

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0125.11
(904 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
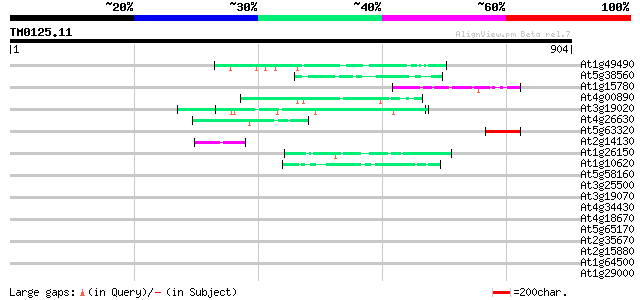
Score E
Sequences producing significant alignments: (bits) Value
At1g49490 hypothetical protein 65 2e-10
At5g38560 putative protein 52 2e-06
At1g15780 unknown protein 49 2e-05
At4g00890 46 1e-04
At3g19020 hypothetical protein 46 1e-04
At4g26630 unknown protein 44 5e-04
At5g63320 putative protein 43 7e-04
At2g14130 putative protein on FARE2.8 43 7e-04
At1g26150 Pto kinase interactor, putative 43 7e-04
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 43 7e-04
At5g58160 strong similarity to unknown protein (gb|AAD23008.1) 43 0.001
At3g25500 formin-like protein AHF1 43 0.001
At3g19070 hypothetical protein 43 0.001
At4g34430 putative protein 42 0.001
At4g18670 extensin-like protein 42 0.001
At5g65170 putative protein 40 0.005
At2g35670 fertilization-independent seed 2 protein (FIS2) 40 0.005
At2g15880 unknown protein 40 0.005
At1g64500 unknown protein 40 0.005
At1g29000 hypothetical protein 40 0.005
>At1g49490 hypothetical protein
Length = 847
Score = 65.1 bits (157), Expect = 2e-10
Identities = 95/415 (22%), Positives = 152/415 (35%), Gaps = 70/415 (16%)
Query: 330 EESSEESDVPLTKKKKADQPKRKHD----------ETSKPDDGNDGDNDGGPSPPKKKKK 379
E+ S + P + K ++P+ KH+ + SKP+D + PK ++
Sbjct: 429 EQPSPKPQPPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEESPKPEQP 488
Query: 380 QVRLVVKPTRVEPAAQ-------------VVRRIETPPRVTRSSV---------RSSSKS 417
Q+ KP AQ RR PP+V + V S
Sbjct: 489 QIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPSP 548
Query: 418 VVDSDADLNS----FDALPISAMLKRSTNPLTLIPESQPAAQTTSQPN----SLRSSFFQ 469
+ ++S + P + P+ P P S P S F
Sbjct: 549 IYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFS 608
Query: 470 PSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTE-PEPRMSDHS 528
P P +P+++ + P SP ++P T +P + P T+ P P
Sbjct: 609 PPP-PSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQ 667
Query: 529 VPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYY 588
VP S +++D S + +P+ PT V S+PS+ + S+ E Y
Sbjct: 668 VPTPS-------SESDQSQILSPVQAPTPVQSSTPSSEPTQV--------PTPSSSESYQ 712
Query: 589 LTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNH 648
SP + P P + + P E++ QA P P+ + P P N
Sbjct: 713 APNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQA-PTPILEPVHAPTP-----NS 766
Query: 649 SSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVP 703
V+SP P +SEP S P Q+ ++ +P+ T SS +SP T+ SI P
Sbjct: 767 KPVQSPTP---SSEP---VSSPE-QSEEVEAPEPTPVNPSSVPSSSPSTDTSIPP 814
Score = 37.7 bits (86), Expect = 0.029
Identities = 65/265 (24%), Positives = 94/265 (34%), Gaps = 52/265 (19%)
Query: 456 TTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIP 515
++ PN R+S +PS E + PSD + P T + + P P+ P
Sbjct: 392 SSPSPNPPRTSEPKPSKPEPVM--------PKPSDSSKPETPKTPEQPSPKPQPPKHESP 443
Query: 516 VQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFME 575
EPE + H +P+ E +P P+ DS K E
Sbjct: 444 KPEEPE---NKHELPKQKE---------------SPKPQPSKPEDSPKPEQP----KPEE 481
Query: 576 VRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQE 635
K + + E P GP P+ DP ++A P+ + P P + E
Sbjct: 482 SPKPEQPQIPEPTKPVSPPNEAQGPTPD---DP---------YDASPVKNRRSPPPPKVE 529
Query: 636 -----PVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGAS--EPHVQT--CDIGSPQGTSEA 686
P QP P S S S + SP P V + P + +S PHV + + SP S
Sbjct: 530 DTRVPPPQP-PMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPP 588
Query: 687 HSSNHPTSPETNLSIVPYTSLRPTS 711
+ P P P S P S
Sbjct: 589 PPVHSPPPPPVFSPPPPVFSPPPPS 613
Score = 36.2 bits (82), Expect = 0.085
Identities = 44/195 (22%), Positives = 71/195 (35%), Gaps = 24/195 (12%)
Query: 516 VQTEPEPRMSDHSVPRASERLVSRTTDTD-------SSSVYTPISFPTNVADSSPSNNSE 568
+Q P + + +P +VSR D S+ +P P ++ PS
Sbjct: 357 LQNRPSQKPAKQCLP-----VVSRPVDCSKDKCSGGSNGGSSPSPNPPRTSEPKPSKPEP 411
Query: 569 SIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQ 628
+ K + K + E P P ++ P+PE + E P + P Q ++
Sbjct: 412 VMPKPSDSSKPETPKTPEQPSPKPQPPKHESPKPEEPENKHE----LPKQKESPKPQPSK 467
Query: 629 PDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHS 688
P+ +P QP PE+S + P P S P +E T D P S +
Sbjct: 468 PED-SPKPEQPKPEES-PKPEQPQIPEPTKPVSPP----NEAQGPTPD--DPYDASPVKN 519
Query: 689 SNHPTSPETNLSIVP 703
P P+ + VP
Sbjct: 520 RRSPPPPKVEDTRVP 534
>At5g38560 putative protein
Length = 681
Score = 51.6 bits (122), Expect = 2e-06
Identities = 61/240 (25%), Positives = 80/240 (32%), Gaps = 62/240 (25%)
Query: 460 PNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTE 519
P S SS P P LQ QPT PS P P+S P + PV +
Sbjct: 12 PPSSNSSTTAPPP--------LQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSS 63
Query: 520 PEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKE 579
P P S P S +++ T +SS P+ +A PS +
Sbjct: 64 PPPSSS----PPPSPPVITSPPPTVASSPPPPVV----IASPPPSTPAT----------- 104
Query: 580 KVSTLEEYYLTCPSPRRY--PGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPV 637
T P+P + P P P+ P P NP + P P P + P
Sbjct: 105 ----------TPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPS 154
Query: 638 QPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPET 697
P P S +P P TS P P TS + S++PT P T
Sbjct: 155 PPKPSPS--------TPTPTTTTSPP---------------PPPATSASPPSSNPTDPST 191
>At1g15780 unknown protein
Length = 1335
Score = 48.5 bits (114), Expect = 2e-05
Identities = 55/214 (25%), Positives = 99/214 (45%), Gaps = 27/214 (12%)
Query: 617 LHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCD 676
+H QQ Q Q +P+Q Q+V + S NP + + + + P Q
Sbjct: 682 MHRPRKPVQQGQLPQSQMQPMQQPQSQTVQDQSHDNQTNPQMQSMS--MQGAGPRAQQSS 739
Query: 677 IGSPQGTSEAHSSNHPTSPETNL-SIVPYTSLRPTSLSECINVFNREASLMLRNVQGQTD 735
+ + Q ++ S ++P+ N+ S +P +SL S N N + + ++Q T
Sbjct: 740 MTNMQ-SNVLSSRPGVSAPQQNIPSSIPASSLE----SGQGNTLNNGQQVAMGSMQQNTS 794
Query: 736 LSENAESVAEKWNSLSTWL-------VAQVPVMMQHLTAERDQRIKAAKQRFARRVALHE 788
N S + + + LST ++ + QHL ++DQ+++ KQ+F +R
Sbjct: 795 QLVNNSSASAQ-SGLSTLQSNVNQPQLSSSLLQHQHLKQQQDQQMQL-KQQFQQR----- 847
Query: 789 QQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQ 822
Q Q+ +L +AR++Q+Q + ARQ AAQ +Q
Sbjct: 848 QMQQQQL-----QARQQQQQQQLQARQQAAQLQQ 876
>At4g00890
Length = 431
Score = 45.8 bits (107), Expect = 1e-04
Identities = 72/319 (22%), Positives = 127/319 (39%), Gaps = 46/319 (14%)
Query: 372 SPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSS--VRSSSKSVVDSDADLNSFD 429
SPP+ K + + ++ EP +Q + + + T + ++S KS +S F
Sbjct: 53 SPPEPKSLPLSPLSPKSKEEPESQTMTPLMSKHVKTHRNLLIQSPPKSPPESTESQQQFL 112
Query: 430 A-LPISAMLKRSTNPLTLIPESQPAAQTTSQPN----SLRSSFFQPSP---------NEA 475
A +P+ + PL+ + ++ SQP+ S+ ++F+PSP +
Sbjct: 113 ASVPLRPLTTEPKTPLSPSSTLKATEESQSQPSPPLESIVETWFRPSPPTSTETGDETQL 172
Query: 476 PLWNMLQNQPTGPSDPTSPITIQYNPESEPLT-LEPELTIPVQTEPEPRMSDHSVPRASE 534
P+ Q T PS P+ + ES+P L P +I P MS S P
Sbjct: 173 PI-PPPQEAKTPPSSPSMMLNATEEFESQPKPPLLPSKSIDETRLRSPLMSQASSPPP-- 229
Query: 535 RLVSRTTD-TDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPS 593
L S++ D ++ S PIS P + + +S + K + + + PS
Sbjct: 230 -LPSKSIDENETRSQSPPISPPKSDKQARSQTHSSPSPPPLLSPKASENHQSKSPMPPPS 288
Query: 594 PR--------RYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSV 645
P + P P P + P P ++PL + P + + P Q +P Q
Sbjct: 289 PTAQISLSSLKSPIPSPATITAPPPPF-SSPLSQTTPSPKPSLP--------QIEPNQ-- 337
Query: 646 SNHSSVRSPNPLVDTSEPH 664
++SP+P + +E H
Sbjct: 338 -----IKSPSPKLTNTESH 351
>At3g19020 hypothetical protein
Length = 951
Score = 45.8 bits (107), Expect = 1e-04
Identities = 80/386 (20%), Positives = 128/386 (32%), Gaps = 52/386 (13%)
Query: 332 SSEESDVPLTKKKKADQPKRKHDE-----TSKPDD--------GNDGDNDGGPSP-PKKK 377
SS+E T ++P +K + S+P D G G ++ P P P K
Sbjct: 352 SSQEKQFDDTSNCLQNRPNQKSAKECLPVVSRPVDCSKDKCAGGGGGGSNPSPKPTPTPK 411
Query: 378 KKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAML 437
+ + + P +E ++ P + + S K S N + P S
Sbjct: 412 APEPKKEINPPNLEEPSK------PKPEESPKPQQPSPKPETPSHEPSNPKEPKPESP-- 463
Query: 438 KRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPN-EAPLWNMLQNQPTGPSD------ 490
+ +P T P+ +P + P QP P E+P + +P P +
Sbjct: 464 -KQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEP 522
Query: 491 --------PTSPITIQYNPESEPLTLEPELTIPVQTE-PEPRMSDHSVPRASERLVSRTT 541
P P PE P P+ P E P+P+ P+ E +
Sbjct: 523 PKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPP 582
Query: 542 DTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVS-TLEEYYLTCP-SPRRYPG 599
+ + +S N+ + R + S + EE T P SP +
Sbjct: 583 KQEQPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSP 642
Query: 600 PRPERLVDPDEPILANP---------LHEADPLAQQAQPDPVQQEP--VQPDPEQSVSNH 648
P P + P P+ + P ++ P P PV P V P S
Sbjct: 643 PPPPPVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPP 702
Query: 649 SSVRSPNPLVDTSEPHLGASEPHVQT 674
V SP P V + P + + P VQ+
Sbjct: 703 PPVHSPPPPVHSPPPPVHSPPPPVQS 728
Score = 43.5 bits (101), Expect = 5e-04
Identities = 84/410 (20%), Positives = 132/410 (31%), Gaps = 50/410 (12%)
Query: 271 SNLVQDMVNAGCVEDLNTIVSDVFTANSLKKMGLVQKKIAPAHEDTSSKIRQRRRKMVLE 330
SN +Q+ N ++ +VS + K G P+ + T + +K +
Sbjct: 362 SNCLQNRPNQKSAKECLPVVSRPVDCSKDKCAGGGGGGSNPSPKPTPTPKAPEPKKEINP 421
Query: 331 ESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKK---KKKQVRLVVKP 387
+ EE P K +++ +P++ + P + P PK+ K +Q + +
Sbjct: 422 PNLEEPSKP--KPEESPKPQQPSPKPETPSHEPSNPKEPKPESPKQESPKTEQPKPKPES 479
Query: 388 TRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDA-----LPISAMLKRSTN 442
+ E Q + E P S + SSK + + P K+ T
Sbjct: 480 PKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETP 539
Query: 443 PLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPE 502
P+ QP Q T +P S QP E P +P P P E
Sbjct: 540 KPEESPKPQPPKQETPKPEE--SPKPQPPKQETP-------KPEESPKPQPP-----KQE 585
Query: 503 SEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSS 562
P T P++ P P P + P R + T+ + +P S P +
Sbjct: 586 QPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPP 645
Query: 563 PSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADP 622
P +S P P + P P P P+H P
Sbjct: 646 PPVHS------------------------PPPPVFSPPPPMHSPPPPVYSPPPPVHSPPP 681
Query: 623 LAQQAQPDPVQQ--EPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEP 670
+ P PV PV P S V SP P V + P + + P
Sbjct: 682 PPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQSPPP 731
Score = 42.0 bits (97), Expect = 0.002
Identities = 97/448 (21%), Positives = 141/448 (30%), Gaps = 56/448 (12%)
Query: 330 EESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTR 389
E S E P K+ K + PK++ +T +P + P K +Q + + +
Sbjct: 446 ETPSHEPSNP--KEPKPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPK 503
Query: 390 VEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPE 449
E + Q + E P+ S K ++ P K+ T P+
Sbjct: 504 QESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEES-PKPQPPKQETPKPEESPK 562
Query: 450 SQPAAQTTSQPNSLRS----SFFQPSPNEAPLWNMLQNQPTGPSDP--TSPITIQYNPES 503
QP Q T +P QP EAP + P+DP SPI +
Sbjct: 563 PQPPKQETPKPEESPKPQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPP 622
Query: 504 EPLTLEPELTIPVQT---EPEPRMSDHSVPRA--SERLVSRTTDTDSSSVYTPISFPTNV 558
P T E + T P P P HS P S + S P+ P
Sbjct: 623 SPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPP 682
Query: 559 ADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLH 618
SP S P P + P P P P+H
Sbjct: 683 PVHSPPPPVHS----------------------PPPPVHSPPPPVHSPPPPVHSPPPPVH 720
Query: 619 EADPLAQQAQPDPVQQEPV-------------QPDPEQSVSNHSSVRSPNPLVDTSEPHL 665
P Q P PV P P P V SP P V + P +
Sbjct: 721 SPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPV 780
Query: 666 GASEP--HVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPTS--LSECINVFNR 721
+ P H + SP S +S P P + P T L P + ++ +
Sbjct: 781 HSPPPPVHSPPPPVHSPPPPSPIYS---PPPPVFSPPPKPVTPLPPATSPMANAPTPSSS 837
Query: 722 EASLMLRNVQGQTDLSENAESVAEKWNS 749
E+ + VQ T SE+ E+ ++ +S
Sbjct: 838 ESGEISTPVQAPTPDSEDIEAPSDSNHS 865
Score = 40.8 bits (94), Expect = 0.003
Identities = 97/454 (21%), Positives = 153/454 (33%), Gaps = 76/454 (16%)
Query: 307 KKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQ---PKRKHDETSKPDDGN 363
K+ AP E K +++ +E + + P + K ++ P+ ET KP++
Sbjct: 486 KQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESP 545
Query: 364 DGDNDGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSV----- 418
P PPK++ + KP +P Q + E P+ K+
Sbjct: 546 K------PQPPKQETPKPEESPKP---QPPKQETPKPEESPKPQPPKQEQPPKTEAPKMG 596
Query: 419 ---VDSDADLNSFDALPISAMLKRSTNPLT-----LIPESQPAAQTTSQP---NSLRSSF 467
++S + +DA PI + +P T P+S P P + F
Sbjct: 597 SPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVF 656
Query: 468 FQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSD- 526
P P +P + P S P P+ P P PV + P P S
Sbjct: 657 SPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPP 716
Query: 527 ---HSVPRASERLVSRT--TDTDSSSVYTP----ISFPTNVADSSPSNNSESIRKFMEVR 577
HS P + + + +Y+P + P S P S +
Sbjct: 717 PPVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSP 776
Query: 578 KEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPV 637
V + P P + P P P PI + P P + P P P+
Sbjct: 777 PPPVHS--------PPPPVHSPPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPM 828
Query: 638 ---------------------QPDPEQ----SVSNHSSV--RSPNPLVDTSEPHLGASEP 670
PD E S SNHS V SP P D SEP + A P
Sbjct: 829 ANAPTPSSSESGEISTPVQAPTPDSEDIEAPSDSNHSPVFKSSPAPSPD-SEPEVEAPVP 887
Query: 671 HVQTCDIGSPQGTSEAHSSNHPTS-PETNLSIVP 703
+ ++ +P+ + SS+ P+S P +++ P
Sbjct: 888 SSEP-EVEAPKQSEATPSSSPPSSNPSPDVTAPP 920
Score = 33.5 bits (75), Expect = 0.55
Identities = 35/133 (26%), Positives = 49/133 (36%), Gaps = 10/133 (7%)
Query: 483 NQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDH---SVPRASERLVSR 539
+ P PS SP ++P +P+T P T P+ P P S+ S P + S
Sbjct: 795 HSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSE 854
Query: 540 TTDTDSSSVYTPI--SFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRY 597
+ S S ++P+ S P DS P + EV K S T S
Sbjct: 855 DIEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEPEVEAPKQSE-----ATPSSSPPS 909
Query: 598 PGPRPERLVDPDE 610
P P+ P E
Sbjct: 910 SNPSPDVTAPPSE 922
>At4g26630 unknown protein
Length = 763
Score = 43.5 bits (101), Expect = 5e-04
Identities = 45/195 (23%), Positives = 77/195 (39%), Gaps = 14/195 (7%)
Query: 295 TANSLKKMGLVQK--KIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRK 352
+A S KK K K + AH D S+ + + EE +EE + ++ + P +
Sbjct: 498 SAKSQKKSEEATKVVKKSLAHSDDESEEEKEEEEKQEEEKAEEKEEKKEEENENGIPDKS 557
Query: 353 HDETSKPDDGNDGDNDGGPSPPK--KKKKQVRLVV----KPTRVEPAAQVVRRIETPPRV 406
DE +P + + D S + KKK+ RL R VV +PP
Sbjct: 558 EDEAPQPSESEEKDESEEHSEEETTKKKRGSRLSAGKKESAGRARNKKAVVAAKSSPPEK 617
Query: 407 TRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSS 466
S+ + D D+D + S+ K+S NP+ P P+ + + R+
Sbjct: 618 ITQKRSSAKRKKTDDDSDTSP----KASSKRKKSENPIKASP--APSKSASKEKPVKRAG 671
Query: 467 FFQPSPNEAPLWNML 481
+ P++ L N +
Sbjct: 672 KGKDKPSDKVLKNAI 686
>At5g63320 putative protein
Length = 569
Score = 43.1 bits (100), Expect = 7e-04
Identities = 22/55 (40%), Positives = 34/55 (61%)
Query: 768 ERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQ 822
E ++R++ K+R E+ +R EAAE+ARR++EQ EAARQA + E+
Sbjct: 105 EFEKRLREEKERLQAEAKAAEEARRKAKAEAAEKARREREQEREAARQALQKMEK 159
>At2g14130 putative protein on FARE2.8
Length = 789
Score = 43.1 bits (100), Expect = 7e-04
Identities = 30/85 (35%), Positives = 42/85 (49%), Gaps = 4/85 (4%)
Query: 298 SLKKMGLVQKKIAPA--HEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDE 355
SL M L K++ P+ ED +Q RK + S + DVP +K+A KRK
Sbjct: 482 SLSFMQLQNKRVKPSVHAEDNLKTRKQVPRKRQKQVDSADVDVPT--RKEAKSKKRKIIG 539
Query: 356 TSKPDDGNDGDNDGGPSPPKKKKKQ 380
+D NDGDND P++K K+
Sbjct: 540 NDGDNDDNDGDNDDFQPAPQRKSKR 564
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 43.1 bits (100), Expect = 7e-04
Identities = 66/276 (23%), Positives = 90/276 (31%), Gaps = 28/276 (10%)
Query: 443 PLTLIPESQPA--AQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYN 500
PL +P QP+ + P ++ P P Q+ P P P S + +
Sbjct: 22 PLMALPPPQPSFPGDNATSPTREPTNGNPPETTNTPA----QSSPP-PETPLSSPPPEPS 76
Query: 501 PESEPLTLEPELTIPVQTEPEPR----MSDHSVPRASERLVSRTTDTDSSSVYTPISFPT 556
P S LT P TIPV PEP + + P A+ VS S P P
Sbjct: 77 PPSPSLTGPPPTTIPVSPPPEPSPPPPLPTEAPPPANP--VSSPPPESSPPPPPPTEAPP 134
Query: 557 NVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANP 616
+SPS + L P P P P P +LV P + P
Sbjct: 135 TTPITSPSPPT----------NPPPPPESPPSLPAPDPPSNPLP-PPKLVPPSH---SPP 180
Query: 617 LHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCD 676
H P A + P P + P P E+ + S P+P P
Sbjct: 181 RHLPSPPASEIPPPP-RHLPSPPASERPSTPPSDSEHPSPPPPGHPKRREQPPPPGSKRP 239
Query: 677 IGSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPTSL 712
SP S++ HP+ P +P P L
Sbjct: 240 TPSPPSPSDSKRPVHPSPPSPPEETLPPPKPSPDPL 275
Score = 39.3 bits (90), Expect = 0.010
Identities = 65/243 (26%), Positives = 89/243 (35%), Gaps = 24/243 (9%)
Query: 431 LPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSD 490
+P+S + S P L E+ P A S P SS P P EAP + T PS
Sbjct: 90 IPVSPPPEPSPPP-PLPTEAPPPANPVSSPPP-ESSPPPPPPTEAPPTTPI----TSPSP 143
Query: 491 PTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYT 550
PT+P +P S P P +P P+ HS PR + + +
Sbjct: 144 PTNPPPPPESPPSLPAPDPPSNPLP---PPKLVPPSHSPPRHLPSPPASEIPPPPRHLPS 200
Query: 551 PISFPTNVADSSPSNNSESIRK---FMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVD 607
P P + S+P ++SE R+E+ T P RP
Sbjct: 201 P---PASERPSTPPSDSEHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPSDSKRPVHPSP 257
Query: 608 PDEP--ILANPLHEADPLAQQAQPDPVQQEP---VQP--DPEQSVS--NHSSVRSPNPLV 658
P P L P DPL + P P V P P +SVS ++ S +P P+
Sbjct: 258 PSPPEETLPPPKPSPDPLPSNSSSPPTLLPPSSVVSPPSPPRKSVSGPDNPSPNNPTPVT 317
Query: 659 DTS 661
D S
Sbjct: 318 DNS 320
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 43.1 bits (100), Expect = 7e-04
Identities = 59/258 (22%), Positives = 95/258 (35%), Gaps = 42/258 (16%)
Query: 440 STNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQY 499
+T P P S P+ T + P ++ QP PN+ P N P+ P+SP
Sbjct: 40 TTQPPATSPPSPPSPDTQTSPPPATAA--QPPPNQPP------NTTPPPTPPSSP----- 86
Query: 500 NPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLV--SRTTDTDSSSVYTPISFPTN 557
P P + P+P+ S P +V S++ P S
Sbjct: 87 ----------PPSITPPPSPPQPQPPPQSTPTGDSPVVIPFPKPQLPPPSLFPPPSLVNQ 136
Query: 558 VADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPL 617
+ D P++N+ +E +S L P + P E P L++ L
Sbjct: 137 LPDPRPNDNN-----ILEPINNPIS------LPSPPSTPFSPPSQENSGSQGSPPLSSLL 185
Query: 618 HEADPLAQQAQPDPVQ--QEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTC 675
PL + +P+Q P+ + + V + SS SP P + S H G S H
Sbjct: 186 PPMLPLNPNSPGNPLQPLDSPLGGESNR-VPSSSSSPSP-PSLSGSNNHSGGSNRH--NA 241
Query: 676 DIGSPQGTSEAHSSNHPT 693
+ GTS+ + ++ T
Sbjct: 242 NSNGDGGTSQQSNESNYT 259
>At5g58160 strong similarity to unknown protein (gb|AAD23008.1)
Length = 1307
Score = 42.7 bits (99), Expect = 0.001
Identities = 63/279 (22%), Positives = 103/279 (36%), Gaps = 35/279 (12%)
Query: 432 PISAMLKRSTNPL--TLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPS 489
P+ A + + L +++ ++P +Q S SL S FQ PNE N++ PT P
Sbjct: 553 PLPAAASKPSEQLQHSVVQATEPLSQGNSWM-SLAGSTFQTVPNEK---NLITLPPTPPL 608
Query: 490 DPTSPITIQYNPES-EPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSV 548
TS + + + ++ L L P+ + T P +S A+ + +D +S++
Sbjct: 609 ASTSHASPEPSSKTTNSLLLSPQASPATPTNPSKTVSVDFFGAATSPHLG-ASDNVASNL 667
Query: 549 YTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVD- 607
P P +++S ++ L P P P P V
Sbjct: 668 GQPARSPPPISNSD----------------------KKPALPRPPPPPPPPPMQHSTVTK 705
Query: 608 -PDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHL- 665
P P A P P+ + P P P P P SN S +P + P L
Sbjct: 706 VPPPPPPAPPAPPT-PIVHTSSPPPPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPPRLP 764
Query: 666 -GASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVP 703
++ P T P G + A S+ P P+ + P
Sbjct: 765 THSASPPPPTAPPPPPLGQTRAPSAPPPPPPKLGTKLSP 803
>At3g25500 formin-like protein AHF1
Length = 1051
Score = 42.7 bits (99), Expect = 0.001
Identities = 72/314 (22%), Positives = 118/314 (36%), Gaps = 39/314 (12%)
Query: 372 SPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDAL 431
SP + + + P R EP V+ E VRS S S+ + L + D +
Sbjct: 293 SPGRSTFISISPSMSPKRSEPKPPVISTPEPAELTDYRFVRSPSLSLASLSSGLKNSDEV 352
Query: 432 PISAMLKRST-NPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSD 490
++ + + T LT PE+ + + P S S+ + PN+ P P+ S
Sbjct: 353 GLNQIFRSPTVTSLTTSPENN---KKENSPLSSTSTSPERRPNDTP--EAYLRSPSHSSA 407
Query: 491 PTSPI-TIQYNPESEPL---TLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSS 546
TSP Q +PE P L L + + P + + L SR+ + SS
Sbjct: 408 STSPYRCFQKSPEVLPAFMSNLRQGLQSQLLSSPSNSHGGQGFLKQLDALRSRSPSSSSS 467
Query: 547 SV---------YTPISFPTNVADSSPSNNSESIRKF---MEVRKEKVSTLEEYYLTCPSP 594
SV +P++ P + +S S +S R F ++V ++S + L P
Sbjct: 468 SVCSSPEKASHKSPVTSPKLSSRNSQSLSSSPDRDFSHSLDV-SPRISNISPQILQSRVP 526
Query: 595 RRYPGPRPERL------------VDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPE 642
P P P L P L P H P ++ PV P++ PE
Sbjct: 527 PPPPPPPPLPLWGRRSQVTTKADTISRPPSLTPPSH---PFVIPSENLPVTSSPME-TPE 582
Query: 643 QSVSNHSSVRSPNP 656
++ ++ +P P
Sbjct: 583 TVCASEAAEETPKP 596
>At3g19070 hypothetical protein
Length = 346
Score = 42.7 bits (99), Expect = 0.001
Identities = 57/285 (20%), Positives = 107/285 (37%), Gaps = 33/285 (11%)
Query: 345 KADQPKRKHDETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPP 404
K D+ +K DE +K D+ D K Q R + + + + PP
Sbjct: 23 KIDEHLKKLDEETKEDEDEDA----------AKIIQDRSTAREAEIALLMDLRQNFLPPP 72
Query: 405 RVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTS---QPN 461
++ SS++ S S + + + P+ + S + P S A ++S QP
Sbjct: 73 SMSSSSLQPPSSSTLAPSSSSSLQPPAPLIDFFRSSVSYSHQPPSSSTLATSSSPSLQPP 132
Query: 462 SLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPE 521
S+ SS QP + + + PS ++ T + P S P ++ P +
Sbjct: 133 SMSSSSLQPPASLREFFTSSVSYSHQPSSSSTLATSSFFPSSMPYSVRPP-----DSSDR 187
Query: 522 PRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKV 581
P + + P + + + S FP+ ++ SSP S + F+ + ++
Sbjct: 188 PSLREF-FPSSPSSSIQPPESSSSKRARLSNIFPSPLS-SSP---SPFVNPFLRPQAQE- 241
Query: 582 STLEEYYLTCPS-----PRRYPGPRPERLVDPDEPILANPLHEAD 621
T+ + P P +P P P +P+ + NP+ D
Sbjct: 242 PTIPNFINPIPQISPGLPALFPNPNP----NPNPNPIPNPIRNPD 282
>At4g34430 putative protein
Length = 827
Score = 42.4 bits (98), Expect = 0.001
Identities = 104/589 (17%), Positives = 208/589 (34%), Gaps = 70/589 (11%)
Query: 311 PAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDETSKPDDGND---GDN 367
P +DT+ + VL+++ EE++ KK+ D+ + E +P+DGN+
Sbjct: 233 PISKDTTDLAVSKDDNSVLKDAPEEAE----NKKRVDEDETMK-EVPEPEDGNEEKVSQE 287
Query: 368 DGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNS 427
P ++ ++ K ++E A + + E + ++ + + V S S
Sbjct: 288 SSKPGDASEETNEMEAEQKTPKLETAIEERCKDEADENIALKALTEAFEDVGHSSTPEAS 347
Query: 428 FDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWN-----MLQ 482
F + + L + S A S R+S N L +L+
Sbjct: 348 FSFADLGNPVMGLAAFLVRLAGSDVATA------SARASIKSLHSNSGMLLATRHCYILE 401
Query: 483 NQPTGPSDPTSPITI--QYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRT 540
+ P DPT + + N ++ +PE E D +P
Sbjct: 402 DPPDNKKDPTKSKSADAEGNDDNSHKDDQPEEKSKKAEEVSLNSDDREMP---------- 451
Query: 541 TDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYY----LTCPSPRR 596
DTD+ S N++ + E R K T + + CPS +
Sbjct: 452 -DTDTGKETQDSVSEEKQPGSRTENSTTKLDAVQEKRSSKPVTTDNSEKPVDIICPSQDK 510
Query: 597 YPGPRPERLVDPDEPIL-ANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRS-- 653
G + EP+ N L + A Q+ + QP+ + V +++S
Sbjct: 511 CSGKELQ------EPLKDGNKLSSENKDASQSTVSQSAADASQPEASRDVEMKDTLQSEK 564
Query: 654 -PNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPT-- 710
P +V T + ++ + +P + ++PE + + SL T
Sbjct: 565 DPEDVVKTVGEKVQLAKEEGANDVLSTPDKSVSQQPIGSASAPENGTAGSLFLSLSDTLI 624
Query: 711 ---SLSECINVFNREASLMLR----NVQG--QTDLSENAESV--AEKWNSLSTWLVAQVP 759
+L + + ++ N++G + D+ E + EK + ++
Sbjct: 625 TLPTLHDSFISYRFVIEMLFAGGNPNIEGKKEKDICEGTKDKYNIEKLKRAAISAISAAA 684
Query: 760 VMMQHLTAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQ 819
V ++L + + +I+ + LH+ + + + AE + + E +RQ
Sbjct: 685 VKAKNLAKQEEDQIRQLSGSLIEKQQLHKLEAKLSIFNEAESLTMRVREQLERSRQ---- 740
Query: 820 AEQARLEAERLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPSNSTPP 868
RL ER + A + P + ++ AS P ++ A N + + P
Sbjct: 741 ----RLYHERAQIIAARLGVPP---SMSSKASLPTNRIAANFANVAQRP 782
>At4g18670 extensin-like protein
Length = 839
Score = 42.4 bits (98), Expect = 0.001
Identities = 64/242 (26%), Positives = 85/242 (34%), Gaps = 41/242 (16%)
Query: 410 SVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPN-SLRSSFF 468
S+ S V S S P S + S T P S P + TT P S SS
Sbjct: 427 SISPSPPITVPSPPTTPSPGGSPPSPSIVPSPPSTTPSPGSPPTSPTTPTPGGSPPSSPT 486
Query: 469 QPSPNEAPLWNMLQNQPTGPSDPTSPITIQ--YNPESEPLTLEPELTIPVQTEPEPRMSD 526
P+P +P + P G S P+SP T +P S ++ P +T+P S
Sbjct: 487 TPTPGGSPPSSPTTPTPGG-SPPSSPTTPSPGGSPPSPSISPSPPITVP---------SP 536
Query: 527 HSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEE 586
S P + S ++ T SS + +P + T SP NS I +
Sbjct: 537 PSTPTSPGSPPSPSSPTPSSPIPSPPTPSTPPTPISPGQNSPPI-------------IPS 583
Query: 587 YYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVS 646
T PSP P P P V P PI+ P P P P P +
Sbjct: 584 PPFTGPSPPSSPSP-PLPPVIPSPPIVG--------------PTPSSPPPSTPTPGTLLH 628
Query: 647 NH 648
H
Sbjct: 629 PH 630
>At5g65170 putative protein
Length = 362
Score = 40.4 bits (93), Expect = 0.005
Identities = 63/262 (24%), Positives = 107/262 (40%), Gaps = 42/262 (16%)
Query: 409 SSVRSSSKSVVDSDAD-LNSFDALPISAMLKRSTN---PLTLIPESQPAAQTTSQPNSLR 464
SS++SSS D + +S ISA+ + N P T+ P +Q ++L
Sbjct: 7 SSMQSSSGGGGGGDQEEYDSRADQSISALFNHNNNNNNPTTVSPN----IAVPTQLDALI 62
Query: 465 SSFFQPSPN-EAPLWNMLQNQPTGPSDPTSP----------ITIQYNPESEPLTLEPELT 513
+++F S + + PLW+ QPT S P P +Q N + P T
Sbjct: 63 ANYFNTSWSADNPLWSTTATQPTDGSRPVPPPISSEQVFFTNPLQQNLRTVPNTNTTSPI 122
Query: 514 IPVQTEPEPRMSDHSVPRASERLVSR------TTDTDS------------SSVYTPI--S 553
V T+ + ++ P+ R+ R TTDT + S+ +T + S
Sbjct: 123 CSVPTDKKNGLATTRNPKKRSRVSRRAPTTVLTTDTSNFRAMVQEFTGNPSTPFTGLSSS 182
Query: 554 FPTNVADSSPSNNSESIRKFMEVRKEKV--STLEEYYLTCPSPRRYPGPRPERLVDPDEP 611
FP + D S++S S R + + STL +YL S + L++ + P
Sbjct: 183 FPRSRFDLFGSSSSSSSRPMKPFPHKLISPSTLNHHYLPPSSEYHHHHQHQNLLLNMNTP 242
Query: 612 ILANPLHEADPLAQQAQPDPVQ 633
++NP + L Q +P ++
Sbjct: 243 NISNPFLN-NSLTDQPKPSSLR 263
>At2g35670 fertilization-independent seed 2 protein (FIS2)
Length = 488
Score = 40.4 bits (93), Expect = 0.005
Identities = 60/245 (24%), Positives = 95/245 (38%), Gaps = 29/245 (11%)
Query: 310 APAHEDTSSKIRQ--RRRKMVLEESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDN 367
+P +S KI ++ + ESSE VP P R H K + + D+
Sbjct: 130 SPPRAHSSEKISDILTTTQLAIAESSEPK-VPHVNDGNVSSPPRAHSSAEKNESTHVNDD 188
Query: 368 DGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNS 427
D SPP+ + + T V I +PP+ SS ++ S + D D
Sbjct: 189 DDVSSPPRAHSLEKN---ESTHVNE-----DNISSPPK-AHSSKKNESTHMNDEDVSFP- 238
Query: 428 FDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTG 487
P + K +++ LT +QPA S+P R S + A + + QP
Sbjct: 239 ----PRTRSSKETSDILT---TTQPAIVEPSEPKVRRVS--RRKQLYAKRYKARETQP-A 288
Query: 488 PSDPTSPITIQYNPES-----EPLTLEPELTIPVQTEPE-PRMSDHSVPRASERLVSRTT 541
++ + P + N E+ E +LE I T+P S+ VP ++ VS T
Sbjct: 289 IAESSEPKVLHVNDENVSSPPEAHSLEKASDILTTTQPAIAESSEPKVPHVNDENVSSTP 348
Query: 542 DTDSS 546
SS
Sbjct: 349 RAHSS 353
Score = 32.7 bits (73), Expect = 0.94
Identities = 44/182 (24%), Positives = 70/182 (38%), Gaps = 13/182 (7%)
Query: 528 SVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEY 587
S PRA + +T + ++ +P ++ + S N E + R K ++
Sbjct: 193 SPPRAHSLEKNESTHVNEDNISSPPKAHSSKKNESTHMNDEDVSFPPRTRSSKETS---D 249
Query: 588 YLTCPSPRRYPGPRPE-RLVDPDEPILAN--PLHEADP-LAQQAQPDP--VQQEPVQPDP 641
LT P P+ R V + + A E P +A+ ++P V E V P
Sbjct: 250 ILTTTQPAIVEPSEPKVRRVSRRKQLYAKRYKARETQPAIAESSEPKVLHVNDENVSSPP 309
Query: 642 EQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSI 701
E HS ++ + L T +SEP V + + T AHSS S N+
Sbjct: 310 EA----HSLEKASDILTTTQPAIAESSEPKVPHVNDENVSSTPRAHSSKKNKSTRKNVDN 365
Query: 702 VP 703
VP
Sbjct: 366 VP 367
Score = 32.0 bits (71), Expect = 1.6
Identities = 39/149 (26%), Positives = 57/149 (38%), Gaps = 20/149 (13%)
Query: 290 VSDVFTANSLKKMGLVQKKI--APAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKAD 347
VS A+S KK +K + P+ T S + + + ES P + D
Sbjct: 344 VSSTPRAHSSKKNKSTRKNVDNVPSPPKTRSSKKTSDILTTTQPTIAESSEPKVRHVNDD 403
Query: 348 QPK---RKHDETSKPDDGNDGDNDGGPSPPK----KKKKQVRLVVKPTRVEPAAQVVRRI 400
R H +SK + ++D PSPPK KK + +P + EP
Sbjct: 404 NVSSTPRAH--SSKKNKSTRKNDDNIPSPPKTRSSKKTSNILATTQPAKAEP-------- 453
Query: 401 ETPPRVTRSSVRSSSKSVVDSDADLNSFD 429
+ P+VTR S R + L SFD
Sbjct: 454 -SEPKVTRVSRRKELHAERCEAKRLFSFD 481
Score = 30.0 bits (66), Expect = 6.1
Identities = 52/260 (20%), Positives = 92/260 (35%), Gaps = 15/260 (5%)
Query: 648 HSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSS---NHPT--SPETNLSIV 702
HSS + + L T +SEP V + G+ AHSS N T + + ++S
Sbjct: 135 HSSEKISDILTTTQLAIAESSEPKVPHVNDGNVSSPPRAHSSAEKNESTHVNDDDDVSSP 194
Query: 703 PYTSLRPTSLSECINVFNREASLMLRNVQGQTDLSENAESVAEKWNSLSTWLVAQVPVMM 762
P + S +N N + + + N E V+ + S+ + +
Sbjct: 195 PRAHSLEKNESTHVNEDNISSPPKAHSSKKNESTHMNDEDVSFPPRTRSSKETSDILTTT 254
Query: 763 QHLTAE----RDQRIKAAKQRFARRVALHEQQ------QRHKLLEAAEEARRKQEQAEEA 812
Q E + +R+ KQ +A+R E Q K+L +E +A
Sbjct: 255 QPAIVEPSEPKVRRVSRRKQLYAKRYKARETQPAIAESSEPKVLHVNDENVSSPPEAHSL 314
Query: 813 ARQAAAQAEQARLEAERLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPSNSTPPSSSR 872
+ + AE E + V TP A +S +NV + +PP +
Sbjct: 315 EKASDILTTTQPAIAESSEPKVPHVNDENVSSTPRAHSSKKNKSTRKNVDNVPSPPKTRS 374
Query: 873 LELMEQRLDTHESMLQDMKQ 892
+ L T + + + +
Sbjct: 375 SKKTSDILTTTQPTIAESSE 394
>At2g15880 unknown protein
Length = 727
Score = 40.4 bits (93), Expect = 0.005
Identities = 46/189 (24%), Positives = 63/189 (32%), Gaps = 30/189 (15%)
Query: 491 PTSPITIQYNPESEPLTLEPELTIPVQ---TEPEPRMSDHSVPRASERLVSRTTDTDSSS 547
PT P+ P+ P +P PV+ + P P+ H V + S T S
Sbjct: 402 PTRPVHKPQPPKESPQPNDPYNQSPVKFRRSPPPPQQPHHHVVHSPPPASSPPTSPPVHS 461
Query: 548 VYTPISFPTNVADSSPSNN--SESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERL 605
+P+ P +S N+ +S KF RR P P P
Sbjct: 462 TPSPVHKPQPPKESPQPNDPYDQSPVKF---------------------RRSPPPPPVHS 500
Query: 606 VDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHL 665
P PI + P P P P P P P S V SP P V + P +
Sbjct: 501 PPPPSPIHSPP----PPPVYSPPPPPPVYSPPPPPPVYSPPPPPPVHSPPPPVHSPPPPV 556
Query: 666 GASEPHVQT 674
+ P V +
Sbjct: 557 HSPPPPVHS 565
Score = 33.1 bits (74), Expect = 0.72
Identities = 67/334 (20%), Positives = 106/334 (31%), Gaps = 47/334 (14%)
Query: 332 SSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVE 391
S S VP T+ QP ++ + + P + + P PP++ V P
Sbjct: 395 SKSPSPVP-TRPVHKPQPPKESPQPNDPYNQSPVKFRRSPPPPQQPHHHVVHSPPPASSP 453
Query: 392 PAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQ 451
P + V +P + S + + +D P+ +RS P
Sbjct: 454 PTSPPVHSTPSPVHKPQPPKESPQPN--------DPYDQSPVK--FRRSPPP-------- 495
Query: 452 PAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPE 511
P + P+ + S P P+++ P P P+ Y+P P P
Sbjct: 496 PPVHSPPPPSPIHS------PPPPPVYSPPPPPPVYSPPPPPPV---YSPPPPPPVHSPP 546
Query: 512 LTI-----PVQTEPEPRMSD----HSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSS 562
+ PV + P P S HS P V++P P V
Sbjct: 547 PPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVYSPPPPPVHSP---PPPVHSPP 603
Query: 563 PSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADP 622
P +S + V + + P P P P + P P+ + P P
Sbjct: 604 PPVHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHSP---PPPVYSPPPPVYSPP----PP 656
Query: 623 LAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNP 656
+ P PV P+ P S + V SP P
Sbjct: 657 PVKSPPPPPVYSPPLLPPKMSSPPTQTPVNSPPP 690
>At1g64500 unknown protein
Length = 368
Score = 40.4 bits (93), Expect = 0.005
Identities = 33/122 (27%), Positives = 56/122 (45%), Gaps = 7/122 (5%)
Query: 354 DETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVE-PAAQVVRRIETPP-RVTRSSV 411
D + DD +D D D P ++ K + +KP V+ P V+ TPP R+ R S
Sbjct: 55 DRLDEDDDDDDDDGDDAPKTWEEVSKSLETKLKPAAVKPPEVDSVKPPATPPRRLPRKSA 114
Query: 412 RSSSKSVVDSDADLNSFDALPISAM-LKRSTNPLTLIPESQPAAQTT----SQPNSLRSS 466
+ ++ A + +P + + LKR+ + L PES ++T S P S++ +
Sbjct: 115 SFHTLDELEVRAKRSIAAQIPTTMVKLKRTESMSKLRPESDDRTESTQSSYSGPRSVKEN 174
Query: 467 FF 468
F
Sbjct: 175 IF 176
>At1g29000 hypothetical protein
Length = 287
Score = 40.4 bits (93), Expect = 0.005
Identities = 25/91 (27%), Positives = 46/91 (50%), Gaps = 11/91 (12%)
Query: 295 TANSLKKMGLVQKKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHD 354
T S K + ++KK+ E SSK + ++K EE D KKKK ++ K+K +
Sbjct: 157 TIESAKLLAYIKKKVHKHAEIISSKTEEEKKK-------EEED----KKKKEEEDKKKKE 205
Query: 355 ETSKPDDGNDGDNDGGPSPPKKKKKQVRLVV 385
+ K ++ + + +KKK++V++ V
Sbjct: 206 DEKKKEEEKKKEEENKKKEGEKKKEEVKVEV 236
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,428,394
Number of Sequences: 26719
Number of extensions: 955795
Number of successful extensions: 5320
Number of sequences better than 10.0: 297
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 244
Number of HSP's that attempted gapping in prelim test: 4456
Number of HSP's gapped (non-prelim): 810
length of query: 904
length of database: 11,318,596
effective HSP length: 108
effective length of query: 796
effective length of database: 8,432,944
effective search space: 6712623424
effective search space used: 6712623424
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0125.11


