits (241), Expect = 6e-20
Identities = 72/303 (23%), Positives = 141/303 (45%), Gaps = 38/303 (12%)
Query: 11 DLWDIIVDGYERPVDEEGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYEKITDRE 70
D+W+I+ G+ P +E + + D +K + KA ++ + + +EK+ +
Sbjct: 33 DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---RDKKALCLIYQGLDEDTFEKVVEAT 89
Query: 71 YAKGIFESLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLN 130
AK +E L+ S++G +VK+ + +L ++E+ M+ E + + FSR + ++
Sbjct: 90 SAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNG 149
Query: 131 KSYTTKYHVIRVIRSLPESWMPLVTSIELTRDVERMSLEELISILKCHELKHSEMLDSDE 190
+ + +V+RSL + +VT IE T+D+E M++E+L+ L+ +E K + D E
Sbjct: 150 EKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIAE 209
Query: 191 DELTL-ISKRLN-----------------------RIWK--HKQSKYRGSGKAKGKSESS 224
L + I+K N R W+ + RG ++G+ +
Sbjct: 210 QVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGH 269
Query: 225 GQKKSSIKEVTCFECKESGHYKSDC--PKLKKDKRPKKHFKTKK------SLMVTFDESE 276
+ + V C+ C + GHY S+C P KK + K H+ +K LM ++ + E
Sbjct: 270 PKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEE-KAHYVEEKIQEEDMLLMASYKKDE 328
Query: 277 SED 279
++
Sbjct: 329 QKE 331
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 193 bits (491), Expect = 6e-49
Identities = 98/257 (38%), Positives = 153/257 (59%)
Query: 1241 IQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTY 1300
I ++ +++ +YVDDI+ S + + +E ++A F++ +G LKYFLG++V +T EG
Sbjct: 1134 IGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYFLGLEVARTSEGIS 1193
Query: 1301 IHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLTASR 1360
+ Q KY ELL +ML+ + PM P L K D +++Y ++G L+YLT +R
Sbjct: 1194 LSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYRRLVGKLMYLTITR 1253
Query: 1361 PDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAG 1420
PDI F+V+ +F S PR HL AV ++L+Y+KGT GL Y + L GY DAD+
Sbjct: 1254 PDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGLFYSAEDDLTLKGYTDADWGT 1313
Query: 1421 DRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQ 1480
R+ST+ F+GS+L+SW SK Q T++ S+AEAEY A+ S +M W+ L +
Sbjct: 1314 CPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALASCEMAWLSTLLLALR 1373
Query: 1481 ILESNISIYCDNTAAIH 1497
+ +Y D+TAA++
Sbjct: 1374 VHSGVPILYSDSTAAVY 1390
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 192 bits (488), Expect = 1e-48
Identities = 100/260 (38%), Positives = 153/260 (58%)
Query: 1238 LKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPE 1297
LK + +IV +YVDDI+ S +++ + +E + F++ +G+LKYFLG++V +T
Sbjct: 1124 LKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTA 1183
Query: 1298 GTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLT 1357
G I Q KY ELL+ ML PM P + K+D + Y ++G L+YLT
Sbjct: 1184 GISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLT 1243
Query: 1358 ASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDAD 1417
+RPDI F+V+ +F S PR THLTA R+L+Y+KGT GL Y + + L G+ D+D
Sbjct: 1244 ITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSD 1303
Query: 1418 YAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLE 1477
+A + R+ST+ F+G +L+SW SK Q T++ S+AEAEY A+ + +M+W+ L
Sbjct: 1304 WASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLV 1363
Query: 1478 DYQILESNISIYCDNTAAIH 1497
Q +Y D+TAAI+
Sbjct: 1364 SLQASPPVPILYSDSTAAIY 1383
>At1g70010 hypothetical protein
Length = 1315
Score = 189 bits (481), Expect = 8e-48
Identities = 99/265 (37%), Positives = 153/265 (57%), Gaps = 1/265 (0%)
Query: 1234 YNSLLKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVD 1293
+ LK L V +Y+DDII S N + M++ F++ +GELKYFLG+++
Sbjct: 981 HTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELKYFLGLEIV 1040
Query: 1294 QTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTL 1353
++ +G +I Q KY +LL + L + PM P+ + + V Y +IG L
Sbjct: 1041 RSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGPYRRLIGRL 1100
Query: 1354 LYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGY 1413
+YL +RPDI F+V+ A+F PR+ HL AV +IL+Y+KGT GL Y T E +L Y
Sbjct: 1101 MYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFYSATSELQLKVY 1160
Query: 1414 CDADYAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMK 1473
+ADY R R+STS C FLG +L+ W S+ Q ++ S+AEAEY ++ + +++W+
Sbjct: 1161 ANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVATDELVWLT 1220
Query: 1474 HQLEDYQI-LESNISIYCDNTAAIH 1497
+ L++ Q+ L ++CDN AAIH
Sbjct: 1221 NFLKELQVPLSKPTLLFCDNEAAIH 1245
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 186 bits (471), Expect = 1e-46
Identities = 99/256 (38%), Positives = 146/256 (56%), Gaps = 3/256 (1%)
Query: 1244 DILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 1303
D+LIV +YVDD+I N L F + M+ EFEM+ +G L YFLG++V+Q G ++ Q
Sbjct: 1023 DVLIVSLYVDDLIITGNNTHLINTFKKNMKDEFEMTDLGLLNYFLGMEVNQDDSGIFLSQ 1082
Query: 1304 SKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGK--VCQKLYHGMIGTLLYLTASRP 1361
KY +L+ KF M ES TP+ P + + K Y ++G LLYL ASRP
Sbjct: 1083 EKYANKLIDKFGMKESKSVSTPLTPQGKRKGVEGDDKEFADPTKYRRIVGGLLYLCASRP 1142
Query: 1362 DILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAGD 1421
D++++ +R+ S P H KR+LRY+KGT+N G+++ +L GY D+D+ G
Sbjct: 1143 DVMYASSYLSRYMSSPSIQHYQEAKRVLRYVKGTSNFGVLFTSKETPRLVGYSDSDWGGS 1202
Query: 1422 RTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQI 1481
++KST+ LG + W S Q T+A STAEAEYI + Q +W++ ED+ +
Sbjct: 1203 LEDKKSTTGYVFTLGLAMFCWQSCKQQTVAQSTAEAEYIAVCAATNQAIWLQRLFEDFGL 1262
Query: 1482 -LESNISIYCDNTAAI 1496
+ I I CDN +AI
Sbjct: 1263 KFKEGIPILCDNKSAI 1278
Score = 80.9 bits (198), Expect = 5e-15
Identities = 62/266 (23%), Positives = 131/266 (48%), Gaps = 25/266 (9%)
Query: 12 LWDIIVDGYE-RPVD-EEGKKIPRSEMTADQKKLYSQHHKARAILLSAISYEEYEKITDR 69
LW ++ +G PV EE + R++ ++ + A IL +A++ + + +I
Sbjct: 33 LWSVVEEGVPVEPVQAEETPETARAKTLREEA--VTNDTMALQILQTAVTDQIFSRIAAA 90
Query: 70 EYAKGIFESLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPL 129
+K ++ LK ++G+ +V+ K SL ++YE+ M N++I+ + +L +
Sbjct: 91 SSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENLKMYDNDNIKTFTDKLIVLEIQLTYH 150
Query: 130 NKSYTTKYHVIRVIRSLPESWMPLVTSIELTRDVERMSLEELISILKCHELKHSEMLDSD 189
+ T + +++ SLP + +V+ +E TRD++ +++ EL+ ILK E + + +S
Sbjct: 151 GEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLDALTMSELLGILKAQEARVTAREEST 210
Query: 190 EDELTLI--------------SKRLNRIWK----HKQSKY-RGSGKAKGKSESSGQKKSS 230
++ + + R+N+ K HK SK+ + K K++ G+ K S
Sbjct: 211 KEGAFYVRSKGRESGFKQDNTNNRVNQDKKWCGFHKSSKHTEEECREKPKNDDHGKNKRS 270
Query: 231 IKEVTCFECKESGHYKSDCPKLKKDK 256
+ C++C + GHY ++C K++
Sbjct: 271 --NIKCYKCGKIGHYANECRSKNKER 294
>At4g17450 retrotransposon like protein
Length = 1433
Score = 182 bits (462), Expect = 1e-45
Identities = 95/265 (35%), Positives = 151/265 (56%), Gaps = 2/265 (0%)
Query: 1235 NSLLKNIQNDILI-VQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVD 1293
++L N +L+ V +YVDDI+ S + +F+ +++ F++ +G KYFLGI++
Sbjct: 1097 HTLFIKYANGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKLRDLGAAKYFLGIEIA 1156
Query: 1294 QTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTL 1353
++ +G I Q KY ELL L S + P+ P+ L KED Y ++G L
Sbjct: 1157 RSEKGISICQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGVPLTDSTSYRKLVGKL 1216
Query: 1354 LYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGY 1413
+YL +RPDI ++V+ +F P HL+AV ++LRYLKGT GL Y ++ L GY
Sbjct: 1217 MYLQITRPDIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYSADDKFDLRGY 1276
Query: 1414 CDADYAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMK 1473
D+D+ R+ + C F+G LVSW SK Q T+++STAEAE+ + + +M+W+
Sbjct: 1277 TDSDFGSCTDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMSQGTKEMIWLS 1336
Query: 1474 HQLEDYQI-LESNISIYCDNTAAIH 1497
+D+++ +YCDNTAA+H
Sbjct: 1337 RLFDDFKVPFIPPAYLYCDNTAALH 1361
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 177 bits (449), Expect = 4e-44
Identities = 86/254 (33%), Positives = 152/254 (58%), Gaps = 1/254 (0%)
Query: 1245 ILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 1304
I++V +YVDD++ S + S + E++++EF++ +G ++FLG+++ ++ EG + Q
Sbjct: 716 IIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVCQR 775
Query: 1305 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLTASRPDIL 1364
KY + LL+ + + PM P L KE + Y ++G LLYL +RPDI
Sbjct: 776 KYAQNLLEDVGLSGCKPSSIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDIT 835
Query: 1365 FSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAGDRTE 1424
F+VH ++F S P + H+ A ++LRYLKG GLMY + E L+G+ DAD+ +
Sbjct: 836 FAVHTLSQFLSAPTDIHMQAAHKVLRYLKGNPGQGLMYSASSELCLNGFSDADWGTCKDS 895
Query: 1425 RKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQI-LE 1483
R+S + C +LG++L++W SK QS ++ S+ E+EY A + +++W++ L+D + +
Sbjct: 896 RRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKDLHVTMT 955
Query: 1484 SNISIYCDNTAAIH 1497
++CDN +A+H
Sbjct: 956 CPAKLFCDNKSALH 969
>At4g23160 putative protein
Length = 1240
Score = 176 bits (445), Expect = 1e-43
Identities = 96/267 (35%), Positives = 145/267 (53%), Gaps = 1/267 (0%)
Query: 1232 SRYNSLLKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQ 1291
S + LK L V +YVDDII S N + E +++ F++ +G LKYFLG++
Sbjct: 264 SDHTYFLKITATLFLCVLVYVDDIIICSNNDAAVDELKSQLKSCFKLRDLGPLKYFLGLE 323
Query: 1292 VDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIG 1351
+ ++ G I Q KY +LL + +L + PM P+ V K Y +IG
Sbjct: 324 IARSAAGINICQRKYALDLLDETGLLGCKPSSVPMDPSVTFSAHSGGDFVDAKAYRRLIG 383
Query: 1352 TLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLS 1411
L+YL +R DI F+V+ ++F PR H AV +IL Y+KGT GL Y E +L
Sbjct: 384 RLMYLQITRLDISFAVNKLSQFSEAPRLAHQQAVMKILHYIKGTVGQGLFYSSQAEMQLQ 443
Query: 1412 GYCDADYAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLW 1471
+ DA + + R+ST+ C FLG++L+SW SK Q ++ S+AEAEY + + +M+W
Sbjct: 444 VFSDASFQSCKDTRRSTNGYCMFLGTSLISWKSKKQQVVSKSSAEAEYRALSFATDEMMW 503
Query: 1472 MKHQLEDYQI-LESNISIYCDNTAAIH 1497
+ + Q+ L ++CDNTAAIH
Sbjct: 504 LAQFFRELQLPLSKPTLLFCDNTAAIH 530
>At2g15870 putative retroelement pol polyprotein
Length = 1264
Score = 174 bits (441), Expect = 4e-43
Identities = 91/260 (35%), Positives = 148/260 (56%)
Query: 1238 LKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPE 1297
LK+ + ++V +YVDDI+ S N++ + S+ +Q F++ +G+LKYFLG+++ +T
Sbjct: 934 LKDCGGEYVVVLVYVDDIVIASTNEAAAVQLSQDLQNLFKLRDLGDLKYFLGLEITRTEA 993
Query: 1298 GTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLT 1357
G + Q KY ELL M+ + PM P L K D ++ Y ++GTL+YLT
Sbjct: 994 GISLCQRKYALELLASTGMINCKLVSVPMVPNLKLMKVDGELLEDREQYRRIVGTLMYLT 1053
Query: 1358 ASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDAD 1417
+RPDI F+V+ +F P HL A R+L+Y+KGT GL Y + + L G+ D+D
Sbjct: 1054 ITRPDITFAVNKLCQFSYAPTTAHLQAAHRVLQYIKGTVGQGLFYSASSDLTLKGFADSD 1113
Query: 1418 YAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLE 1477
+A R ST+ F+G +L+S SK Q ++ S+AEAEY A+ + +++W+ L
Sbjct: 1114 WASCPDSRHSTTGFTMFVGDSLISLRSKKQHVVSRSSAEAEYRALALATCELVWLHTLLA 1173
Query: 1478 DYQILESNISIYCDNTAAIH 1497
+ ++ D+TAAI+
Sbjct: 1174 SLTAATTIPILFSDSTAAIY 1193
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 170 bits (430), Expect = 7e-42
Identities = 94/264 (35%), Positives = 149/264 (55%), Gaps = 2/264 (0%)
Query: 1236 SLLKNIQNDI-LIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQ 1294
SL +N+I L V IYVDD++ + + ++F + + F M +G+LKYFLGI+V +
Sbjct: 1168 SLFSYTRNNIELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIEVSR 1227
Query: 1295 TPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLL 1354
PEG ++ Q KY +++ L S A TP+ L +D K Y ++G LL
Sbjct: 1228 GPEGIFLSQRKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVGRLL 1287
Query: 1355 YLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYC 1414
YL +RP++ +SVH+ A+F +PRE H A R++RYLKG+ G++ + L YC
Sbjct: 1288 YLLHTRPELSYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILLNADPDLTLEVYC 1347
Query: 1415 DADYAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKH 1474
D+D+ R+S S LG + +SW +K Q T++ S+AEAEY + ++ W++
Sbjct: 1348 DSDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSYALKEIKWLRK 1407
Query: 1475 QLEDYQILESN-ISIYCDNTAAIH 1497
L++ I +S +YCD+ AAIH
Sbjct: 1408 LLKELGIEQSTPARLYCDSKAAIH 1431
>At1g47650 hypothetical protein
Length = 1409
Score = 170 bits (430), Expect = 7e-42
Identities = 89/260 (34%), Positives = 146/260 (55%)
Query: 1238 LKNIQNDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPE 1297
+K + +IV +YVDDI S ++ + ++ +Q F++ +G+LKYFLG+++ +T
Sbjct: 915 IKEYDGEFVIVLVYVDDIAIASTSEGAAIQLTQDLQQRFKLRDLGDLKYFLGLEIARTEA 974
Query: 1298 GTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLT 1357
G I Q KY ELL M+ PM P + K D ++ Y ++G L+YLT
Sbjct: 975 GISICQRKYALELLASTGMVNCKPVSVPMIPNVKMMKTDGDLLDDREQYRRIVGKLMYLT 1034
Query: 1358 ASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDAD 1417
+R DI F+V+ +F S PR +HL A R+L+Y+KG+ GL Y + + L G+ D+D
Sbjct: 1035 ITRLDITFAVNKLCQFSSAPRTSHLQAAYRVLQYIKGSVGQGLFYSASADLTLKGFADSD 1094
Query: 1418 YAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLE 1477
+A R+ST+ F+G +L+SW SK Q ++ S+AEAEY A+ + +++W+ L
Sbjct: 1095 WASCPDSRRSTTGFSMFVGDSLISWRSKKQHVVSRSSAEAEYRALALVTCELVWLHTLLM 1154
Query: 1478 DYQILESNISIYCDNTAAIH 1497
+ +Y D+T AI+
Sbjct: 1155 SLKASYLVPVLYSDSTTAIY 1174
>At4g19780 putative LTR retrotransposon (fragment)
Length = 306
Score = 168 bits (425), Expect = 3e-41
Identities = 85/232 (36%), Positives = 134/232 (57%), Gaps = 1/232 (0%)
Query: 1267 EFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPM 1326
+F+ +++ F++ +G KYFLGI++ ++ +G I Q KY ELL L S + P+
Sbjct: 3 QFTAELKSYFKLRDLGAAKYFLGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPL 62
Query: 1327 HPTCILEKEDASGKVCQKLYHGMIGTLLYLTASRPDILFSVHLCARFQSDPRETHLTAVK 1386
P+ L KED Y ++G L+YL +RPDI ++V+ +F P HL AV
Sbjct: 63 DPSVKLNKEDGVPLTDSTSYRKLVGKLMYLQITRPDIAYAVNTLCQFSHAPTSVHLNAVH 122
Query: 1387 RILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAGDRTERKSTSENCQFLGSNLVSWASKW 1446
++LRYLKGT GL Y ++ L GY D+D+ R+ + C F+G +LVSW SK
Sbjct: 123 KVLRYLKGTVGQGLFYPADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDSLVSWKSKK 182
Query: 1447 QSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQI-LESNISIYCDNTAAIH 1497
Q T+++STAEAE+ + + +M+W+ L+D+++ +Y DNTAA+H
Sbjct: 183 QDTVSMSTAEAEFRAMSQGTKEMIWLSRLLDDFKVPFIPPAYLYSDNTAALH 234
>At1g26990 polyprotein, putative
Length = 1436
Score = 166 bits (419), Expect = 1e-40
Identities = 87/253 (34%), Positives = 147/253 (57%), Gaps = 1/253 (0%)
Query: 1246 LIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSK 1305
L+V +YVDDI+ S ++ E + + + F++ +GE K+FLGI++ + +G + Q K
Sbjct: 1113 LVVLVYVDDILIASTTEAASAELTSQLSSFFQLRDLGEPKFFLGIEIARNADGISLCQRK 1172
Query: 1306 YTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLTASRPDILF 1365
Y +LL + + + PM P L K+ + K Y ++G L YL +RPDI F
Sbjct: 1173 YVLDLLASSDFSDCKPSSIPMEPNQKLSKDTGTLLEDGKQYRRILGKLQYLCLTRPDINF 1232
Query: 1366 SVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAGDRTER 1425
+V A++ S P + HL A+ +ILRYLKGT GL Y + L G+ D+D+ R
Sbjct: 1233 AVSKLAQYSSAPTDIHLQALHKILRYLKGTIGQGLFYGADTNFDLRGFSDSDWQTCPDTR 1292
Query: 1426 KSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQILESN 1485
+ + F+G++LVSW SK Q +++S+AEAEY ++ + +++W+ + L ++I ++
Sbjct: 1293 RCVTGFAIFVGNSLVSWRSKKQDVVSMSSAEAEYRAMSVATKELIWLGYILTAFKIPFTH 1352
Query: 1486 IS-IYCDNTAAIH 1497
+ +YCDN AA+H
Sbjct: 1353 PAYLYCDNEAALH 1365
>At3g25450 hypothetical protein
Length = 1343
Score = 164 bits (414), Expect = 5e-40
Identities = 88/254 (34%), Positives = 148/254 (57%), Gaps = 1/254 (0%)
Query: 1244 DILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 1303
+IL+V +YVDD++ +N + F + M +FEMS +G+L Y+LGI+V Q+ +G + Q
Sbjct: 1006 NILVVAVYVDDLLVTGSNLDIILNFKKGMVGKFEMSDLGKLTYYLGIEVLQSKDGITLKQ 1065
Query: 1304 SKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLTASRPDI 1363
+Y K++L++ M + TPM + L K ++ + Y IG L YL +RPD+
Sbjct: 1066 ERYAKKILEEAGMSKCNTVNTPMIASLELSKAQDEKRIDETDYRRNIGCLRYLLHTRPDL 1125
Query: 1364 LFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAGDRT 1423
++V + +R+ +PRE+H A+K+ILRYL+GTT+ GL +KK L GY D+ + D
Sbjct: 1126 SYNVGILSRYLQEPRESHGAALKQILRYLQGTTSHGLYFKKGENAGLIGYSDSSHNVDLD 1185
Query: 1424 ERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQILE 1483
+ KST + +L ++W S+ Q + LS+ EAE++ + Q +W++ L + E
Sbjct: 1186 DGKSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEFMAATEAAKQAIWLQELLAEVIGTE 1245
Query: 1484 -SNISIYCDNTAAI 1496
++I DN +AI
Sbjct: 1246 CEKVTIRVDNKSAI 1259
Score = 55.5 bits (132), Expect = 2e-07
Identities = 35/163 (21%), Positives = 89/163 (54%), Gaps = 5/163 (3%)
Query: 37 TADQKKLYSQHHKARAILLSAISYEEYEKITDREYAKGIFESLKMSHEGNKKVKESKALS 96
+AD++K + ARA+L +I ++ ++ + ++E++K + G ++VKE++ +
Sbjct: 54 SADEEK----NDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQT 109
Query: 97 LIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKYHVIRVIRSLP-ESWMPLVT 155
L+ +++ M+ +E+I++ R + L + V + ++SLP + ++ +V
Sbjct: 110 LMAEFDKLKMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVA 169
Query: 156 SIELTRDVERMSLEELISILKCHELKHSEMLDSDEDELTLISK 198
++E D++ + E++ +K +E + + DS ED+ L+++
Sbjct: 170 ALEQVLDLKTTTFEDIAGRIKTYEDRVWDDDDSHEDQGKLMTE 212
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 164 bits (414), Expect = 5e-40
Identities = 88/264 (33%), Positives = 149/264 (56%), Gaps = 2/264 (0%)
Query: 1236 SLLKNIQNDILI-VQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQ 1294
SL ++ + I + IYVDD+I +Q ++F E + + F M +G LKYFLGI+V +
Sbjct: 1000 SLFTLVKGSVRIKILIYVDDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVAR 1059
Query: 1295 TPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLL 1354
+ G YI Q KY +++ + +L A P+ L + + Y ++G L+
Sbjct: 1060 STTGIYICQRKYALDIISETGLLGVKPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLI 1119
Query: 1355 YLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYC 1414
YL +R D+ FSVH+ ARF +PRE H A R++RYLK G+ +++ +++++G+C
Sbjct: 1120 YLAVTRLDLAFSVHILARFMQEPREDHWAAALRVVRYLKADPGQGVFLRRSGDFQITGWC 1179
Query: 1415 DADYAGDRTERKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKH 1474
D+D+AGD R+S + G + +SW +K Q T++ S+AEAEY + ++++LW+K
Sbjct: 1180 DSDWAGDPMSRRSVTGYFVQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQ 1239
Query: 1475 QLEDYQILESNISIY-CDNTAAIH 1497
L + I CD+ +AI+
Sbjct: 1240 LLFSLGVSHVQPMIMCCDSKSAIY 1263
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 164 bits (414), Expect = 5e-40
Identities = 87/254 (34%), Positives = 144/254 (56%), Gaps = 3/254 (1%)
Query: 1245 ILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 1304
IL + +YVDDI+ ++QSL ++ + ++ F M +G +YFLGIQ++ G ++HQ+
Sbjct: 1047 ILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFLGIQIEDYANGLFLHQT 1106
Query: 1305 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLTASRPDIL 1364
Y ++L++ M + TP+ L+ ++ + + G L YLT +RPDI
Sbjct: 1107 AYATDILQQAGMSDCNPMPTPLPQQ--LDNLNSELFAEPTYFRSLAGKLQYLTITRPDIQ 1164
Query: 1365 FSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAGDRTE 1424
F+V+ + P + +KRILRY+KGT +GL K+ LS Y D+D+AG +
Sbjct: 1165 FAVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMGLPIKRNSTLTLSAYSDSDHAGCKNT 1224
Query: 1425 RKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQILE- 1483
R+ST+ C LGSNL+SW++K Q T++ S+ EAEY + ++ W+ L D I +
Sbjct: 1225 RRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAAREITWISFLLRDLGIPQY 1284
Query: 1484 SNISIYCDNTAAIH 1497
+YCDN +A++
Sbjct: 1285 LPTQVYCDNLSAVY 1298
>At1g59265 polyprotein, putative
Length = 1466
Score = 162 bits (410), Expect = 1e-39
Identities = 93/254 (36%), Positives = 139/254 (54%), Gaps = 1/254 (0%)
Query: 1245 ILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 1304
I+ + +YVDDI+ + +L + + F + EL YFLGI+ + P G ++ Q
Sbjct: 1144 IVYMLVYVDDILITGNDPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQR 1203
Query: 1305 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDASGKVCQKLYHGMIGTLLYLTASRPDIL 1364
+Y +LL + NM+ + TPM P+ L + Y G++G+L YL +RPDI
Sbjct: 1204 RYILDLLARTNMITAKPVTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDIS 1263
Query: 1365 FSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKT*EYKLSGYCDADYAGDRTE 1424
++V+ ++F P E HL A+KRILRYL GT N G+ KK L Y DAD+AGD+ +
Sbjct: 1264 YAVNRLSQFMHMPTEEHLQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDD 1323
Query: 1425 RKSTSENCQFLGSNLVSWASKWQSTIALSTAEAEYILTAICSTQMLWMKHQLEDYQI-LE 1483
ST+ +LG + +SW+SK Q + S+ EAEY A S++M W+ L + I L
Sbjct: 1324 YVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLT 1383
Query: 1484 SNISIYCDNTAAIH 1497
IYCDN A +
Sbjct: 1384 RPPVIYCDNVGATY 1397
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.347 0.151 0.513
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,094,034
Number of Sequences: 26719
Number of extensions: 1252949
Number of successful extensions: 9042
Number of sequences better than 10.0: 260
Number of HSP's better than 10.0 without gapping: 117
Number of HSP's successfully gapped in prelim test: 150
Number of HSP's that attempted gapping in prelim test: 8314
Number of HSP's gapped (non-prelim): 660
length of query: 1564
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1452
effective length of database: 8,326,068
effective search space: 12089450736
effective search space used: 12089450736
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0134.10


