

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
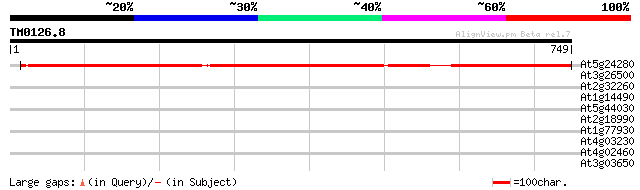
Query= TM0126.8
(749 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g24280 putative protein 704 0.0
At3g26500 unknown protein 30 4.9
At2g32260 putative phospholipid cytidylyltransferase 30 4.9
At1g14490 hypothetical protein 30 4.9
At5g44030 cellulose synthase catalytic subunit-like protein 30 6.5
At2g18990 putative ATP binding protein 30 6.5
At1g77930 dnaJ like protein 30 6.5
At4g03230 putative receptor kinase 29 8.4
At4g02460 DNA mismatch repair protein (PMS1) 29 8.4
At3g03650 unknown protein 29 8.4
>At5g24280 putative protein
Length = 1634
Score = 704 bits (1816), Expect = 0.0
Identities = 371/740 (50%), Positives = 489/740 (65%), Gaps = 45/740 (6%)
Query: 15 NAKRKLVLRKDDDDTGKAYRFKILLPNGTSVELTLRDPQPQMPLPDFLQLVEDAYNGARE 74
+ KR LVL DDDD Y FK+LLPNGTSV+LTL++P+P++ + F+ LV+ Y+ AR+
Sbjct: 15 SVKRSLVL-DDDDDEDIFYNFKVLLPNGTSVKLTLKNPEPEISMQSFVNLVKKEYDNARK 73
Query: 75 HNQSMKRKKDINWIGAPLFLHDSTDAKIKNVIDFHNYKPHKCHILQLHGYFDSV*NAMQN 134
M ++ ++W F +S K+K ++ F +KP CHI++L +N
Sbjct: 74 DCLLMSKRMKVDWNSGGKFHLESNGGKMKGIVRFAAFKPDLCHIIRLDDGSGIASTMYEN 133
Query: 135 MWDLTPDIDLLLELPEEYSFQTALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDN 194
+WDLTPD DLL ELPE YSF+TALADLIDNSLQAVW +RKLI V++ D I++FD
Sbjct: 134 LWDLTPDTDLLKELPENYSFETALADLIDNSLQAVWPYREGARKLISVDISGDHITVFDT 193
Query: 195 GPGMDDTDDNSLVKWGKLGASLHRNSKLQAIGGKPPYLMPYFGMFGYGGPIASMHLGR** 254
G GMD ++ NS+ KWGK+GASLHR+ K AIGG PPYL PYFGMFGYGGP ASM LGR
Sbjct: 194 GRGMDSSEGNSIDKWGKIGASLHRSQKTGAIGGNPPYLKPYFGMFGYGGPYASMFLGRCD 253
Query: 255 HSFPTSVQALPQNYIYLFIAPSHVLSHNRRASVSSKTKHVKKVYMLHLEREALLNTCSEV 314
SF LP + + L RR VSSKTK KKV+ L ++EAL++ S V
Sbjct: 254 FSF-----CLP--ILICIVLLRLTLFSVRRTLVSSKTKESKKVFTLQFKKEALIDNRSIV 306
Query: 315 --KWKTDGGIRDPSKDEIK-DCHGSFTMVEIFDPKVKDFDINRLQCHLKDIYFPYIQFLI 371
WKTDGG+RDPS++E+K HGSFT VEIF+ + I +LQC LKDIYFPYIQF +
Sbjct: 307 GKNWKTDGGMRDPSEEEMKLSPHGSFTKVEIFESEFDISKIYQLQCRLKDIYFPYIQFCL 366
Query: 372 VVFIQCDELSERGKTITPIKFQVNSVDLTEIQGGEVAITNLHSCNGPEFQFQLRLSFFSG 431
CDELS+ G+T P+ FQVN DL EI GGEVAITNLHS G F FQ+R + F G
Sbjct: 367 ATIFLCDELSKTGRTERPVAFQVNGEDLAEIAGGEVAITNLHS-KGQFFSFQIRFTLFGG 425
Query: 432 SRE--IQVANARLRVVYFPFTKGKENIERILEKLADDGCVIRENFLNFSRVSVRRLGRLL 489
R+ Q ANARL+ VYFP +GKE+IE+IL+ L ++GC + E+F F RVS+RRLGRLL
Sbjct: 426 KRKGTAQEANARLKFVYFPIVQGKESIEKILQSLEEEGCKVSESFQTFGRVSLRRLGRLL 485
Query: 490 PDARWTLLPFMDIRNKKGNRANILKRCSLRVKCFIETDAGFKPTQTKTDLAHHSPLTTAL 549
P+ RW +PFM ++GNRA+ L++ RVKCF++ DAGF PT +KTDLA +P + AL
Sbjct: 486 PEVRWDSIPFM----QRGNRASTLQKSCRRVKCFVDLDAGFSPTPSKTDLASQNPFSVAL 541
Query: 550 KNIGNKISDNQKGILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLE 609
+N G+K ++ +K DV I I ++ K ++ LE
Sbjct: 542 RNFGSKSTEKEK---------------------------DDDVNIVIHREGKSVSYAHLE 574
Query: 610 KEYVEWLLQMHGQYDEEADSGEDQAVIIVNPANKKELRISSDVIRVHKVLKRKEKLWKHG 669
++Y EW+L+MH +DEEA SG D+AV+IV +KK L I D +RVHK ++RKEK WK G
Sbjct: 575 EKYQEWVLEMHNTHDEEAASGLDEAVLIVGSLDKKALGILRDAVRVHKEVRRKEKTWKRG 634
Query: 670 ERIKVQKGACAGCHKNTVFATIEYFLLEGFEGDAGGDSRIICRSMDIPDKNGCLLSVTDE 729
+ IK+ +GA AG H N V+ATI+YFL+EGFE +AGGD+RI+CR +D P+ GC LS+ D
Sbjct: 635 QNIKILRGAYAGIHNNNVYATIDYFLIEGFEDEAGGDTRILCRPIDRPENEGCKLSIIDG 694
Query: 730 NASLEIRGSMSIPIGVIDSG 749
+ LE++ S+S+PI +IDSG
Sbjct: 695 ISKLEVQSSLSLPITIIDSG 714
>At3g26500 unknown protein
Length = 471
Score = 30.0 bits (66), Expect = 4.9
Identities = 21/91 (23%), Positives = 49/91 (53%), Gaps = 3/91 (3%)
Query: 104 NVIDFHNYKPHKCHILQLHGYFDSV*NAMQ-NMWDLTPDIDLLLELPEEYSFQTALA--D 160
++ + +N K H+ ++HG +S+ + + +L+ + + L+ +P+ + T L D
Sbjct: 294 SISEMYNLKYLDAHMNEIHGIPNSIGRLTKLEVLNLSSNFNNLMGVPDTITDLTNLRELD 353
Query: 161 LIDNSLQAVWSNGGNSRKLIRVNVGPDKISI 191
L +N +QA+ + RKL ++N+ + + I
Sbjct: 354 LSNNQIQAIPDSFYRLRKLEKLNLDQNPLEI 384
>At2g32260 putative phospholipid cytidylyltransferase
Length = 332
Score = 30.0 bits (66), Expect = 4.9
Identities = 29/109 (26%), Positives = 49/109 (44%), Gaps = 23/109 (21%)
Query: 569 YTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEYVEWLLQMHGQYDEEAD 628
Y + RF +TQ T + ++D+ + I KD L++ Y
Sbjct: 143 YEFVKKVGRFKETQRTEGISTSDIIMRIVKDYNQYVMRNLDRGY---------------- 186
Query: 629 SGEDQAVIIVNPANKKELRISSDVIRVHKVLKR-KEKLWKHGERIKVQK 676
S ED V V +K LR++ +R+ K+ +R KE+ + GE+I+ K
Sbjct: 187 SREDLGVSFV---KEKRLRVN---MRLKKLQERVKEQQERVGEKIQTVK 229
>At1g14490 hypothetical protein
Length = 206
Score = 30.0 bits (66), Expect = 4.9
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 6/59 (10%)
Query: 1 VSPP--PYVLETDMDGNAKRKLVLRKDDDDTGKAYRFKILLPNGTSVELTLRDPQPQMP 57
+ PP PY+LE + L + GKA F +L +G+ ++TLR P P P
Sbjct: 26 IDPPMSPYILEVPSGNDVVEAL----NRFCRGKAIGFCVLSGSGSVADVTLRQPSPAAP 80
>At5g44030 cellulose synthase catalytic subunit-like protein
Length = 1043
Score = 29.6 bits (65), Expect = 6.5
Identities = 14/41 (34%), Positives = 25/41 (60%)
Query: 179 LIRVNVGPDKISIFDNGPGMDDTDDNSLVKWGKLGASLHRN 219
L + + G KI+ + G DD+DD +K+ + G+S+H+N
Sbjct: 66 LYKRHKGSPKIAGDEENNGPDDSDDELNIKYRQDGSSIHQN 106
>At2g18990 putative ATP binding protein
Length = 211
Score = 29.6 bits (65), Expect = 6.5
Identities = 14/63 (22%), Positives = 31/63 (48%)
Query: 563 ILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEYVEWLLQMHGQ 622
IL TV + +D ++ S K + +E+ ++R++ ++ ++ W+ HG+
Sbjct: 8 ILEKQVLTVAKAMEDKIDDEIASLEKLDEDDLEVLRERRLKQMKKMAEKKKRWISLGHGE 67
Query: 623 YDE 625
Y E
Sbjct: 68 YSE 70
>At1g77930 dnaJ like protein
Length = 271
Score = 29.6 bits (65), Expect = 6.5
Identities = 20/97 (20%), Positives = 47/97 (47%), Gaps = 6/97 (6%)
Query: 573 TLLSRFLDTQLT-SYLKSADVAIEIFKDRKVLTPLQLEKEYVEWLLQMHGQYDEEAD--- 628
T +RF+ Q L ++ ++ D +V P++ + ++EWL++ +D+ D
Sbjct: 119 TAEARFIKIQAAYELLMDSEKKVQYDMDNRV-NPMKASQAWMEWLMKKRKAFDQRGDMAV 177
Query: 629 -SGEDQAVIIVNPANKKELRISSDVIRVHKVLKRKEK 664
+ +Q + +N ++ R D K+L++++K
Sbjct: 178 AAWAEQQQLDINLRARRLSRSKVDPEEERKILEKEKK 214
>At4g03230 putative receptor kinase
Length = 852
Score = 29.3 bits (64), Expect = 8.4
Identities = 25/79 (31%), Positives = 34/79 (42%), Gaps = 14/79 (17%)
Query: 449 FTKGKENIERILEKLADDGCVIRENFLNFSRVSVRRLGRLLPDARWTLLPFMDIRNKKGN 508
F+ G RI K DG V+ + FLN S V V PD+++ D N+K
Sbjct: 328 FSGGCSRESRICGK---DGVVVGDMFLNLSVVEVGS-----PDSQF------DAHNEKEC 373
Query: 509 RANILKRCSLRVKCFIETD 527
RA L C + + E D
Sbjct: 374 RAECLNNCQCQAYSYEEVD 392
>At4g02460 DNA mismatch repair protein (PMS1)
Length = 923
Score = 29.3 bits (64), Expect = 8.4
Identities = 21/68 (30%), Positives = 34/68 (49%), Gaps = 11/68 (16%)
Query: 156 TALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDNGPGMDDTDDNSLVKWGKLGAS 215
+A+ +L++NSL A G S ++ + G D + DNG G+ T+ K+ A
Sbjct: 39 SAVKELVENSLDA----GATSIEINLRDYGEDYFQVIDNGCGISPTN-------FKVLAL 87
Query: 216 LHRNSKLQ 223
H SKL+
Sbjct: 88 KHHTSKLE 95
>At3g03650 unknown protein
Length = 499
Score = 29.3 bits (64), Expect = 8.4
Identities = 28/96 (29%), Positives = 40/96 (41%), Gaps = 16/96 (16%)
Query: 279 LSHNRRASVSSKTKHVKKVYMLHLEREALLNTCSEVKWKTDGGIRDPSKDEIKDCHGSFT 338
LS+NR + V+ K K K L+ + S+ +WKT GG KD + H +
Sbjct: 211 LSYNRFSKVNQKQK---KSQDKELQENVVKYVTSQKEWKTSGG-----KDHVIMAHHPNS 262
Query: 339 MV----EIFDPKVKDFDINRLQCHL----KDIYFPY 366
M ++F D R H+ KDI PY
Sbjct: 263 MSTARHKLFPAMFVVADFGRYSPHVANVDKDIVAPY 298
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,764,324
Number of Sequences: 26719
Number of extensions: 799539
Number of successful extensions: 1689
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1677
Number of HSP's gapped (non-prelim): 10
length of query: 749
length of database: 11,318,596
effective HSP length: 107
effective length of query: 642
effective length of database: 8,459,663
effective search space: 5431103646
effective search space used: 5431103646
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0126.8


