

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0127b.1
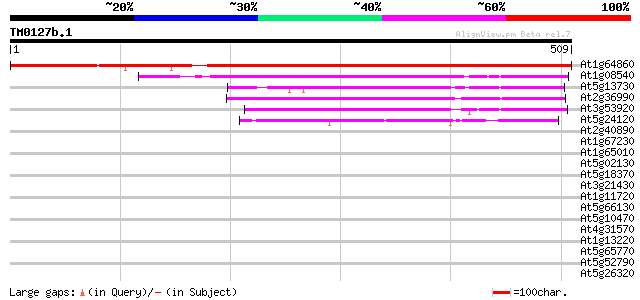
(509 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g64860 plastid RNA polymerase sigma-subunit (SIG2) 669 0.0
At1g08540 plastid RNA polymerase sigma-subunit (SIG1) 170 1e-42
At5g13730 sigma-like factor (SIG4) 148 8e-36
At2g36990 putative RNA polymerase sigma-70 factor 147 1e-35
At3g53920 sigma factor SigC 108 9e-24
At5g24120 sigma-like factor (emb|CAA77213.1) 106 3e-23
At2g40890 cytochrome P450 like protein 33 0.49
At1g67230 unknown protein 32 1.1
At1g65010 hypothetical protein 31 1.4
At5g02130 putative protein 31 1.8
At5g18370 disease resistance protein -like 30 3.2
At3g21430 unknown protein 30 3.2
At1g11720 glycogen synthase like protein 30 4.1
At5g66130 AtRAD17 (dbj|BAA90479.1) 29 5.4
At5g10470 TH65 protein (th65) 29 5.4
At4g31570 putative protein 29 5.4
At1g13220 putative nuclear matrix constituent protein 29 5.4
At5g65770 nuclear matrix constituent protein 1 (NMCP1)-like 29 7.0
At5g52790 unknown protein 29 7.0
At5g26320 putative protein 29 7.0
>At1g64860 plastid RNA polymerase sigma-subunit (SIG2)
Length = 502
Score = 669 bits (1725), Expect = 0.0
Identities = 353/515 (68%), Positives = 414/515 (79%), Gaps = 19/515 (3%)
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQA 60
MATAAVIGL+ GKRLLSSS+Y+SD+ EK +D S+ Y I KS AKK+S+Y+ +
Sbjct: 1 MATAAVIGLNTGKRLLSSSFYHSDVTEKFLSVNDHCSSQYHIASTKSGITAKKASNYSPS 60
Query: 61 FPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSD----LGYSVEALLLLQK 116
FP+S+R QS KALKE VD V + + W +LE E D + +SVEA+LLLQK
Sbjct: 61 FPSSNRHTQSAKALKESVD-VASTEKPWLPNGTDKELEEECYDDDDLISHSVEAILLLQK 119
Query: 117 SMLEKQWSLSCEREVLTEHPRQEKSSKKV--AVTCSGVSARQRRINTKRKIPGKTGSAMQ 174
SMLEK W+LS E+ V +E+P + KK +TCSG+SARQRRI K+K
Sbjct: 120 SMLEKSWNLSFEKAVSSEYPGKGTIRKKKIPVITCSGISARQRRIGAKKKTN-------- 171
Query: 175 ACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKE 234
M +S + +GYVKGV+SE +LSH EVV+LS+KIK GL LD+HKSRLK+
Sbjct: 172 ----MTHVKAVSDVSSGKQVRGYVKGVISEDVLSHVEVVRLSKKIKSGLRLDDHKSRLKD 227
Query: 235 KLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMAD 294
+LGCEPSD+Q+A SLKISR+EL+A +EC LARE+LAMSNVRLVMSIAQRYDN+GAEM+D
Sbjct: 228 RLGCEPSDEQLAVSLKISRAELQAWLMECHLAREKLAMSNVRLVMSIAQRYDNLGAEMSD 287
Query: 295 LVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLI 354
LVQGGLIGLLRGIEKFDSSKGF+ISTYVYWWIRQGVSRALV+NSRTLRLPTHLHERL LI
Sbjct: 288 LVQGGLIGLLRGIEKFDSSKGFRISTYVYWWIRQGVSRALVDNSRTLRLPTHLHERLGLI 347
Query: 355 RTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHH 414
R AK RL+E+GITP+IDRIA+SLNMSQKKVRNATEA+SKVFSLDR+AFPSLNGLPG+THH
Sbjct: 348 RNAKLRLQEKGITPSIDRIAESLNMSQKKVRNATEAVSKVFSLDRDAFPSLNGLPGETHH 407
Query: 415 CYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKR 474
YIAD RLEN PWHG D+ LK+EV+KLI+ TL ERE+EIIRLYYGLDKECLTWEDISKR
Sbjct: 408 SYIADTRLENNPWHGYDDLALKEEVSKLISATLGEREKEIIRLYYGLDKECLTWEDISKR 467
Query: 475 IGLSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
IGLSRERVRQVGLVALEKLKHAARKR+MEAM+LK+
Sbjct: 468 IGLSRERVRQVGLVALEKLKHAARKRKMEAMILKN 502
>At1g08540 plastid RNA polymerase sigma-subunit (SIG1)
Length = 572
Score = 170 bits (431), Expect = 1e-42
Identities = 122/393 (31%), Positives = 214/393 (54%), Gaps = 27/393 (6%)
Query: 118 MLEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSAMQACD 177
+LE+Q S+S + R+ + +K + T SG+ + KTGS+ +
Sbjct: 201 LLEEQPSVSLAVRSTRQTERKARRAKGLEKTASGIPSV------------KTGSSPKK-- 246
Query: 178 AMQMRSLISPELLQNRSKGYVKGVVSE-QLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKL 236
+ L++ E+ N Y++ S +LL+ E +LS I+ L L+ ++ L E+
Sbjct: 247 ----KRLVAQEVDHNDPLRYLRMTTSSSKLLTVREEHELSAGIQDLLKLERLQTELTERS 302
Query: 237 GCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLV 296
G +P+ Q A++ + + LR + TL ++++ SN+RLV+SIA+ Y G + DLV
Sbjct: 303 GRQPTFAQWASAAGVDQKSLRQRIHHGTLCKDKMIKSNIRLVISIAKNYQGAGMNLQDLV 362
Query: 297 QGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRT 356
Q G GL+RG EKFD++KGFK STY +WWI+Q V ++L + SR +RLP H+ E ++
Sbjct: 363 QEGCRGLVRGAEKFDATKGFKFSTYAHWWIKQAVRKSLSDQSRMIRLPFHMVEATYRVKE 422
Query: 357 AKFRL-EERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHC 415
A+ +L E G P + IA++ +S K++ + SLD++ + N P +
Sbjct: 423 ARKQLYSETGKHPKNEEIAEATGLSMKRLMAVLLSPKPPRSLDQKIGMNQNLKPSEV--- 479
Query: 416 YIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGL-DKECLTWEDISKR 474
IAD + E+ ++ +++K+++ +L RE+++IR +G+ D T ++I +
Sbjct: 480 -IADPEAVTSEDILIKEF-MRQDLDKVLD-SLGTREKQVIRWRFGMEDGRMKTLQEIGEM 536
Query: 475 IGLSRERVRQVGLVALEKLKHAARKREMEAMLL 507
+G+SRERVRQ+ A KLK+ R ++ L+
Sbjct: 537 MGVSRERVRQIESSAFRKLKNKKRNNHLQQYLV 569
>At5g13730 sigma-like factor (SIG4)
Length = 419
Score = 148 bits (373), Expect = 8e-36
Identities = 109/313 (34%), Positives = 163/313 (51%), Gaps = 21/313 (6%)
Query: 198 VKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKIS---RS 254
V G LS E V+L +K G L E LG ++++ + L S +
Sbjct: 114 VVGASRSGFLSRLEEVQLCLYLKEGAKL--------ENLGTSVEENEMVSVLLASGRGKK 165
Query: 255 ELRAKTIECTL--ARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDS 312
+ A I C ARE++ RLV+SIA Y G + DL+Q G IGLLRG E+FD
Sbjct: 166 KRSANEILCRRKEAREKITRCYRRLVVSIATGYQGKGLNLQDLIQEGSIGLLRGAERFDP 225
Query: 313 SKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLEER-GITPTID 371
+G+K+STYVYWWI+Q + RA+ SR ++LP + E + + A L + P+ +
Sbjct: 226 DRGYKLSTYVYWWIKQAILRAIAHKSRLVKLPGSMWELTAKVAEASNVLTRKLRRQPSCE 285
Query: 372 RIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVD 431
IA+ LN++ VR A E SLDR A S NG I E P V
Sbjct: 286 EIAEHLNLNVSAVRLAVERSRSPVSLDRVA--SQNGRMTLQE---IVRGPDETRPEEMVK 340
Query: 432 EWTLKDEVNKLINVTLVEREREIIRLYYGLDKEC-LTWEDISKRIGLSRERVRQVGLVAL 490
+K E+ +L+ +L RE ++ LY+GL+ E +++E+I K + LSRERVRQ+ +AL
Sbjct: 341 REHMKHEIEQLLG-SLTARESRVLGLYFGLNGETPMSFEEIGKSLKLSRERVRQINGIAL 399
Query: 491 EKLKHAARKREME 503
+KL++ +++
Sbjct: 400 KKLRNVHNVNDLK 412
>At2g36990 putative RNA polymerase sigma-70 factor
Length = 547
Score = 147 bits (372), Expect = 1e-35
Identities = 96/310 (30%), Positives = 169/310 (53%), Gaps = 8/310 (2%)
Query: 197 YVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSEL 256
++ G ++QLL+ E +L I+ L L++ K++L+ + GCEP+ + A ++ IS L
Sbjct: 238 FLWGPETKQLLTAKEEAELISHIQHLLKLEKVKTKLESQNGCEPTIGEWAEAMGISSPVL 297
Query: 257 RAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGF 316
++ +RE+L +N+RLV+ IA++Y N G DL+Q G +GL++ +EKF G
Sbjct: 298 KSDIHRGRSSREKLITANLRLVVHIAKQYQNRGLNFQDLLQEGSMGLMKSVEKFKPQSGC 357
Query: 317 KISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTA-KFRLEERGITPTIDRIAK 375
+ +TY YWWIRQ + +++ +NSRT+RLP +++ L + A K ++E P+ + +A
Sbjct: 358 RFATYAYWWIRQSIRKSIFQNSRTIRLPENVYMLLGKVSEARKTCVQEGNYRPSKEELAG 417
Query: 376 SLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTL 435
+ +S +K+ S+ + + DT I + P V + +
Sbjct: 418 HVGVSTEKLDKLLYNTRTPLSMQQPIWSD-----QDTTFQEITPDSGIETPTMSVGKQLM 472
Query: 436 KDEVNKLINVTLVEREREIIRLYYGLD-KECLTWEDISKRIGLSRERVRQVGLVALEKLK 494
++ V L+NV L +ER II+L +G+D + + +I + GLS+ERVRQ+ AL +LK
Sbjct: 473 RNHVRNLLNV-LSPKERRIIKLRFGIDGGKQRSLSEIGEIYGLSKERVRQLESRALYRLK 531
Query: 495 HAARKREMEA 504
+ A
Sbjct: 532 QNMNSHGLHA 541
>At3g53920 sigma factor SigC
Length = 571
Score = 108 bits (269), Expect = 9e-24
Identities = 81/300 (27%), Positives = 154/300 (51%), Gaps = 17/300 (5%)
Query: 214 KLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMS 273
++S +K+ ++ +++L+E+ G S AA+ ++ L R+ L S
Sbjct: 280 EMSTGVKIVADMERIRTQLEEESGKVASLSCWAAAAGMNEKLLMRNLHYGWYCRDELVKS 339
Query: 274 NVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRA 333
LV+ +A+ Y +G DL+Q G +G+L+G E+FD ++G+K STYV +WIR+ +S
Sbjct: 340 TRSLVLFLARNYRGLGIAHEDLIQAGYVGVLQGAERFDHTRGYKFSTYVQYWIRKSMSTM 399
Query: 334 LVENSRTLRLPTHLHERLSLIRTAKFRLE-ERGITPTID-RIAKSLNMSQKKVRNATEAI 391
+ ++R + +P+ + ++ I+ A+ L+ GI D IAK S KK+R A + +
Sbjct: 400 VSRHARGVHIPSSIIRTINHIQKARKTLKTSHGIKYAADEEIAKLTGHSVKKIRAANQCL 459
Query: 392 SKVFSLDREAFPSLNGLPGDTHHC----YIADNRLENIPWHGVDEWTLKDEVNKLINVTL 447
V S+D++ GD + D +E+ P V + + +++ L+ L
Sbjct: 460 KVVGSIDKKV--------GDCFTTKFLEFTPDTTMES-PEEAVMRQSARRDIHDLLE-GL 509
Query: 448 VEREREIIRLYYGL-DKECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEAML 506
RE++++ L YGL D + E+I K + +S+E +R++ A+ KL+ ++ L
Sbjct: 510 EPREKQVMVLRYGLQDYRPKSLEEIGKLLKVSKEWIRKIERRAMAKLRDQPNAEDLRYYL 569
>At5g24120 sigma-like factor (emb|CAA77213.1)
Length = 517
Score = 106 bits (265), Expect = 3e-23
Identities = 92/301 (30%), Positives = 152/301 (49%), Gaps = 28/301 (9%)
Query: 209 HAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARE 268
HA + KL + +K +L E K L++ LG EP + +IA + ++ E++ K AR
Sbjct: 225 HAWLFKLMQPMK---ALLEVKDVLQKSLGREPREAEIAGEINMTAGEVKKKIEIGRAARN 281
Query: 269 RLAMSNVRLVMSIAQRYDNM---GAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWW 325
+L N+RLV+ + +Y + G + DL Q G+ GL+ I++F+ + F++STY +W
Sbjct: 282 KLIKHNLRLVLFVMNKYFHEFTNGPKFQDLCQAGMRGLITAIDRFEPKRKFRLSTYGLFW 341
Query: 326 IRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRL-EERGITPTIDRIAKSLNMSQKKV 384
IR + R++ ++ T R+P L I AK L E G PT D + + L ++ ++
Sbjct: 342 IRHAIIRSMTTSNFT-RVPFGLESVRVEIYKAKTELWFEMGRPPTEDEVVERLKITPERY 400
Query: 385 RNATEAISKVFSLD------REAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDE 438
R A V SL+ +E F +NG+ D ++R + +D+
Sbjct: 401 REVLRAAKPVLSLNSKHSVTQEEF--INGIT-DVDGVGANNSRQPALLRLALDD------ 451
Query: 439 VNKLINVTLVEREREIIRLYYGLD-KECLTWEDISKRIGLSRERVRQVGLVALEKLKHAA 497
+ +L +E +IR YGLD K T +I+ + +SRE VR+ + AL KLKH A
Sbjct: 452 ----VLDSLKPKESLVIRQRYGLDGKGDRTLGEIAGNLNISREMVRKHEVKALMKLKHQA 507
Query: 498 R 498
R
Sbjct: 508 R 508
>At2g40890 cytochrome P450 like protein
Length = 508
Score = 32.7 bits (73), Expect = 0.49
Identities = 33/107 (30%), Positives = 49/107 (44%), Gaps = 9/107 (8%)
Query: 199 KGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSEL-R 257
+GVV EQ L +V K+ LS+ EH L+ P+D++ A R L R
Sbjct: 195 EGVVDEQGLEFKAIVSNGLKLGASLSIAEHIPWLRWMF---PADEKAFAEHGARRDRLTR 251
Query: 258 AKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLL 304
A E TLAR++ + + V ++ D DL + +IGLL
Sbjct: 252 AIMEEHTLARQKSSGAKQHFVDALLTLKDQY-----DLSEDTIIGLL 293
>At1g67230 unknown protein
Length = 1132
Score = 31.6 bits (70), Expect = 1.1
Identities = 47/245 (19%), Positives = 98/245 (39%), Gaps = 23/245 (9%)
Query: 49 TVAKKSSDYTQAFPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSDLGYSV 108
T K ++ + + + ++A VD A + + EVE+ +
Sbjct: 150 TADSKLTEANALVRSVEEKSLEVEAKLRAVDAKLAEVSRKSSDVERKAKEVEARESSLQR 209
Query: 109 EALLLLQKSMLEKQWSLSCEREVLTEHPRQ-EKSSKKVAVTCSGVSARQRRINTKRKIPG 167
E + + ++ +LS +RE L E R+ ++ ++VA + V R+ R N KI
Sbjct: 210 ERFSYIAEREADEA-TLSKQREDLREWERKLQEGEERVAKSQMIVKQREDRANESDKIIK 268
Query: 168 KTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVV----------------SEQLLSHAE 211
+ G ++ + ++ + L++ +K + + +L + E
Sbjct: 269 QKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALREQETDVLKKSIETKARELQALQE 328
Query: 212 VVKLSEKIKVGLSLDEHKSRLKE-----KLGCEPSDDQIAASLKISRSELRAKTIECTLA 266
++ EK+ V +DEH+++L +L E I SLK +E+ + E
Sbjct: 329 KLEAREKMAVQQLVDEHQAKLDSTQREFELEMEQKRKSIDDSLKSKVAEVEKREAEWKHM 388
Query: 267 RERLA 271
E++A
Sbjct: 389 EEKVA 393
>At1g65010 hypothetical protein
Length = 1318
Score = 31.2 bits (69), Expect = 1.4
Identities = 23/78 (29%), Positives = 39/78 (49%), Gaps = 6/78 (7%)
Query: 70 SIKALKE--HVDGVPAAAETWFQECDSNDLEVESSDL--GYSVEALLLLQKSMLEKQWSL 125
S+K + E H++ E+ FQ +LE+++ D +E L L++S+LEK+ L
Sbjct: 1071 SLKKIDELLHLEQSWLEKESEFQRVTQENLELKTQDALAAKKIEELSKLKESLLEKETEL 1130
Query: 126 SCEREVLTEHPRQEKSSK 143
C E + E+ SK
Sbjct: 1131 KCREAAALE--KMEEPSK 1146
>At5g02130 putative protein
Length = 420
Score = 30.8 bits (68), Expect = 1.8
Identities = 28/97 (28%), Positives = 48/97 (48%), Gaps = 10/97 (10%)
Query: 207 LSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIA----ASLKISRSELRAKTIE 262
LSHA K E G+ + L++ LG +P+DDQ++ A++ ++ S+L ++
Sbjct: 58 LSHARSQKSDESYAQGMLV------LEQCLGNQPNDDQVSHDSKATVLLAMSDLLYESGN 111
Query: 263 CTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGG 299
+ A ERL S+A R + A + L+Q G
Sbjct: 112 SSEAIERLKQVMTLTHSSLAIRVVAVEALVGLLIQSG 148
>At5g18370 disease resistance protein -like
Length = 1210
Score = 30.0 bits (66), Expect = 3.2
Identities = 19/61 (31%), Positives = 35/61 (57%), Gaps = 9/61 (14%)
Query: 5 AVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGST----HYQIVPAKSVTVAKKSSDYTQA 60
A++ LS K SSS+ ++++E ++C + G T YQ+ P+ V K++ D+ +A
Sbjct: 117 AIVLLS--KNYASSSWCLNELVEIMNCREEIGQTVMTVFYQVDPS---DVRKQTGDFGKA 171
Query: 61 F 61
F
Sbjct: 172 F 172
>At3g21430 unknown protein
Length = 961
Score = 30.0 bits (66), Expect = 3.2
Identities = 25/99 (25%), Positives = 42/99 (42%), Gaps = 12/99 (12%)
Query: 34 DFGSTHYQIVPAKSVTVAKKSSDYTQAFPASDRPNQSIKALKEHVDGVPAAAETWFQECD 93
DF + I + +V + Q P++ P S A H++ + D
Sbjct: 837 DFVNNQLSIDQTEGSSVQQTQGGQDQRLPSTPNPPSSTPANDSHLN-----------QPD 885
Query: 94 SNDLEVESSDLGYSVEALLLLQKSMLEKQWSLSCEREVL 132
NDL+V S + + LL++QK E+Q+ S +VL
Sbjct: 886 QNDLQVPSDLVSRCIATLLMIQK-CTERQFPPSEVAQVL 923
>At1g11720 glycogen synthase like protein
Length = 1025
Score = 29.6 bits (65), Expect = 4.1
Identities = 21/62 (33%), Positives = 30/62 (47%), Gaps = 3/62 (4%)
Query: 225 LDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQR 284
+++ RL +K DD + LK+ R LR K IE TLA E LA + V + +
Sbjct: 78 IEDGSDRLDKKT--TDDDDLLEQKLKLERENLRRKEIE-TLAAENLARGDRMFVYPVIVK 134
Query: 285 YD 286
D
Sbjct: 135 PD 136
>At5g66130 AtRAD17 (dbj|BAA90479.1)
Length = 599
Score = 29.3 bits (64), Expect = 5.4
Identities = 28/108 (25%), Positives = 45/108 (40%), Gaps = 4/108 (3%)
Query: 232 LKEKLGCEPSDDQIAASLKISRSELRAK-TIECTLARERLAMSNVRLVMSIAQRYDNMGA 290
LK+KL E DD + +L+ SRS ++K A A RL + A + D +
Sbjct: 2 LKKKLSLEDEDDDRSYNLRSSRSNAKSKPRSSAGTATNPRASKRARLSGASATQED---S 58
Query: 291 EMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENS 338
+ D ++ + F S G++ S W+ + R L E S
Sbjct: 59 SLVDKIRLSFEDFDEALSGFKVSSGYERSKNTDLWVDKYRPRTLEELS 106
>At5g10470 TH65 protein (th65)
Length = 1273
Score = 29.3 bits (64), Expect = 5.4
Identities = 46/216 (21%), Positives = 83/216 (38%), Gaps = 26/216 (12%)
Query: 54 SSDYTQAFPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVE---------SSDL 104
+SD + +R NQ+ LK+ V G+ A + +C EV+ SDL
Sbjct: 473 ASDARKELLEKERENQN---LKQEVVGLKKALKDANDQCVLLYSEVQRAWKVSFTLQSDL 529
Query: 105 GYSVEALLLLQKSMLEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRK 164
E ++L+ K LEK+ + ++ ++ ++ S + Q +I
Sbjct: 530 --KSENIMLVDKHRLEKEQNSQLRNQIAQFLQLDQEQKLQMQQQDSAIQNLQAKITDLES 587
Query: 165 IPGKT--GSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVG 222
+ + DA+Q + + SP + V S + KL E++K
Sbjct: 588 QVSEAVRSDTTRTGDALQSQDIFSP----------IPKAVEGTTDSSSVTKKLEEELKKR 637
Query: 223 LSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRA 258
+L E EKL ++ +A S ++ LRA
Sbjct: 638 DALIERLHEENEKLFDRLTERSMAVSTQVLSPSLRA 673
>At4g31570 putative protein
Length = 2712
Score = 29.3 bits (64), Expect = 5.4
Identities = 46/206 (22%), Positives = 79/206 (38%), Gaps = 63/206 (30%)
Query: 214 KLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMS 273
KL++++ L+L EH S ++E+ R+ L + E ++LA
Sbjct: 730 KLTQEL---LTLQEHMSTVEEE-----------------RTHLEVELREAIARLDKLAEE 769
Query: 274 NVRLVMSI----AQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQG 329
N L SI A+ DN A+++ L+ + EK S +S
Sbjct: 770 NTSLTSSIMVEKARMVDNGSADVSGLINQEIS------EKLGRSSEIGVSKQ-------- 815
Query: 330 VSRALVENSRTLRLPTHLHERLSLIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATE 389
S + +EN++ L E R T + K+L +K V+N E
Sbjct: 816 -SASFLENTQYTNLE-----------------EVREYTSEFSALMKNLEKGEKMVQNLEE 857
Query: 390 AISKVF-------SLDREAFPSLNGL 408
AI ++ S D+ A P+++ L
Sbjct: 858 AIKQILTDSSVSKSSDKGATPAVSKL 883
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 29.3 bits (64), Expect = 5.4
Identities = 25/122 (20%), Positives = 59/122 (47%), Gaps = 4/122 (3%)
Query: 377 LNMSQKKVRNATEAISKVFSLDREAFPSLN--GLPGDTHHCYIADNRLENIPWHGVDEWT 434
LN + + ++ + K+ +L++E + + GL + ++ + N + E
Sbjct: 72 LNEASMEKKDQEALLEKISTLEKELYGYQHNMGLLLMENKELVSKHEQLNQAFQEAQEIL 131
Query: 435 LKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIGLSRERVRQVGLVALEKLK 494
+++ + L +T VE+ E +R GL+K+C+ +++ K + +E ++ L + KL
Sbjct: 132 KREQSSHLYALTTVEQREENLRKALGLEKQCV--QELEKALREIQEENSKIRLSSEAKLV 189
Query: 495 HA 496
A
Sbjct: 190 EA 191
>At5g65770 nuclear matrix constituent protein 1 (NMCP1)-like
Length = 1042
Score = 28.9 bits (63), Expect = 7.0
Identities = 53/251 (21%), Positives = 100/251 (39%), Gaps = 25/251 (9%)
Query: 46 KSVTVAKKSSDYTQAFPASDR-PNQSIKALKEHVDGVPAAAETWFQECDSNDLEV----- 99
K + S D +AF + + I++LKE + + + D+ LE+
Sbjct: 662 KREELENSSRDREKAFEQEKKLEEERIQSLKEMAEKELEHVQVELKRLDAERLEIKLDRE 721
Query: 100 ----ESSDLGYSVEALLLLQKSMLEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVS-A 154
E ++L SVE L + ++ + ++ L ER+ + + K + + V +S A
Sbjct: 722 RREREWAELKDSVEELKVQREKLETQRHMLRAERDEIRHEIEELKKLENLKVALDDMSMA 781
Query: 155 RQRRINTKR---KIPGKTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAE 211
+ + N +R K+ + D + +++ +S + N GY + + L+ +
Sbjct: 782 KMQLSNLERSWEKVSALKQKVVSRDDELDLQNGVS--TVSNSEDGYNSSMERQNGLTPSS 839
Query: 212 VVKLS---EKIKVGLSLDEHKSRLK---EKLGCEPSDDQIAASLKISRSELRAKTIECTL 265
S + KS L E+ G PS+ L+ SR E +A T ++
Sbjct: 840 ATPFSWIKRCTNLIFKTSPEKSTLMHHYEEEGGVPSE---KLKLESSRREEKAYTEGLSI 896
Query: 266 ARERLAMSNVR 276
A ERL R
Sbjct: 897 AVERLEAGRKR 907
>At5g52790 unknown protein
Length = 500
Score = 28.9 bits (63), Expect = 7.0
Identities = 12/26 (46%), Positives = 20/26 (76%)
Query: 373 IAKSLNMSQKKVRNATEAISKVFSLD 398
I+ +L+MSQK ++A +S++FSLD
Sbjct: 196 ISGALDMSQKSAKDAMTPVSQIFSLD 221
>At5g26320 putative protein
Length = 352
Score = 28.9 bits (63), Expect = 7.0
Identities = 18/62 (29%), Positives = 37/62 (59%), Gaps = 3/62 (4%)
Query: 155 RQRRINTKRKIPGKTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVK 214
+ +R + ++ + A+++ ++ S+I E LQN SKGY +V++ ++ AE+VK
Sbjct: 287 KNQRSSNHIQLYSEAWCAIRSGYGIEGNSIILLEDLQNSSKGY---LVNDAIIFEAELVK 343
Query: 215 LS 216
+S
Sbjct: 344 VS 345
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,295,704
Number of Sequences: 26719
Number of extensions: 411438
Number of successful extensions: 1323
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 1292
Number of HSP's gapped (non-prelim): 39
length of query: 509
length of database: 11,318,596
effective HSP length: 104
effective length of query: 405
effective length of database: 8,539,820
effective search space: 3458627100
effective search space used: 3458627100
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0127b.1


