

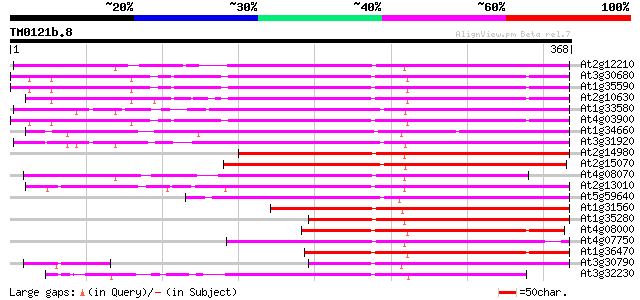
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0121b.8
(368 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g12210 putative TNP2-like transposon protein 239 1e-63
At3g30680 hypothetical protein 233 1e-61
At1g35590 CACTA-element, putative 231 6e-61
At2g10630 En/Spm transposon hypothetical protein 1 227 9e-60
At1g33580 En/Spm-like transposon protein, putative 222 3e-58
At4g03900 putative transposon protein 221 4e-58
At1g34660 hypothetical protein 216 2e-56
At3g31920 hypothetical protein 216 2e-56
At2g14980 pseudogene 202 2e-52
At2g15070 En/Spm-like transposon protein 200 1e-51
At4g08070 190 1e-48
At2g13010 pseudogene 190 1e-48
At5g59640 serine/threonine-specific protein kinase - like 189 2e-48
At1g31560 putative protein 189 2e-48
At1g35280 hypothetical protein 179 2e-45
At4g08000 putative transposon protein 172 3e-43
At4g07750 putative transposon protein 167 6e-42
At1g36470 hypothetical protein 165 4e-41
At3g30790 hypothetical protein 135 5e-32
At3g32230 hypothetical protein 133 1e-31
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 239 bits (611), Expect = 1e-63
Identities = 139/375 (37%), Positives = 202/375 (53%), Gaps = 37/375 (9%)
Query: 3 RMWMDYDRVSKEYLDGVESFL-DHAFRESNGETIPCPCKKCVLHYVVNRAMAYEHLMVNG 61
+ W+ R Y G + F+ D A + + I CPCK C + + +HL+ G
Sbjct: 4 KAWVRLCRADPGYEKGAQKFVTDVAVALGDIDMIVCPCKDCRNVERKSGSDVVDHLVRRG 63
Query: 62 IIPSYDT---WFCHGESINVLNRKEETNCREEAFRGDDMVGLIHDAFEGFNQSMGNNISD 118
+ +Y W+ HG+ +V +C R + + ++ A +GF+ +G I++
Sbjct: 64 MDEAYKLRADWYHHGDVDSV------ADCESNFSRWNAEILELYQAAQGFDADLGE-IAE 116
Query: 119 GGDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELYSGCKKFSKLSFVLHLYHIKRL 178
DI ++F + DA LY C SKLS ++ L+ IK
Sbjct: 117 DEDIRE------------------DEFLAKLADAEIPLYPSCLNHSKLSAIVTLFRIKTK 158
Query: 179 FKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNEL 238
W+++SF+ LLG L + LP L +S YE KK + + YEKIHAC NDC LFR
Sbjct: 159 NGWSDKSFNELLGTLPEMLPADNVLHTSLYEVKKFLRSFDMGYEKIHACVNDCCLFRKRY 218
Query: 239 ADKNVNKCIKCGASRWKND------EKKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHD 292
KN+ C KC ASRWK++ +K VP KVLRYFP+ R +R+F S E ++ +RWH
Sbjct: 219 --KNLENCPKCNASRWKSNMQTGEAKKGVPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHY 276
Query: 293 KGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVI 352
++ DG LRHP DS W K +D++ EFA + RN+RLGL++DGFNP++ S WPV+
Sbjct: 277 SNKSTDGKLRHPVDSVTWDKMNDKYPEFAIEERNLRLGLSTDGFNPFNMKNTRYSCWPVL 336
Query: 353 LVPYNLPPWLCMKQD 367
LV YNLPP LCMK++
Sbjct: 337 LVNYNLPPDLCMKKE 351
>At3g30680 hypothetical protein
Length = 1116
Score = 233 bits (594), Expect = 1e-61
Identities = 140/395 (35%), Positives = 211/395 (52%), Gaps = 41/395 (10%)
Query: 1 RSRMWMDYDRV----SKEYLDGVESFLDHA----FRESNGETIPCPCKKCVLH-YVVNRA 51
R M+ +D V S EY+ GVE F+ A +S CPC C ++++
Sbjct: 15 RDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIISGR 74
Query: 52 MAYEHLMVNGIIPSYDTWFCHGESINV-------------LNRKEETNCREEAFRGDDMV 98
HL G +P Y W+ HGE +N+ N +E N E+ + V
Sbjct: 75 RVSSHLFSQGFMPDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPY-----V 129
Query: 99 GLIHDAFEGFNQSMGNNIS-DGGDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELY 157
+++DAF FN +N+ D +N+ RN S+ KF L++ AN LY
Sbjct: 130 DMVNDAFN-FNVGYDDNVGHDDCYHHDGSYQNVEEPVRNHSN----KFYDLLEGANNPLY 184
Query: 158 SGCKKF-SKLSFVLHLYHIKRLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGR 216
GC++ S+LS L H K + + + D++ + D LPEG + +SHY+T+K++
Sbjct: 185 DGCREGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMRN 244
Query: 217 LGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWK-NDEK---KVPAKVLRYFPLK 272
LGL Y I C N+CMLF E D+ ++C CGA RWK D++ KVP + Y P+
Sbjct: 245 LGLPYHTIDVCKNNCMLFWKE--DEKEDQCRFCGAQRWKPKDDRRRTKVPYSRMWYLPIA 302
Query: 273 SRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLA 332
+L+R++ S +T+ MRWH + ++K+G++ HP+D+ W+ F + H FA++ RNV LGL
Sbjct: 303 DQLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLGLC 362
Query: 333 SDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
+DGFNP+ SM S WPVIL PYNLPP +CM +
Sbjct: 363 TDGFNPF-SMSRNHSLWPVILTPYNLPPRMCMNTE 396
>At1g35590 CACTA-element, putative
Length = 1180
Score = 231 bits (588), Expect = 6e-61
Identities = 139/395 (35%), Positives = 209/395 (52%), Gaps = 41/395 (10%)
Query: 1 RSRMWMDYDRV----SKEYLDGVESFLDHA----FRESNGETIPCPCKKCVLH-YVVNRA 51
R M+ +D V S EY+ GVE F+ A +S CPC C ++++
Sbjct: 15 RDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIISGR 74
Query: 52 MAYEHLMVNGIIPSYDTWFCHGESINV-------------LNRKEETNCREEAFRGDDMV 98
HL G +P Y W+ HGE +N+ N +E N E+ + V
Sbjct: 75 RVSSHLFSQGFMPDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPY-----V 129
Query: 99 GLIHDAFEGFNQSMGNNIS-DGGDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELY 157
+++DAF FN +N+ D +N+ RN S+ KF L++ AN LY
Sbjct: 130 DMVNDAFN-FNVGYDDNVGHDDCYHHDGSYQNVEEPVRNHSN----KFYDLLEGANNLLY 184
Query: 158 SGCKKF-SKLSFVLHLYHIKRLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGR 216
GC++ S+LS L H K + + + D++ + D LPEG + +SHY+T+K++
Sbjct: 185 DGCREGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMHN 244
Query: 217 LGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWK-NDEK---KVPAKVLRYFPLK 272
LGL Y I C N+CMLF E D+ ++C CGA RWK D++ KVP + Y P+
Sbjct: 245 LGLPYHTIDVCKNNCMLFWKE--DEKEDQCRFCGAQRWKPKDDRRRTKVPYSRMWYLPIA 302
Query: 273 SRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLA 332
RL+R++ +T+ MRWH + ++K+G++ HP+D+ W+ F + H FA++ RNV LGL
Sbjct: 303 DRLKRMYQCHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLGLC 362
Query: 333 SDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
+DGFNP+ M S WPVIL PYNLPP +CM +
Sbjct: 363 TDGFNPF-GMSRNHSLWPVILTPYNLPPGMCMNTE 396
>At2g10630 En/Spm transposon hypothetical protein 1
Length = 515
Score = 227 bits (578), Expect = 9e-60
Identities = 136/375 (36%), Positives = 203/375 (53%), Gaps = 31/375 (8%)
Query: 11 VSKEYLDGVESFLDHA----FRESNGETIPCPCKKCVLH-YVVNRAMAYEHLMVNGIIPS 65
+S EY+ GVE F+ A +S+ CPC C ++++ HL +G +
Sbjct: 29 LSAEYVAGVEQFMTFANSQPIVQSSRGKFHCPCSVCKNEKHIISGRRVSSHLFSHGFMHD 88
Query: 66 YDTWFCHGESINV------LNRKEETNCREEAFR--GDDMVGLIHDAFEGFNQSMGNNIS 117
Y W+ HGE +N+ NR + EE D V +++DAF FN +N
Sbjct: 89 YYVWYKHGEEMNMDLGTSYTNRTYFSENHEEVGNIVEDPYVDMVNDAFN-FNVGYDDNYR 147
Query: 118 DGGDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELYSGCK-KFSKLSFVLHLYHIK 176
D N++E + RN S+ KF L++ AN LY GC+ + S+LS L H K
Sbjct: 148 HD-DSYQNVEEPV----RNHSN----KFYDLLEGANNPLYDGCRERQSQLSLASRLMHNK 198
Query: 177 RLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRN 236
+ + D++ D LPE + +SHY+T+K++ LGL Y I C N+CMLF
Sbjct: 199 AEYNMNEKFVDSVCETFTDFLPEENQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLFWK 258
Query: 237 ELADKNVNKCIKCGASRWK-NDEK---KVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHD 292
E ++ ++C CGA RWK D++ KVP + Y P+ RL+R++ S +T+ MRWH
Sbjct: 259 E--EEKEDQCRFCGAKRWKPKDDRRRTKVPYSCMWYLPIGDRLKRMYQSHKTAAAMRWHA 316
Query: 293 KGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVI 352
+ ++K+G++ HP+D+ W+ F H +FA+D RNV LGL +DGFNP+ M S WPVI
Sbjct: 317 EHQSKEGEMNHPSDAAEWRYFQGLHPQFAEDPRNVYLGLCTDGFNPF-GMSRNHSLWPVI 375
Query: 353 LVPYNLPPWLCMKQD 367
L PYNLPP +CM +
Sbjct: 376 LTPYNLPPGMCMNTE 390
>At1g33580 En/Spm-like transposon protein, putative
Length = 428
Score = 222 bits (565), Expect = 3e-58
Identities = 135/377 (35%), Positives = 199/377 (51%), Gaps = 35/377 (9%)
Query: 3 RMWMDYDRVSKEYLDGVESFLDHAFRE-SNGETIPCPCKKC--VLHYVVNRAMAYEHLMV 59
+ W+ R S EY G F+ + R + + CPC C + H ++ + EHL++
Sbjct: 13 KSWVWLPRNSIEYEKGATKFVYASSRRLGDPSEMFCPCTDCRNLCHQPIDTVL--EHLVI 70
Query: 60 NGIIPSYDT---WFCHGESINVLNRKEETNCREEAFRGDDMVGLIHDAFEGFNQSMGNNI 116
G+ Y W HGE I + E++ A+ LI A+
Sbjct: 71 KGMNHKYKRNGCWSKHGE-IRADKLEAESSSEFGAYE------LIRTAY----------- 112
Query: 117 SDGGDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELYSGCKKFSKLSFVLHLYHIK 176
DG D + ++N + S+ S E F + +KDA LY GC K++K+S ++ LY IK
Sbjct: 113 FDGEDHSDSQNQN-ENDSKEPSTKEESDFREKLKDAETPLYYGCPKYTKVSAIMRLYRIK 171
Query: 177 RLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRN 236
+ FD LL L+ D LP L +S E KK + G Y+ IHACPNDC+L+R
Sbjct: 172 VKSGMSENYFDQLLSLVHDILPGENVLLTSTNEIKKFLKMFGFGYDIIHACPNDCILYRK 231
Query: 237 ELADKNVNKCIKCGASRWKNDE------KKVPAKVLRYFPLKSRLQRLFMSSETSRLMRW 290
E ++ C +C A RW+ D+ K +PAKVLRYFP+K R +R+F S + + +RW
Sbjct: 232 EYELRDT--CPRCSAFRWERDKHTGEEKKGIPAKVLRYFPIKDRFKRMFRSKKMAEDLRW 289
Query: 291 HDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWP 350
H + DG +RHP DS AW + + + +FA + RN+RLGL++DG NP+S STWP
Sbjct: 290 HFSNASDDGTMRHPVDSLAWAQVNYKWPQFAAEPRNLRLGLSTDGMNPFSIQNTKYSTWP 349
Query: 351 VILVPYNLPPWLCMKQD 367
V+LV YN+ P +CMK +
Sbjct: 350 VLLVNYNMAPTMCMKAE 366
>At4g03900 putative transposon protein
Length = 817
Score = 221 bits (564), Expect = 4e-58
Identities = 136/395 (34%), Positives = 207/395 (51%), Gaps = 41/395 (10%)
Query: 1 RSRMWMDYDRV----SKEYLDGVESFLDHA----FRESNGETIPCPCKKCVLH-YVVNRA 51
R M+ +D V S EY+ GVE F+ A +S CPC C ++++
Sbjct: 15 RDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFYCPCSVCKNEKHIISGR 74
Query: 52 MAYEHLMVNGIIPSYDTWFCHGESINV-------------LNRKEETNCREEAFRGDDMV 98
HL + +P Y W+ HGE +N+ N +E N E+ + V
Sbjct: 75 RVSSHLFSHEFMPDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPY-----V 129
Query: 99 GLIHDAFEGFNQSMGNNIS-DGGDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELY 157
+++DAF FN +N+ D +N+ RN S+ KF L++ AN LY
Sbjct: 130 DMVNDAFN-FNVGYDDNVGHDDNYHHDGSYQNVEEPVRNHSN----KFYDLLEGANNPLY 184
Query: 158 SGCKKF-SKLSFVLHLYHIKRLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGR 216
GC++ S+LS L H K + + + D++ + LPEG + +SHY+T+K++
Sbjct: 185 DGCREGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTAFLPEGNQATTSHYQTEKLMRN 244
Query: 217 LGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWK-NDEK---KVPAKVLRYFPLK 272
LGL Y I N+CMLF E D+ ++C CGA RWK D++ KVP + Y P+
Sbjct: 245 LGLPYHTIDVYKNNCMLFWKE--DEKEDQCRFCGAQRWKPKDDRRRTKVPYSRMWYLPIA 302
Query: 273 SRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLA 332
RL+R++ S +T+ MRWH + ++K+G++ HP+D+ W+ F + H FA++ NV LGL
Sbjct: 303 DRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPHNVYLGLY 362
Query: 333 SDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
+DGFNP+ M S WPVIL PYNLPP +CM +
Sbjct: 363 TDGFNPF-GMSRNHSLWPVILTPYNLPPGMCMNTE 396
>At1g34660 hypothetical protein
Length = 501
Score = 216 bits (550), Expect = 2e-56
Identities = 124/370 (33%), Positives = 193/370 (51%), Gaps = 30/370 (8%)
Query: 11 VSKEYLDGVESFLDHAFRESNGETIP-------CPCKKCVLHYVVNRAMAYEHLMVNGII 63
++KE+ +G+ F+ R + + IP CPC C + ++HL G +
Sbjct: 12 LTKEFKEGLVDFM----RFAANQEIPLECGKMYCPCSTCGNEKFLPVDSIWKHLYSRGFM 67
Query: 64 PSYDTWFCHGESINVLNRKEETNCREEAFRGDDMVGLIHDAFEGFNQSMGNNISDGGDI- 122
P Y WF HG+ L E DD + + N+ N I D +
Sbjct: 68 PGYYIWFSHGKDFETLANSNE----------DDEIHGLEPEDNNSNEEEENIIRDPTGVI 117
Query: 123 --ESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELYSGCKK-FSKLSFVLHLYHIKRLF 179
+N T + + + E +KF ++ A + +Y GCK+ S+LS L +IK +
Sbjct: 118 QMVTNAFRETTESGIEEPNLEAKKFFDMLDAAKKPIYDGCKEGHSRLSAATRLMNIKTEY 177
Query: 180 KWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELA 239
+ + DA+ +KD LPE P S+YE +K+V LGL Y+ I C ++CM+F +
Sbjct: 178 NLSEDCVDAIADFVKDLLPEPNLAPGSYYEIQKLVSSLGLPYQMIDVCVDNCMIFWRDTV 237
Query: 240 DKNVNKCIKCGASRWK--NDEKKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNK 297
N++ C C R++ + + +VP K + Y P+ RL+RL+ S T+ MRWH + +
Sbjct: 238 --NLDACTFCQKPRFQVTSGKSRVPFKRMWYLPITDRLKRLYQSERTAGPMRWHAE-HSS 294
Query: 298 DGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVILVPYN 357
DG++ HP+D+EAWK F+ +++FA + RNV LGL +DGFNP+ S WPVIL PYN
Sbjct: 295 DGNISHPSDAEAWKHFNSMYKDFANEHRNVYLGLCTDGFNPFGKSGRKYSLWPVILTPYN 354
Query: 358 LPPWLCMKQD 367
LPP LCMK++
Sbjct: 355 LPPSLCMKRE 364
>At3g31920 hypothetical protein
Length = 725
Score = 216 bits (549), Expect = 2e-56
Identities = 132/378 (34%), Positives = 193/378 (50%), Gaps = 34/378 (8%)
Query: 3 RMWMDYDRVSKEYLDGVESFLDHAFRESNGETIP--CPCKKC--VLHYVVNRAMAYEHLM 58
+ W+ R + EY G +F+ +A S G+ CPC C + H + + EHL+
Sbjct: 3 KSWVWLPRNNIEYEKGATTFV-YASSRSLGDPAEMFCPCIDCRNLCHQPIETIL--EHLV 59
Query: 59 VNGIIPSYDT---WFCHGESINVLNRKEETNCREEAFRGDDMVGLIHDAFEGFNQSMGNN 115
+ G+ Y W HGE E ++ E + LI A+
Sbjct: 60 MKGMDHKYKRNRCWSKHGEIRADKLEVEPSSTTSEC----EAYELIRTAYFD-------- 107
Query: 116 ISDGGDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELYSGCKKFSKLSFVLHLYHI 175
GD+ S+ S+ E F + +K+A LY GC K++K+S ++ LY I
Sbjct: 108 ----GDVHSDSQNQNEDDSKEPETKEEADFREKLKEAETPLYHGCPKYTKVSAIMGLYRI 163
Query: 176 KRLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFR 235
K + FD LL L+ D LP LP+S E KK + G Y+ IHAC NDC+L+R
Sbjct: 164 KVKSGMSENYFDQLLSLVHDILPGENVLPTSTNEIKKFLKMFGFGYDIIHACQNDCILYR 223
Query: 236 NELADKNVNKCIKCGASRWKNDE------KKVPAKVLRYFPLKSRLQRLFMSSETSRLMR 289
E ++ C +C ASRW+ D+ K +PAKVL YFP+K R +R+F S + + R
Sbjct: 224 KEYELRDT--CPRCNASRWERDKHTGEEKKGIPAKVLGYFPIKDRFKRMFRSKKMAEDQR 281
Query: 290 WHDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTW 349
WH + DG +RH DS AW + +D+ EFA + RN+RLGL++DG NP+S STW
Sbjct: 282 WHFSNASDDGTMRHHVDSLAWAQVNDKWPEFAAEPRNLRLGLSADGMNPFSIQNTKYSTW 341
Query: 350 PVILVPYNLPPWLCMKQD 367
P++LV YN+ P +CMK +
Sbjct: 342 PILLVNYNMAPTMCMKAE 359
>At2g14980 pseudogene
Length = 665
Score = 202 bits (515), Expect = 2e-52
Identities = 101/223 (45%), Positives = 140/223 (62%), Gaps = 8/223 (3%)
Query: 151 DANEELYSGCKKFSKLSFVLHLYHIKRLFKWTNESFDALLGLLKDALPEGEKLPSSHYET 210
DA LY C SKLS ++ L+ +K W+++SF+ LL L + LPE L +S YE
Sbjct: 24 DAETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSFNDLLETLPEMLPEDNVLHTSLYEV 83
Query: 211 KKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWKND------EKKVPAK 264
KK + + Y+KIHAC NDC LFR + K + C+KC ASRWK + +K VP K
Sbjct: 84 KKFLKSFDMGYQKIHACVNDCCLFRKKY--KKLQNCLKCNASRWKTNMHTGEVKKGVPQK 141
Query: 265 VLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDS 324
VLRYF + RL+R+F S E +R +RWH ++ DG L HP DS W + + ++ FA +
Sbjct: 142 VLRYFSIVPRLKRMFRSEEMARDLRWHFHNKSTDGKLCHPVDSVTWDQMNAKYPLFASEE 201
Query: 325 RNVRLGLASDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
RN+RLGL+++GFNP++ S WPV+LV YNLPP LCMK++
Sbjct: 202 RNLRLGLSTNGFNPFNMKNTRYSCWPVLLVNYNLPPDLCMKKE 244
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 200 bits (508), Expect = 1e-51
Identities = 100/231 (43%), Positives = 140/231 (60%), Gaps = 8/231 (3%)
Query: 141 EVEKFEQLMKDANEELYSGCKKFSKLSFVLHLYHIKRLFKWTNESFDALLGLLKDALPEG 200
E F + +KD LY GC K++K+S ++ LY IK + FD LL L+ D LP
Sbjct: 54 EESYFREKLKDVETPLYYGCPKYTKVSAIMGLYRIKVKSGMSENYFDQLLSLVHDILPGE 113
Query: 201 EKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWKNDE-- 258
LP+S E KK + G Y+ IHACPNDC+L++ E ++ C+ C ASRWK D+
Sbjct: 114 NVLPTSTNEIKKFLKMFGFGYDIIHACPNDCILYKKEYELRDT--CLGCSASRWKRDKHT 171
Query: 259 ----KKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFD 314
K +PAKVL YF +K R +R+F S + ++WH + DG +RHP DS AW + +
Sbjct: 172 GEEKKGIPAKVLWYFSIKDRFKRMFRSKKMVEDLQWHFSNASVDGTMRHPIDSLAWAQVN 231
Query: 315 DQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMK 365
D+ +FA +SRN+RLGL++D NP+S STWPV+LV YN+ P +CMK
Sbjct: 232 DKWPQFAAESRNLRLGLSTDKMNPFSIQNTKYSTWPVLLVNYNMAPTMCMK 282
>At4g08070
Length = 767
Score = 190 bits (482), Expect = 1e-48
Identities = 113/340 (33%), Positives = 173/340 (50%), Gaps = 30/340 (8%)
Query: 10 RVSKEYLDGVESFLDHAFRESNGETIPCPCKKCVLHYVVNRAMAYEHLMVNGIIPSYDT- 68
R Y G F+ + I CPC C + + +HL+ G+ +Y
Sbjct: 27 RADPTYERGAIKFVRDVAAALGDDMIVCPCIDCRNIDRHSGCVVVDHLVTRGMDEAYKRR 86
Query: 69 --WFCHGESINVLNRKEETNCREEAFRGDDMVGLIHDAFEGFNQSMGNNISDGGDIESNL 126
W+ HGE ++ + + + + +D V ++ A E ++ + G E
Sbjct: 87 CDWYHHGELVSGGESESKVS------QWNDEVVELYQAAEYLDEEFDAMVQLGEIAE--- 137
Query: 127 DENLTHTSRNQSHPEVEKFEQLMKDANEELYSGCKKFSKLSFVLHLYHIKRLFKWTNESF 186
+ + ++F + DA LY C SKLS ++ L+ +K W+++SF
Sbjct: 138 ----------RDDKKEDEFLAKLADAETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSF 187
Query: 187 DALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKC 246
+ LL L + LPE L +S YE KK + + Y+KIHAC NDC LFR + K + C
Sbjct: 188 NDLLETLSEMLPEDNVLHTSLYEVKKFLKSFDMGYQKIHACVNDCCLFRKKY--KKLQNC 245
Query: 247 IKCGASRWKND------EKKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGD 300
KC ASRWK + +K VP KVLRYFP+ RL+R+F S E +R +RWH ++ DG
Sbjct: 246 PKCNASRWKTNMHTGEVKKGVPQKVLRYFPIIPRLKRMFRSEEMARDLRWHFHNKSTDGK 305
Query: 301 LRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYS 340
LRHP DS W + + ++ FA + RN+RLGL++DGFNP++
Sbjct: 306 LRHPVDSVTWDQMNAKYPLFASEERNLRLGLSTDGFNPFN 345
>At2g13010 pseudogene
Length = 1040
Score = 190 bits (482), Expect = 1e-48
Identities = 119/374 (31%), Positives = 187/374 (49%), Gaps = 28/374 (7%)
Query: 11 VSKEYLDGVESFL----DHAFRESNGETIPCPCKKCVLHYVVNRAMAYEHLMVNGIIPSY 66
V+ EY D + F+ + F NG TI CPC+KC+ + + +HL G +P+Y
Sbjct: 184 VTIEYYDELRGFMYQTTNEPFARENG-TILCPCRKCLNEKYLEFNVVKKHLYNRGFMPNY 242
Query: 67 DTWFCHGESINVLNRKEETNCREEAFRGDDMVGLIH-------DAFEGFNQSMGNNISDG 119
W HG+ + + + + ++V LI D F G+ + N D
Sbjct: 243 YVWLRHGDVVEIAHELVI-----QIMVSSNLVILIMMRIMQVPDQF-GYGYNQENRFHDM 296
Query: 120 GDIESNLDENLTHTSRNQSHP---EVEKFEQLMKDANEELYSGCKKF-SKLSFVLHLYHI 175
+ + E + N S + + F ++ AN+ +Y GC++ SKLS + I
Sbjct: 297 --VTNAFHETIASFPENISEEPNVDAQHFYDMLDAANQPIYEGCREGNSKLSLASRMMTI 354
Query: 176 KRLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFR 235
K + + D+ L+K+ LP S+YE +K+V GL E I C + CM+F
Sbjct: 355 KADNNLSEKCMDSWAELIKEYLPPDNISAESYYEIQKLVSSHGLPSEMIDVCIDYCMIFW 414
Query: 236 NELADKNVNKCIKCGASRWKND--EKKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDK 293
+ D N+ +C CG R++ +VP K + Y P+ RL+RL+ S T+ MRWH +
Sbjct: 415 GD--DVNLQECRFCGKPRYQTTGGRTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQ 472
Query: 294 GRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVIL 353
DG++ HP+D++AWK F + +FA + RNV LGL +DGF+P+ S WPVIL
Sbjct: 473 HTTADGEITHPSDAKAWKHFQTVYPDFANECRNVYLGLCTDGFSPFGLCGRQYSLWPVIL 532
Query: 354 VPYNLPPWLCMKQD 367
PY LPP +CM+ +
Sbjct: 533 TPYKLPPDMCMQSE 546
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 189 bits (481), Expect = 2e-48
Identities = 100/258 (38%), Positives = 145/258 (55%), Gaps = 12/258 (4%)
Query: 116 ISDG-GDIESNLDENLTHTSRNQSHPEVEKFEQLMKDANEELYSGCKK-FSKLSFVLHLY 173
+SD G ES D+ + E +KF ++ A + +Y GCK+ SKLS L
Sbjct: 31 VSDAFGSTESEFDQ----MREEDPNFEAKKFYDILDAAKQPIYDGCKEGLSKLSLAARLM 86
Query: 174 HIKRLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCML 233
+K + D++ ++++ LPEG P S+YE KK++ LGL Y+KI C ++CM+
Sbjct: 87 SLKTDNNLSQNCMDSIAQIMQEYLPEGNNSPKSYYEIKKLMRSLGLPYQKIDVCQDNCMI 146
Query: 234 FRNELADKNVNKCIKCGASRW----KNDEKKVPAKVLRYFPLKSRLQRLFMSSETSRLMR 289
F E + C+ C R+ K +K +P + + Y P+ RL+RL+ S T++ MR
Sbjct: 147 FWKETEKEEY--CLFCKKDRYRPTQKIGQKSIPYRQMFYLPIADRLKRLYQSHNTAKHMR 204
Query: 290 WHDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTW 349
WH + DG++ HP+D EAWK F H +FA RNV LGL +DGFNP+ S W
Sbjct: 205 WHAEHLASDGEMGHPSDGEAWKHFHKVHPDFASKPRNVYLGLCTDGFNPFGMSGHNYSLW 264
Query: 350 PVILVPYNLPPWLCMKQD 367
PVIL PYNLPP +CMKQ+
Sbjct: 265 PVILTPYNLPPEMCMKQE 282
>At1g31560 putative protein
Length = 1431
Score = 189 bits (480), Expect = 2e-48
Identities = 96/202 (47%), Positives = 125/202 (61%), Gaps = 8/202 (3%)
Query: 172 LYHIKRLFKWTNESFDALLGLLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDC 231
L IK W+++SF+ LLG L + LP L +S YE KK + + YEKIHAC NDC
Sbjct: 83 LAKIKTKNGWSDKSFNELLGTLPEMLPADNVLRTSLYEVKKFLKSFDMGYEKIHACVNDC 142
Query: 232 MLFRNELADKNVNKCIKCGASRWKN------DEKKVPAKVLRYFPLKSRLQRLFMSSETS 285
LFR KN+ C KC ASRWKN +K VP KVLRYF + R +++F S E +
Sbjct: 143 CLFRKRY--KNLENCPKCNASRWKNYMQTGEAKKGVPQKVLRYFLIIPRFKKMFRSKEMA 200
Query: 286 RLMRWHDKGRNKDGDLRHPADSEAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVF 345
+ +RWH ++ DG RHP DS W K D++ EFA + RN+RLGL++DGFNP++
Sbjct: 201 KDLRWHYSNKSTDGKHRHPVDSVTWDKMIDKYPEFAAEERNMRLGLSTDGFNPFNMKNTR 260
Query: 346 DSTWPVILVPYNLPPWLCMKQD 367
S WPV+LV YNLPP CMK +
Sbjct: 261 YSCWPVLLVNYNLPPDQCMKNE 282
>At1g35280 hypothetical protein
Length = 557
Score = 179 bits (454), Expect = 2e-45
Identities = 88/177 (49%), Positives = 115/177 (64%), Gaps = 8/177 (4%)
Query: 197 LPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWKN 256
LP L +S YE KK + + YEKIHAC NDC LFR KN+ C KC ASRWK+
Sbjct: 2 LPADNVLHTSLYEVKKFLKSFDMGYEKIHACVNDCCLFRKRY--KNLENCPKCNASRWKS 59
Query: 257 D------EKKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAW 310
+ +K VP KVLRYFP+ R +R+F S E ++ +RWH ++ DG LRHP DS W
Sbjct: 60 NMQTGEAKKGVPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHYSNKSIDGKLRHPVDSVTW 119
Query: 311 KKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
K +D++ EFA + RN+RLGL++DGFNP++ S WPV+LV YNLP LCMK++
Sbjct: 120 DKMNDKYPEFAAEERNLRLGLSTDGFNPFNMKNTRYSCWPVLLVNYNLPLDLCMKKE 176
>At4g08000 putative transposon protein
Length = 609
Score = 172 bits (435), Expect = 3e-43
Identities = 83/177 (46%), Positives = 117/177 (65%), Gaps = 7/177 (3%)
Query: 192 LLKDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGA 251
LL++ LPEG + SSHY+T+K++ LGL Y I C N+CMLF E D+ + C CGA
Sbjct: 74 LLEEFLPEGNQATSSHYQTEKLMRNLGLPYHTIDVCSNNCMLFWKE--DEQEDHCRFCGA 131
Query: 252 SRWK-NDEK---KVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADS 307
RWK D++ KVP + Y P+ RL+R++ S +T+ MRWH + ++K+G++ HP+D+
Sbjct: 132 QRWKPKDDRRRTKVPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDA 191
Query: 308 EAWKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVILVPYNLPPWLCM 364
W+ F + H FA++ RNV LGL +DGFNP+ M S WPVIL PYNLPP +CM
Sbjct: 192 AEWRYFQELHPRFAEEPRNVYLGLCTDGFNPF-GMSRNHSLWPVILTPYNLPPSMCM 247
>At4g07750 putative transposon protein
Length = 569
Score = 167 bits (424), Expect = 6e-42
Identities = 88/231 (38%), Positives = 130/231 (56%), Gaps = 18/231 (7%)
Query: 143 EKFEQLMKDANEELYSGCKKFSKLSFVLHLYHIKRLFKWTNESFDALLGLLKDALPEGEK 202
++F + DA LY C SKLS ++ L+ + W+++SF+ LL +L + LP+
Sbjct: 64 DEFLAKLADAETPLYPSCVNHSKLSAIVSLFRLNTKNGWSDKSFNDLLEILPEMLPKDNV 123
Query: 203 LPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWKND----- 257
L +S Y+ KK + + Y+KIHAC NDC LFR K ++ C KC ASRWK +
Sbjct: 124 LHTSLYKVKKFLKSFDMGYQKIHACVNDCCLFRKNY--KKLDSCPKCNASRWKTNLHIGE 181
Query: 258 -EKKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFDDQ 316
+K VP KVLRYFP+ RL+ +F S E + +RWH ++ D L HP D W + + +
Sbjct: 182 IKKGVPQKVLRYFPIIPRLKGMFRSKEMDKDLRWHFSNKSSDEKLCHPVDLVTWDQMNAK 241
Query: 317 HQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
+ FA + RN+R GL++DGFNP++ S WP LCMK++
Sbjct: 242 YPSFAGEERNLRHGLSTDGFNPFNMKNTRYSCWPD----------LCMKKE 282
>At1g36470 hypothetical protein
Length = 753
Score = 165 bits (417), Expect = 4e-41
Identities = 80/178 (44%), Positives = 115/178 (63%), Gaps = 7/178 (3%)
Query: 194 KDALPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASR 253
+D LPEG + +SHY+T+K++ LGL Y I C N+CML E D+ ++C CGA R
Sbjct: 54 RDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLSWKE--DEKEDQCRFCGAKR 111
Query: 254 WK-NDEK---KVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEA 309
WK D++ KVP + Y P+ RL+R++ S +T+ MRWH + ++K+G++ H +D+
Sbjct: 112 WKPKDDRRRTKVPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHLSDAAE 171
Query: 310 WKKFDDQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
W+ F H EFA++ RNV LGL +DGFNP+ M S WPVIL PYNLPP +CM +
Sbjct: 172 WRYFQGLHPEFAEEPRNVYLGLCTDGFNPF-GMSRNHSLWPVILTPYNLPPGMCMNTE 228
>At3g30790 hypothetical protein
Length = 953
Score = 135 bits (339), Expect = 5e-32
Identities = 68/173 (39%), Positives = 103/173 (59%), Gaps = 4/173 (2%)
Query: 197 LPEGEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWK- 255
LP S++E +K+V LGL E I C + CM+F + D N+ +C CG R++
Sbjct: 126 LPPDNISAESYFEIQKLVLSLGLPSEMIDVCIDYCMIFWGD--DVNLQECRFCGKPRYQT 183
Query: 256 -NDEKKVPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFD 314
+ +VP K + Y P+ RL+RL+ S T+ MRWH + DG+++HP+D++AWK F
Sbjct: 184 TSGRTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTTADGEIKHPSDTKAWKHFQ 243
Query: 315 DQHQEFAKDSRNVRLGLASDGFNPYSSMKVFDSTWPVILVPYNLPPWLCMKQD 367
+ +FA + RNV LGL +DGF+P+ S W +IL PYNLPP + M+ +
Sbjct: 244 TVYPDFASECRNVYLGLCTDGFSPFVLSGRQYSLWHIILTPYNLPPDMYMQSE 296
Score = 42.7 bits (99), Expect = 3e-04
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query: 10 RVSKEYLDGVESFLDHAFRE----SNGETIPCPCKKCVLHYVVNRAMAYEHLMVNGIIPS 65
+V++EY DG+ F+ A E NG TI CPC+KC+ + + +HL G +P+
Sbjct: 63 KVTREYYDGLRGFMYQAINEPFARENG-TIFCPCRKCLNEKYLEFNVVKKHLYNRGFMPN 121
Query: 66 Y 66
Y
Sbjct: 122 Y 122
>At3g32230 hypothetical protein
Length = 1169
Score = 133 bits (335), Expect = 1e-31
Identities = 99/322 (30%), Positives = 156/322 (47%), Gaps = 47/322 (14%)
Query: 24 DHAFRESNGETIPCPCKKCVLHYVVNRAMAYEHLMVNGIIPS---YDTWFCHGESINVLN 80
+ +F +NG +I CPC+ C R A M G++ Y+ + H ES N
Sbjct: 175 NESFARANG-SIFCPCR-C-------RDSAGSSNMNYGVVEPTNYYENYGHHQESDQGYN 225
Query: 81 RKEETNCREEAFRGDDMVGLIHDAFEGFNQSMGNNISDGGDIESNLDENLTHTSRNQSHP 140
+ R DMV DAF + NI + E N+D
Sbjct: 226 HEN---------RFHDMVT---DAFHETTAAFHENIIE----EPNVD------------- 256
Query: 141 EVEKFEQLMKDANEELYSGCKKF-SKLSFVLHLYHIKRLFKWTNESFDALLGLLKDALPE 199
++F ++ AN+ +Y C++ SKLS + IK + D+ L+K+ L
Sbjct: 257 -AQRFYDMLDAANKPIYEECREGQSKLSLASRMMTIKADNNLSENCMDSWAELIKEYLIA 315
Query: 200 GEKLPSSHYETKKIVGRLGLKYEKIHACPNDCMLFRNELADKNVNKCIKCGASRWKNDEK 259
S+YE +K+V LGL E I C + CM+F + D+N+ +C CG R++ +
Sbjct: 316 DNISAESYYEIQKLVSSLGLPSEMIDVCIDYCMIFWKD--DENLQECRFCGKPRYQPTGR 373
Query: 260 K--VPAKVLRYFPLKSRLQRLFMSSETSRLMRWHDKGRNKDGDLRHPADSEAWKKFDDQH 317
+ VP K + Y P+ RL+RL+ S T+ MRWH + +G++ HP+D++AWK F +
Sbjct: 374 RTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAEHSTVNGEVTHPSDAKAWKHFQTVY 433
Query: 318 QEFAKDSRNVRLGLASDGFNPY 339
+FA + RNV LGL +DGF+P+
Sbjct: 434 PDFASECRNVYLGLCTDGFSPF 455
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,295,598
Number of Sequences: 26719
Number of extensions: 426318
Number of successful extensions: 1150
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1044
Number of HSP's gapped (non-prelim): 48
length of query: 368
length of database: 11,318,596
effective HSP length: 101
effective length of query: 267
effective length of database: 8,619,977
effective search space: 2301533859
effective search space used: 2301533859
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0121b.8


