

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0119.8
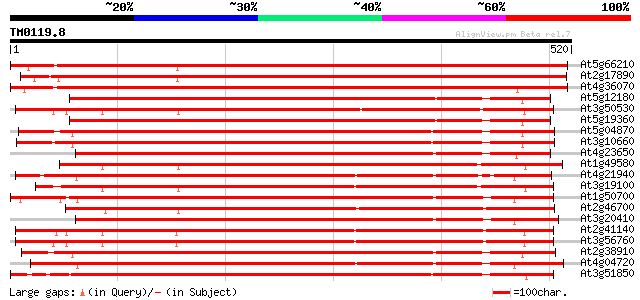
(520 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66210 calcium-dependent protein kinase 821 0.0
At2g17890 putative calcium-dependent ser/thr protein kinase 816 0.0
At4g36070 Calcium-dependent serine/threonine protein kinase 786 0.0
At5g12180 calcium-dependent protein kinase 435 e-122
At3g50530 calcium/calmodulin-dependent protein kinase CaMK1 431 e-121
At5g19360 calcium-dependent protein kinase - like 430 e-120
At5g04870 calcium-dependent protein kinase 429 e-120
At3g10660 calmodulin-domain protein kinase CDPK isoform 2 428 e-120
At4g23650 calcium-dependent protein kinase (CDPK6) 422 e-118
At1g49580 CDPK-related protein kinase, putative 422 e-118
At4g21940 calcium-dependent protein kinase like protein 414 e-116
At3g19100 CDPK-related kinase 412 e-115
At1g50700 hypothetical protein 412 e-115
At2g46700 calcium/calmodulin-dependent protein kinase CaMK4 409 e-114
At3g20410 calmodulin-domain protein kinase CDPK isoform 9 409 e-114
At2g41140 CDPK-related kinase 1 (CRK1) 409 e-114
At3g56760 calcium-dependent protein kinase-like 408 e-114
At2g38910 putative calcium-dependent protein kinase 408 e-114
At4g04720 putative calcium dependent protein kinase 405 e-113
At3g51850 calcium-dependent like protein kinase 402 e-112
>At5g66210 calcium-dependent protein kinase
Length = 523
Score = 821 bits (2120), Expect = 0.0
Identities = 410/522 (78%), Positives = 456/522 (86%), Gaps = 7/522 (1%)
Query: 1 MGSCFSTNTATKPQQS--TAAQKKQPPPNRHQRQRAQRHVAGSRHVPSGMRADFGYDKDF 58
MG CFS T S ++ K + P + + + GS +P G R DFGY KDF
Sbjct: 1 MGVCFSAIRVTGASSSRRSSQTKSKAAPTPIDTKASTKRRTGS--IPCGKRTDFGYSKDF 58
Query: 59 DERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVA 118
+ Y++GKLLGHGQFGYTYV + + +GDRVAVKRL+KSKMVLP AVEDVK+EV IL A++
Sbjct: 59 HDHYTIGKLLGHGQFGYTYVAIHRPNGDRVAVKRLDKSKMVLPIAVEDVKREVQILIALS 118
Query: 119 GHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILA---NRYTEKDAAVVVRQMLKVAAE 175
GHENVVQF+NAF+DD YVYIVMELCEGGELLDRIL+ NRY+EKDAAVVVRQMLKVA E
Sbjct: 119 GHENVVQFHNAFEDDDYVYIVMELCEGGELLDRILSKKGNRYSEKDAAVVVRQMLKVAGE 178
Query: 176 CHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLK 235
CHL GLVHRDMKPENFLFKS + DSPLKATDFGLSDFIKPGK+FHDIVGSAYYVAPEVLK
Sbjct: 179 CHLHGLVHRDMKPENFLFKSAQLDSPLKATDFGLSDFIKPGKRFHDIVGSAYYVAPEVLK 238
Query: 236 RKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDF 295
R+SGP+SDVWSIGVITYILLCGRRPFWD+TEDGIFKEVLRNKPDF RKPW TIS++AKDF
Sbjct: 239 RRSGPESDVWSIGVITYILLCGRRPFWDRTEDGIFKEVLRNKPDFSRKPWATISDSAKDF 298
Query: 296 VKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRAL 355
VKKLL+KDPRARLTAAQALSH WVREGG A++IP+DISVLNNLRQFV YSR KQFALRAL
Sbjct: 299 VKKLLVKDPRARLTAAQALSHAWVREGGNATDIPVDISVLNNLRQFVRYSRLKQFALRAL 358
Query: 356 ASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNT 415
AS L+E E++DL+DQFDAIDVDKNG ISLEEMRQALAKDL WKLK+SRV EIL+AIDSNT
Sbjct: 359 ASTLDEAEISDLRDQFDAIDVDKNGVISLEEMRQALAKDLPWKLKDSRVAEILEAIDSNT 418
Query: 416 DGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRGS 475
DGLVDFTEFVAA LHVHQLEEHDS+KWQLRS+AAFEKFDLDKDGYITPEELRMHTGLRGS
Sbjct: 419 DGLVDFTEFVAAALHVHQLEEHDSEKWQLRSRAAFEKFDLDKDGYITPEELRMHTGLRGS 478
Query: 476 IDPLLEEADIDKDGKISLSEFRRLLRTASMGSQTVTTPTGYQ 517
IDPLL+EADID+DGKISL EFRRLLRTAS+ SQ +P G++
Sbjct: 479 IDPLLDEADIDRDGKISLHEFRRLLRTASISSQRAPSPAGHR 520
>At2g17890 putative calcium-dependent ser/thr protein kinase
Length = 571
Score = 816 bits (2108), Expect = 0.0
Identities = 407/515 (79%), Positives = 459/515 (89%), Gaps = 10/515 (1%)
Query: 11 TKPQQSTAAQK---KQPPPNRHQRQRAQRHVAGSRH---VPSGMRADFGYDKDFDERYSL 64
T+P+++ A+K + PP+ R++ + G RH +P G R DFGY KDFD RY++
Sbjct: 52 TQPRRNATAKKTPTRHTPPHGKVREKVISN-NGRRHGETIPYGKRVDFGYAKDFDHRYTI 110
Query: 65 GKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVV 124
GKLLGHGQFGYTYV DK +GDRVAVK+++K+KM +P AVEDVK+EV IL+A+ GHENVV
Sbjct: 111 GKLLGHGQFGYTYVATDKKTGDRVAVKKIDKAKMTIPIAVEDVKREVKILQALTGHENVV 170
Query: 125 QFYNAFDDDSYVYIVMELCEGGELLDRILA---NRYTEKDAAVVVRQMLKVAAECHLRGL 181
+FYNAF+D + VYIVMELCEGGELLDRILA +RY+E+DAAVVVRQMLKVAAECHLRGL
Sbjct: 171 RFYNAFEDKNSVYIVMELCEGGELLDRILARKDSRYSERDAAVVVRQMLKVAAECHLRGL 230
Query: 182 VHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQ 241
VHRDMKPENFLFKST EDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKR+SGP+
Sbjct: 231 VHRDMKPENFLFKSTEEDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRRSGPE 290
Query: 242 SDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLI 301
SDVWSIGVI+YILLCGRRPFWDKTEDGIFKEVL+NKPDFRRKPWPTISN+AKDFVKKLL+
Sbjct: 291 SDVWSIGVISYILLCGRRPFWDKTEDGIFKEVLKNKPDFRRKPWPTISNSAKDFVKKLLV 350
Query: 302 KDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNE 361
KDPRARLTAAQALSHPWVREGG+ASEIPIDISVLNN+RQFV +SR KQFALRALA+ L+E
Sbjct: 351 KDPRARLTAAQALSHPWVREGGDASEIPIDISVLNNMRQFVKFSRLKQFALRALATTLDE 410
Query: 362 EELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDF 421
EELADL+DQFDAIDVDKNG ISLEEMRQALAKD WKLK++RV EILQAIDSNTDG VDF
Sbjct: 411 EELADLRDQFDAIDVDKNGVISLEEMRQALAKDHPWKLKDARVAEILQAIDSNTDGFVDF 470
Query: 422 TEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRGSIDPLLE 481
EFVAA LHV+QLEEHDS+KWQ RS+AAFEKFD+D DG+IT EELRMHTGL+GSI+PLLE
Sbjct: 471 GEFVAAALHVNQLEEHDSEKWQQRSRAAFEKFDIDGDGFITAEELRMHTGLKGSIEPLLE 530
Query: 482 EADIDKDGKISLSEFRRLLRTASMGSQTVTTPTGY 516
EADID DGKISL EFRRLLRTAS+ S+ V +P GY
Sbjct: 531 EADIDNDGKISLQEFRRLLRTASIKSRNVRSPPGY 565
>At4g36070 Calcium-dependent serine/threonine protein kinase
Length = 536
Score = 786 bits (2029), Expect = 0.0
Identities = 390/534 (73%), Positives = 451/534 (84%), Gaps = 20/534 (3%)
Query: 1 MGSCFSTNTATK------------PQQSTAAQKKQPPPNRHQRQRAQRHVAGSRHVPSGM 48
MG CFS+ AT+ P Q A++K ++ ++ RH G +P G
Sbjct: 1 MGLCFSSPKATRRGTGSRNPNPDSPTQGKASEKVSNKNKKNTKKIQLRHQGG---IPYGK 57
Query: 49 RADFGYDKDFDERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVK 108
R DFGY KDFD RY++GKLLGHGQFG+TYV D +G+RVAVKR++K+KM P VEDVK
Sbjct: 58 RIDFGYAKDFDNRYTIGKLLGHGQFGFTYVATDNNNGNRVAVKRIDKAKMTQPIEVEDVK 117
Query: 109 QEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYTEKDAAVVVRQ 168
+EV IL+A+ GHENVV F+NAF+D +Y+YIVMELC+GGELLDRILANRYTEKDAAVVVRQ
Sbjct: 118 REVKILQALGGHENVVGFHNAFEDKTYIYIVMELCDGGELLDRILANRYTEKDAAVVVRQ 177
Query: 169 MLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYY 228
MLKVAAECHLRGLVHRDMKPENFLFKST E S LKATDFGLSDFIKPG KF DIVGSAYY
Sbjct: 178 MLKVAAECHLRGLVHRDMKPENFLFKSTEEGSSLKATDFGLSDFIKPGVKFQDIVGSAYY 237
Query: 229 VAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTI 288
VAPEVLKR+SGP+SDVWSIGVITYILLCGRRPFWDKT+DGIF EV+R KPDFR PWPTI
Sbjct: 238 VAPEVLKRRSGPESDVWSIGVITYILLCGRRPFWDKTQDGIFNEVMRKKPDFREVPWPTI 297
Query: 289 SNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFK 348
SN AKDFVKKLL+K+PRARLTAAQALSH WV+EGGEASE+PIDISVLNN+RQFV +SR K
Sbjct: 298 SNGAKDFVKKLLVKEPRARLTAAQALSHSWVKEGGEASEVPIDISVLNNMRQFVKFSRLK 357
Query: 349 QFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEIL 408
Q ALRALA +NE+EL DL+DQFDAID+DKNGSISLEEMRQALAKD+ WKLK++RV EIL
Sbjct: 358 QIALRALAKTINEDELDDLRDQFDAIDIDKNGSISLEEMRQALAKDVPWKLKDARVAEIL 417
Query: 409 QAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRM 468
QA DSNTDGLVDFTEFV A LHV+QLEEHDS+KWQ RS+AAF+KFD+D DG+ITPEELR+
Sbjct: 418 QANDSNTDGLVDFTEFVVAALHVNQLEEHDSEKWQQRSRAAFDKFDIDGDGFITPEELRL 477
Query: 469 H-----TGLRGSIDPLLEEADIDKDGKISLSEFRRLLRTASMGSQTVTTPTGYQ 517
+ TGL+GSI+PLLEEAD+D+DG+IS++EFRRLLR+AS+ S+ V +P GYQ
Sbjct: 478 NQCLQQTGLKGSIEPLLEEADVDEDGRISINEFRRLLRSASLKSKNVKSPPGYQ 531
>At5g12180 calcium-dependent protein kinase
Length = 528
Score = 435 bits (1118), Expect = e-122
Identities = 217/452 (48%), Positives = 308/452 (68%), Gaps = 13/452 (2%)
Query: 56 KDFDERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILK 115
+D YSLGK LG GQFG T++ KA+G + A K + K K+V +EDV++EV I+
Sbjct: 67 EDVKASYSLGKELGRGQFGVTHLCTQKATGHQFACKTIAKRKLVNKEDIEDVRREVQIMH 126
Query: 116 AVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAA 174
+ G N+V+ A++D V++VMELC GGEL DRI+A Y+E+ AA ++R ++++
Sbjct: 127 HLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKGHYSERAAASLLRTIVQIVH 186
Query: 175 ECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVL 234
CH G++HRD+KPENFL + E+SPLKATDFGLS F KPG+ F DIVGSAYY+APEVL
Sbjct: 187 TCHSMGVIHRDLKPENFLLLNKDENSPLKATDFGLSVFYKPGEVFKDIVGSAYYIAPEVL 246
Query: 235 KRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKD 294
KRK GP++D+WSIGV+ YILLCG PFW ++E+GIF +LR DF PWP+IS AKD
Sbjct: 247 KRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPQAKD 306
Query: 295 FVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRA 354
VKK+L DP+ RLTAAQ L+HPW++E GEA ++P+D +V++ L+QF + FK+ ALR
Sbjct: 307 LVKKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDNAVMSRLKQFKAMNNFKKVALRV 366
Query: 355 LASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSN 414
+A L+EEE+ LK+ F +D D +G+I+LEE+RQ LAK +L E V ++++A D++
Sbjct: 367 IAGCLSEEEIMGLKEMFKGMDTDSSGTITLEELRQGLAKQ-GTRLSEYEVQQLMEAADAD 425
Query: 415 TDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRG 474
+G +D+ EF+AAT+H+++L+ + +AF+ FD D GYIT EEL G
Sbjct: 426 GNGTIDYGEFIAATMHINRLDREE------HLYSAFQHFDKDNSGYITMEELEQALREFG 479
Query: 475 -----SIDPLLEEADIDKDGKISLSEFRRLLR 501
I ++ E D D DG+I+ EF ++R
Sbjct: 480 MNDGRDIKEIISEVDGDNDGRINYDEFVAMMR 511
>At3g50530 calcium/calmodulin-dependent protein kinase CaMK1
Length = 601
Score = 431 bits (1108), Expect = e-121
Identities = 238/519 (45%), Positives = 332/519 (63%), Gaps = 23/519 (4%)
Query: 6 STNTATKPQQSTAAQKKQPPPNRHQRQ-RAQRHVA---GSRHVPSGMRAD---------F 52
+ +T + P++ P P +H R A+RH + S +P G A+ F
Sbjct: 79 TNSTNSTPKRFFKRPFPPPSPAKHIRAVLARRHGSVKPNSSAIPEGSEAEGGGVGLDKSF 138
Query: 53 GYDKDFDERYSLGKLLGHGQFGYTYVGVDKAS---GDRVAVKRLEKSKMVLPAAVEDVKQ 109
G+ K F +Y LG +G G FGYT K G +VAVK + K+KM A+EDV++
Sbjct: 139 GFSKSFASKYELGDEVGRGHFGYTCAAKFKKGDNKGQQVAVKVIPKAKMTTAIAIEDVRR 198
Query: 110 EVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN--RYTEKDAAVVVR 167
EV IL+A++GH N+ FY+A++D VYIVMELCEGGELLDRIL+ +YTE+DA V+
Sbjct: 199 EVKILRALSGHNNLPHFYDAYEDHDNVYIVMELCEGGELLDRILSRGGKYTEEDAKTVMI 258
Query: 168 QMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAY 227
Q+L V A CHL+G+VHRD+KPENFLF S + S LKA DFGLSD+++P ++ +DIVGSAY
Sbjct: 259 QILNVVAFCHLQGVVHRDLKPENFLFTSKEDTSQLKAIDFGLSDYVRPDERLNDIVGSAY 318
Query: 228 YVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPT 287
YVAPEVL R ++D+WS+GVI YILLCG RPFW +TE GIF+ VL+ P F PWP
Sbjct: 319 YVAPEVLHRSYSTEADIWSVGVIVYILLCGSRPFWARTESGIFRAVLKADPSFDDPPWPL 378
Query: 288 ISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRF 347
+S+ A+DFVK+LL KDPR RLTAAQALSHPW+++ +A ++P+DI V +R ++ S
Sbjct: 379 LSSEARDFVKRLLNKDPRKRLTAAQALSHPWIKDSNDA-KVPMDILVFKLMRAYLRSSSL 437
Query: 348 KQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEI 407
++ ALRAL+ L +EL L++QF ++ KNG+ISLE ++ AL K + +K+SR+ E
Sbjct: 438 RKAALRALSKTLTVDELFYLREQFALLEPSKNGTISLENIKSALMKMATDAMKDSRIPEF 497
Query: 408 LQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELR 467
L + + +DF EF AA L VHQLE D+W+ ++ A+E F+ + + I +EL
Sbjct: 498 LGQLSALQYRRMDFEEFCAAALSVHQLEA--LDRWEQHARCAYELFEKEGNRPIMIDELA 555
Query: 468 MHTGLRGS--IDPLLEEADIDKDGKISLSEFRRLLRTAS 504
GL S + +L + DGK+S F +LL S
Sbjct: 556 SELGLGPSVPVHAVLHDWLRHTDGKLSFLGFVKLLHGVS 594
>At5g19360 calcium-dependent protein kinase - like
Length = 523
Score = 430 bits (1105), Expect = e-120
Identities = 214/452 (47%), Positives = 307/452 (67%), Gaps = 13/452 (2%)
Query: 56 KDFDERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILK 115
+D Y+LGK LG GQFG T++ KA+G + A K + K K+V +EDV++EV I+
Sbjct: 62 EDVKSSYTLGKELGRGQFGVTHLCTQKATGLQFACKTIAKRKLVNKEDIEDVRREVQIMH 121
Query: 116 AVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAA 174
+ G N+V+ A++D V++VMELC GGEL DRI+A Y+E+ AA ++R ++++
Sbjct: 122 HLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKGHYSERAAASLLRTIVQIIH 181
Query: 175 ECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVL 234
CH G++HRD+KPENFL S E+SPLKATDFGLS F KPG+ F DIVGSAYY+APEVL
Sbjct: 182 TCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKPGEVFKDIVGSAYYIAPEVL 241
Query: 235 KRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKD 294
+RK GP++D+WSIGV+ YILLCG PFW ++E+GIF +L + DF PWP IS AKD
Sbjct: 242 RRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILSGQVDFSSDPWPVISPQAKD 301
Query: 295 FVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRA 354
V+K+L DP+ RLTAAQ L+HPW++E GEA ++P+D +V++ L+QF + FK+ ALR
Sbjct: 302 LVRKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDNAVMSRLKQFKAMNNFKKVALRV 361
Query: 355 LASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSN 414
+A L+EEE+ LK+ F +D D +G+I+LEE+RQ LAK +L E V ++++A D++
Sbjct: 362 IAGCLSEEEIMGLKEMFKGMDTDNSGTITLEELRQGLAKQ-GTRLSEYEVQQLMEAADAD 420
Query: 415 TDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRG 474
+G +D+ EF+AAT+H+++L+ + +AF+ FD D GYIT EEL G
Sbjct: 421 GNGTIDYGEFIAATMHINRLDREE------HLYSAFQHFDKDNSGYITTEELEQALREFG 474
Query: 475 -----SIDPLLEEADIDKDGKISLSEFRRLLR 501
I ++ E D D DG+I+ EF ++R
Sbjct: 475 MNDGRDIKEIISEVDGDNDGRINYEEFVAMMR 506
>At5g04870 calcium-dependent protein kinase
Length = 610
Score = 429 bits (1102), Expect = e-120
Identities = 224/505 (44%), Positives = 323/505 (63%), Gaps = 20/505 (3%)
Query: 9 TATKPQQSTAAQKKQPPPNRHQRQRAQRHVAGSRHVPSGMRADFGYDK---DFDERYSLG 65
T + + T + K PP + ++ + + V+ + G+R + + +F E YSLG
Sbjct: 99 TKQETKSETKPESKPDPPAKPKKPKHMKRVSSA-----GLRTESVLQRKTENFKEFYSLG 153
Query: 66 KLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVVQ 125
+ LG GQFG T++ V+K +G A K + K K++ VEDV++E+ I+ +AGH NV+
Sbjct: 154 RKLGQGQFGTTFLCVEKTTGKEFACKSIAKRKLLTDEDVEDVRREIQIMHHLAGHPNVIS 213
Query: 126 FYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRGLVHR 184
A++D V++VME C GGEL DRI+ YTE+ AA + R ++ V CH G++HR
Sbjct: 214 IKGAYEDVVAVHLVMECCAGGELFDRIIQRGHYTERKAAELTRTIVGVVEACHSLGVMHR 273
Query: 185 DMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDV 244
D+KPENFLF S EDS LK DFGLS F KP F D+VGS YYVAPEVL+++ GP++DV
Sbjct: 274 DLKPENFLFVSKHEDSLLKTIDFGLSMFFKPDDVFTDVVGSPYYVAPEVLRKRYGPEADV 333
Query: 245 WSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDP 304
WS GVI YILL G PFW +TE GIF++VL DF PWP+IS +AKD V+K+L++DP
Sbjct: 334 WSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLDFSSDPWPSISESAKDLVRKMLVRDP 393
Query: 305 RARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEEL 364
+ RLTA Q L HPWV+ G A + P+D +VL+ ++QF ++FK+ ALR +A L+EEE+
Sbjct: 394 KKRLTAHQVLCHPWVQVDGVAPDKPLDSAVLSRMKQFSAMNKFKKMALRVIAESLSEEEI 453
Query: 365 ADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEF 424
A LK+ F+ ID DK+G I+ EE++ L K + LKES +L+++QA D + G +D+ EF
Sbjct: 454 AGLKEMFNMIDADKSGQITFEELKAGL-KRVGANLKESEILDLMQAADVDNSGTIDYKEF 512
Query: 425 VAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRG----SIDPLL 480
+AATLH++++E D AAF FD D GYITP+EL+ G I+ L+
Sbjct: 513 IAATLHLNKIERED------HLFAAFTYFDKDGSGYITPDELQQACEEFGVEDVRIEELM 566
Query: 481 EEADIDKDGKISLSEFRRLLRTASM 505
+ D D DG+I +EF +++ S+
Sbjct: 567 RDVDQDNDGRIDYNEFVAMMQKGSI 591
>At3g10660 calmodulin-domain protein kinase CDPK isoform 2
Length = 646
Score = 428 bits (1101), Expect = e-120
Identities = 223/507 (43%), Positives = 324/507 (62%), Gaps = 17/507 (3%)
Query: 7 TNTATKPQQSTAAQKKQPPPNRHQRQRAQRHVAGSRHVPSGMRADFGYDK---DFDERYS 63
T + +KP+ + + P + +H+ R +G+R + + +F E YS
Sbjct: 130 TKSESKPETTKPETTSETKPETKAEPQKPKHMR--RVSSAGLRTESVLQRKTENFKEFYS 187
Query: 64 LGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENV 123
LG+ LG GQFG T++ ++K +G+ A K + K K++ VEDV++E+ I+ +AGH NV
Sbjct: 188 LGRKLGQGQFGTTFLCLEKGTGNEYACKSISKRKLLTDEDVEDVRREIQIMHHLAGHPNV 247
Query: 124 VQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRGLV 182
+ A++D V++VMELC GGEL DRI+ YTE+ AA + R ++ V CH G++
Sbjct: 248 ISIKGAYEDVVAVHLVMELCSGGELFDRIIQRGHYTERKAAELARTIVGVLEACHSLGVM 307
Query: 183 HRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQS 242
HRD+KPENFLF S EDS LK DFGLS F KP + F D+VGS YYVAPEVL+++ GP+S
Sbjct: 308 HRDLKPENFLFVSREEDSLLKTIDFGLSMFFKPDEVFTDVVGSPYYVAPEVLRKRYGPES 367
Query: 243 DVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIK 302
DVWS GVI YILL G PFW +TE GIF++VL DF PWP+IS +AKD V+K+L++
Sbjct: 368 DVWSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLDFSSDPWPSISESAKDLVRKMLVR 427
Query: 303 DPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEE 362
DP+ RLTA Q L HPWV+ G A + P+D +VL+ ++QF ++FK+ ALR +A L+EE
Sbjct: 428 DPKRRLTAHQVLCHPWVQIDGVAPDKPLDSAVLSRMKQFSAMNKFKKMALRVIAESLSEE 487
Query: 363 ELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFT 422
E+A LK F ID D +G I+ EE++ L K + LKES +L+++QA D + G +D+
Sbjct: 488 EIAGLKQMFKMIDADNSGQITFEELKAGL-KRVGANLKESEILDLMQAADVDNSGTIDYK 546
Query: 423 EFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRG----SIDP 478
EF+AATLH++++E D AAF FD D+ G+ITP+EL+ G I+
Sbjct: 547 EFIAATLHLNKIERED------HLFAAFSYFDKDESGFITPDELQQACEEFGVEDARIEE 600
Query: 479 LLEEADIDKDGKISLSEFRRLLRTASM 505
++ + D DKDG+I +EF +++ S+
Sbjct: 601 MMRDVDQDKDGRIDYNEFVAMMQKGSI 627
>At4g23650 calcium-dependent protein kinase (CDPK6)
Length = 529
Score = 422 bits (1085), Expect = e-118
Identities = 211/446 (47%), Positives = 301/446 (67%), Gaps = 13/446 (2%)
Query: 62 YSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHE 121
Y G+ LG GQFG TY+ K + +VA K + ++V +EDV++EV I+ ++GH
Sbjct: 78 YEFGRELGRGQFGVTYLVTHKETKQQVACKSIPTRRLVHKDDIEDVRREVQIMHHLSGHR 137
Query: 122 NVVQFYNAFDDDSYVYIVMELCEGGELLDRILANR-YTEKDAAVVVRQMLKVAAECHLRG 180
N+V A++D V ++MELCEGGEL DRI++ Y+E+ AA + RQM+ V CH G
Sbjct: 138 NIVDLKGAYEDRHSVNLIMELCEGGELFDRIISKGLYSERAAADLCRQMVMVVHSCHSMG 197
Query: 181 LVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGP 240
++HRD+KPENFLF S E+SPLKATDFGLS F KPG KF D+VGSAYYVAPEVLKR GP
Sbjct: 198 VMHRDLKPENFLFLSKDENSPLKATDFGLSVFFKPGDKFKDLVGSAYYVAPEVLKRNYGP 257
Query: 241 QSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLL 300
++D+WS GVI YILL G PFW + E GIF +L+ + DF PWP +S+ AKD V+K+L
Sbjct: 258 EADIWSAGVILYILLSGVPPFWGENETGIFDAILQGQLDFSADPWPALSDGAKDLVRKML 317
Query: 301 IKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILN 360
DP+ RLTAA+ L+HPW+RE GEAS+ P+D +VL+ ++QF ++ K+ AL+ +A L+
Sbjct: 318 KYDPKDRLTAAEVLNHPWIREDGEASDKPLDNAVLSRMKQFRAMNKLKKMALKVIAENLS 377
Query: 361 EEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVD 420
EEE+ LK+ F ++D D NG ++LEE+R L K L K+ E+ + ++++A D + DG +D
Sbjct: 378 EEEIIGLKEMFKSLDTDNNGIVTLEELRTGLPK-LGSKISEAEIRQLMEAADMDGDGSID 436
Query: 421 FTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRM-----HTGLRGS 475
+ EF++AT+H++++E D AF+ FD D GYIT EEL + + G S
Sbjct: 437 YLEFISATMHMNRIERED------HLYTAFQFFDNDNSGYITMEELELAMKKYNMGDDKS 490
Query: 476 IDPLLEEADIDKDGKISLSEFRRLLR 501
I ++ E D D+DGKI+ EF +++
Sbjct: 491 IKEIIAEVDTDRDGKINYEEFVAMMK 516
>At1g49580 CDPK-related protein kinase, putative
Length = 606
Score = 422 bits (1084), Expect = e-118
Identities = 228/474 (48%), Positives = 312/474 (65%), Gaps = 11/474 (2%)
Query: 47 GMRADFGYDKDFDERYSLGKLLGHGQFGYTYVGVDKAS---GDRVAVKRLEKSKMVLPAA 103
G+ FG+ K+F R LG+ +G G FGYT K G VAVK + KSKM A
Sbjct: 135 GLDKRFGFSKEFHSRVELGEEIGRGHFGYTCSAKFKKGELKGQVVAVKIIPKSKMTTAIA 194
Query: 104 VEDVKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN--RYTEKD 161
+EDV++EV IL+A++GH+N+VQFY+AF+D++ VYI MELCEGGELLDRILA +Y+E D
Sbjct: 195 IEDVRREVKILQALSGHKNLVQFYDAFEDNANVYIAMELCEGGELLDRILARGGKYSEND 254
Query: 162 AAVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHD 221
A V+ Q+L V A CH +G+VHRD+KPENFL+ S E+S LKA DFGLSDF++P ++ +D
Sbjct: 255 AKPVIIQILNVVAFCHFQGVVHRDLKPENFLYTSKEENSQLKAIDFGLSDFVRPDERLND 314
Query: 222 IVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFR 281
IVGSAYYVAPEVL R ++DVWSIGVI YILLCG RPFW +TE GIF+ VL+ P F
Sbjct: 315 IVGSAYYVAPEVLHRSYTTEADVWSIGVIAYILLCGSRPFWARTESGIFRAVLKADPSFD 374
Query: 282 RKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQF 341
PWP +S+ AKDFVK+LL KDPR R++A+QAL HPW+R IP DI + ++ +
Sbjct: 375 EPPWPFLSSDAKDFVKRLLFKDPRRRMSASQALMHPWIRAYNTDMNIPFDILIFRQMKAY 434
Query: 342 VTYSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKE 401
+ S ++ ALRAL+ L ++E+ LK QF + +K+G I+++ +R ALA + + +KE
Sbjct: 435 LRSSSLRKAALRALSKTLIKDEILYLKTQFSLLAPNKDGLITMDTIRMALASNATEAMKE 494
Query: 402 SRVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDS-DKWQLRSQAAFEKFDLDKDGY 460
SR+ E L ++ +DF EF AA ++VHQ H+S D W+ + A+E FD + +
Sbjct: 495 SRIPEFLALLNGLQYRGMDFEEFCAAAINVHQ---HESLDCWEQSIRHAYELFDKNGNRA 551
Query: 461 ITPEELRMHTGLRGSI--DPLLEEADIDKDGKISLSEFRRLLRTASMGSQTVTT 512
I EEL G+ SI +L + DGK+S F +LL S+ + TT
Sbjct: 552 IVIEELASELGVGPSIPVHSVLHDWIRHTDGKLSFFGFVKLLHGVSVRASGKTT 605
>At4g21940 calcium-dependent protein kinase like protein
Length = 554
Score = 414 bits (1065), Expect = e-116
Identities = 224/507 (44%), Positives = 326/507 (64%), Gaps = 21/507 (4%)
Query: 6 STNTATKPQQSTAAQKKQPPPNRHQRQRAQRHVAGSRHVPSGMR-ADFGYDKDFDE---R 61
S T Q + Q++ P N+ Q ++HV P R + K F+E
Sbjct: 45 SPQIPTTTQSNHHHQQESKPVNQ---QIEKKHVLTQPLKPIVFRETETILGKPFEEIRKL 101
Query: 62 YSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHE 121
Y+LGK LG GQFG TY + ++G+ A K + K K+ ++DVK+E+ I++ ++G E
Sbjct: 102 YTLGKELGRGQFGITYTCKENSTGNTYACKSILKRKLTRKQDIDDVKREIQIMQYLSGQE 161
Query: 122 NVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRG 180
N+V+ A++D +++VMELC G EL DRI+A Y+EK AA V+R +L V CH G
Sbjct: 162 NIVEIKGAYEDRQSIHLVMELCGGSELFDRIIAQGHYSEKAAAGVIRSVLNVVQICHFMG 221
Query: 181 LVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGP 240
++HRD+KPENFL ST E++ LKATDFGLS FI+ GK + DIVGSAYYVAPEVL+R G
Sbjct: 222 VIHRDLKPENFLLASTDENAMLKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLRRSYGK 281
Query: 241 QSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLL 300
+ D+WS G+I YILLCG PFW +TE GIF E+++ + DF +PWP+IS +AKD V+KLL
Sbjct: 282 EIDIWSAGIILYILLCGVPPFWSETEKGIFNEIIKGEIDFDSQPWPSISESAKDLVRKLL 341
Query: 301 IKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILN 360
KDP+ R++AAQAL HPW+R GGEA + PID +VL+ ++QF ++ K+ AL+ +A L+
Sbjct: 342 TKDPKQRISAAQALEHPWIR-GGEAPDKPIDSAVLSRMKQFRAMNKLKKLALKVIAESLS 400
Query: 361 EEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVD 420
EEE+ LK F +D DK+G+I+ EE++ LAK L KL E+ V ++++A D + +G +D
Sbjct: 401 EEEIKGLKTMFANMDTDKSGTITYEELKNGLAK-LGSKLTEAEVKQLMEAADVDGNGTID 459
Query: 421 FTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL-----RMHTGLRGS 475
+ EF++AT+H ++ D D+ + AF+ FD D G+IT +EL G S
Sbjct: 460 YIEFISATMHRYRF---DRDEHVFK---AFQYFDKDNSGFITMDELESAMKEYGMGDEAS 513
Query: 476 IDPLLEEADIDKDGKISLSEFRRLLRT 502
I ++ E D D DG+I+ EF ++R+
Sbjct: 514 IKEVIAEVDTDNDGRINYEEFCAMMRS 540
>At3g19100 CDPK-related kinase
Length = 599
Score = 412 bits (1060), Expect = e-115
Identities = 228/488 (46%), Positives = 314/488 (63%), Gaps = 19/488 (3%)
Query: 25 PPNRHQRQRAQRHVAGSRHVPSGMRADFGYDKDFDERYSLGKLLGHGQFGYTYVGVDKAS 84
P R Q++ +R G + FG+ K+ R LG+ +G G FGYT K
Sbjct: 114 PAARQQKEEEEREEVG-------LDKRFGFSKELQSRIELGEEIGRGHFGYTCSAKFKKG 166
Query: 85 ---GDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVME 141
VAVK + KSKM ++EDV++EV IL+A++GH+N+VQFY+AF+D++ VYIVME
Sbjct: 167 ELKDQEVAVKVIPKSKMTSAISIEDVRREVKILRALSGHQNLVQFYDAFEDNANVYIVME 226
Query: 142 LCEGGELLDRILAN--RYTEKDAAVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTRED 199
LC GGELLDRILA +Y+E DA V+ Q+L V A CHL+G+VHRD+KPENFL+ S E+
Sbjct: 227 LCGGGELLDRILARGGKYSEDDAKAVLIQILNVVAFCHLQGVVHRDLKPENFLYTSKEEN 286
Query: 200 SPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRR 259
S LK DFGLSDF++P ++ +DIVGSAYYVAPEVL R ++DVWSIGVI YILLCG R
Sbjct: 287 SMLKVIDFGLSDFVRPDERLNDIVGSAYYVAPEVLHRSYTTEADVWSIGVIAYILLCGSR 346
Query: 260 PFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWV 319
PFW +TE GIF+ VL+ P F PWP++S AKDFVK+LL KDPR R+TA+QAL HPW+
Sbjct: 347 PFWARTESGIFRAVLKADPSFDEPPWPSLSFEAKDFVKRLLYKDPRKRMTASQALMHPWI 406
Query: 320 REGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAIDVDKN 379
G + +IP DI + ++ ++ S ++ AL AL+ L +EL LK QF + +KN
Sbjct: 407 -AGYKKIDIPFDILIFKQIKAYLRSSSLRKAALMALSKTLTTDELLYLKAQFAHLAPNKN 465
Query: 380 GSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDS 439
G I+L+ +R ALA + + +KESR+ + L ++ +DF EF AA++ VHQ H+S
Sbjct: 466 GLITLDSIRLALATNATEAMKESRIPDFLALLNGLQYKGMDFEEFCAASISVHQ---HES 522
Query: 440 -DKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRGSI--DPLLEEADIDKDGKISLSEF 496
D W+ + A+E F+++ + I EEL G+ SI +L + DGK+S F
Sbjct: 523 LDCWEQSIRHAYELFEMNGNRVIVIEELASELGVGSSIPVHTILNDWIRHTDGKLSFLGF 582
Query: 497 RRLLRTAS 504
+LL S
Sbjct: 583 VKLLHGVS 590
>At1g50700 hypothetical protein
Length = 521
Score = 412 bits (1059), Expect = e-115
Identities = 221/523 (42%), Positives = 328/523 (62%), Gaps = 28/523 (5%)
Query: 1 MGSCFSTN--TATKPQQSTAAQKKQPPPNRHQRQRAQRHVAGSRHVP--------SGMRA 50
MG+C + KPQQ+ + R Q + G+ P SG A
Sbjct: 1 MGNCLAKKYGLVMKPQQNGERSVEIENRRRSTHQDPSKISTGTNQPPPWRNPAKHSGAAA 60
Query: 51 DFGYDKDFDER---YSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDV 107
+K +++ Y+L K LG GQFG TY+ +K++G R A K + K K+V ED+
Sbjct: 61 IL--EKPYEDVKLFYTLSKELGRGQFGVTYLCTEKSTGKRFACKSISKKKLVTKGDKEDM 118
Query: 108 KQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVV 166
++E+ I++ ++G N+V+F A++D+ V +VMELC GGEL DRILA Y+E+ AA V
Sbjct: 119 RREIQIMQHLSGQPNIVEFKGAYEDEKAVNLVMELCAGGELFDRILAKGHYSERAAASVC 178
Query: 167 RQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSA 226
RQ++ V CH G++HRD+KPENFL S E + +KATDFGLS FI+ G+ + DIVGSA
Sbjct: 179 RQIVNVVNICHFMGVMHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGRVYKDIVGSA 238
Query: 227 YYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWP 286
YYVAPEVLKR+ G + D+WS G+I YILL G PFW +TE GIF +L + DF +PWP
Sbjct: 239 YYVAPEVLKRRYGKEIDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGEIDFESQPWP 298
Query: 287 TISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSR 346
+ISN+AKD V+++L +DP+ R++AA+ L HPW+REGGEAS+ PID +VL+ ++QF ++
Sbjct: 299 SISNSAKDLVRRMLTQDPKRRISAAEVLKHPWLREGGEASDKPIDSAVLSRMKQFRAMNK 358
Query: 347 FKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLE 406
K+ AL+ +A ++ EE+ LK F ID D +G+I+ EE+++ LAK L +L E+ V +
Sbjct: 359 LKKLALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKEGLAK-LGSRLTEAEVKQ 417
Query: 407 ILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL 466
++ A D + +G +D+ EF+ AT+H H+LE +++ AF+ FD D GYIT +EL
Sbjct: 418 LMDAADVDGNGSIDYIEFITATMHRHRLESNEN------VYKAFQHFDKDGSGYITTDEL 471
Query: 467 -----RMHTGLRGSIDPLLEEADIDKDGKISLSEFRRLLRTAS 504
G +I +L + D D DG+I+ EF ++R+ +
Sbjct: 472 EAALKEYGMGDDATIKEILSDVDADNDGRINYDEFCAMMRSGN 514
>At2g46700 calcium/calmodulin-dependent protein kinase CaMK4
Length = 595
Score = 409 bits (1052), Expect = e-114
Identities = 216/459 (47%), Positives = 309/459 (67%), Gaps = 9/459 (1%)
Query: 52 FGYDKDFDERYSLGKLLGHGQFGYTYVGVDKASGDR---VAVKRLEKSKMVLPAAVEDVK 108
FGY K+F +Y LGK +G G FG+T G K + +AVK + K+KM A+EDV+
Sbjct: 133 FGYGKNFGAKYELGKEVGRGHFGHTCSGRGKKGDIKDHPIAVKIISKAKMTTAIAIEDVR 192
Query: 109 QEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN--RYTEKDAAVVV 166
+EV +LK+++GH+ ++++Y+A +D + VYIVMELC+GGELLDRILA +Y E DA +V
Sbjct: 193 REVKLLKSLSGHKYLIKYYDACEDANNVYIVMELCDGGELLDRILARGGKYPEDDAKAIV 252
Query: 167 RQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSA 226
Q+L V + CHL+G+VHRD+KPENFLF S+REDS LK DFGLSDFI+P ++ +DIVGSA
Sbjct: 253 VQILTVVSFCHLQGVVHRDLKPENFLFTSSREDSDLKLIDFGLSDFIRPDERLNDIVGSA 312
Query: 227 YYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWP 286
YYVAPEVL R ++D+WSIGVITYILLCG RPFW +TE GIF+ VLR +P++ PWP
Sbjct: 313 YYVAPEVLHRSYSLEADIWSIGVITYILLCGSRPFWARTESGIFRTVLRTEPNYDDVPWP 372
Query: 287 TISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSR 346
+ S+ KDFVK+LL KD R R++A QAL+HPW+R+ ++ IP+DI + ++ ++ +
Sbjct: 373 SCSSEGKDFVKRLLNKDYRKRMSAVQALTHPWLRD--DSRVIPLDILIYKLVKAYLHATP 430
Query: 347 FKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLE 406
++ AL+ALA L E EL L+ QF + +K+GS+SLE + AL ++ + ++ESRV E
Sbjct: 431 LRRAALKALAKALTENELVYLRAQFMLLGPNKDGSVSLENFKTALMQNATDAMRESRVPE 490
Query: 407 ILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL 466
IL ++S + F EF AA + +HQLE D+ W+ + A F+ F+ + + IT EEL
Sbjct: 491 ILHTMESLAYRKMYFEEFCAAAISIHQLEAVDA--WEEIATAGFQHFETEGNRVITIEEL 548
Query: 467 RMHTGLRGSIDPLLEEADIDKDGKISLSEFRRLLRTASM 505
+ S L + DGK+S F + L ++
Sbjct: 549 ARELNVGASAYGHLRDWVRSSDGKLSYLGFTKFLHGVTL 587
>At3g20410 calmodulin-domain protein kinase CDPK isoform 9
Length = 541
Score = 409 bits (1050), Expect = e-114
Identities = 206/453 (45%), Positives = 302/453 (66%), Gaps = 13/453 (2%)
Query: 62 YSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHE 121
Y+LGK LG GQFG TY+ + ++G + A K + K K+V A +D+++E+ I++ ++G
Sbjct: 91 YTLGKELGRGQFGVTYLCTENSTGKKYACKSISKKKLVTKADKDDMRREIQIMQHLSGQP 150
Query: 122 NVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRG 180
N+V+F A++D+ V +VMELC GGEL DRI+A YTE+ AA V RQ++ V CH G
Sbjct: 151 NIVEFKGAYEDEKAVNLVMELCAGGELFDRIIAKGHYTERAAASVCRQIVNVVKICHFMG 210
Query: 181 LVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGP 240
++HRD+KPENFL S E + +KATDFGLS FI+ GK + DIVGSAYYVAPEVL+R+ G
Sbjct: 211 VLHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLRRRYGK 270
Query: 241 QSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLL 300
+ D+WS G+I YILL G PFW +TE GIF +L DF +PWP+IS++AKD V+++L
Sbjct: 271 EVDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGHIDFESQPWPSISSSAKDLVRRML 330
Query: 301 IKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILN 360
DP+ R++AA L HPW+REGGEAS+ PID +VL+ ++QF ++ K+ AL+ +A ++
Sbjct: 331 TADPKRRISAADVLQHPWLREGGEASDKPIDSAVLSRMKQFRAMNKLKKLALKVIAENID 390
Query: 361 EEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVD 420
EE+ LK F ID D +G+I+ EE+++ LAK L KL E+ V +++ A D + +G +D
Sbjct: 391 TEEIQGLKAMFANIDTDNSGTITYEELKEGLAK-LGSKLTEAEVKQLMDAADVDGNGSID 449
Query: 421 FTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL-----RMHTGLRGS 475
+ EF+ AT+H H+LE +++ AF+ FD D GYIT +EL G +
Sbjct: 450 YIEFITATMHRHRLESNEN------LYKAFQHFDKDSSGYITIDELESALKEYGMGDDAT 503
Query: 476 IDPLLEEADIDKDGKISLSEFRRLLRTASMGSQ 508
I +L + D D DG+I+ EF ++R+ + Q
Sbjct: 504 IKEVLSDVDSDNDGRINYEEFCAMMRSGNPQQQ 536
>At2g41140 CDPK-related kinase 1 (CRK1)
Length = 576
Score = 409 bits (1050), Expect = e-114
Identities = 229/516 (44%), Positives = 317/516 (61%), Gaps = 20/516 (3%)
Query: 6 STNTATKPQQSTAAQKKQPPPNRHQRQRAQRHVAGSR----HVPSGMRAD------FGYD 55
S++ ++ P + P P +H R R + +P G + FG+
Sbjct: 57 SSSVSSTPLRIFKRPFPPPSPAKHIRAFLARRYGSVKPNEVSIPEGKECEIGLDKSFGFS 116
Query: 56 KDFDERYSLGKLLGHGQFGYTYVGVDKAS---GDRVAVKRLEKSKMVLPAAVEDVKQEVN 112
K F Y + +G G FGYT K G VAVK + KSKM A+EDV +EV
Sbjct: 117 KQFASHYEIDGEVGRGHFGYTCSAKGKKGSLKGQEVAVKVIPKSKMTTAIAIEDVSREVK 176
Query: 113 ILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRIL--ANRYTEKDAAVVVRQML 170
+L+A+ GH+N+VQFY+AF+DD VYIVMELC+GGELLD+IL +Y+E DA V+ Q+L
Sbjct: 177 MLRALTGHKNLVQFYDAFEDDENVYIVMELCKGGELLDKILQRGGKYSEDDAKKVMVQIL 236
Query: 171 KVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVA 230
V A CHL+G+VHRD+KPENFLF + E SPLKA DFGLSD++KP ++ +DIVGSAYYVA
Sbjct: 237 SVVAYCHLQGVVHRDLKPENFLFSTKDETSPLKAIDFGLSDYVKPDERLNDIVGSAYYVA 296
Query: 231 PEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISN 290
PEVL R G ++D+WSIGVI YILLCG RPFW +TE GIF+ VL+ +P+F PWP++S
Sbjct: 297 PEVLHRTYGTEADMWSIGVIAYILLCGSRPFWARTESGIFRAVLKAEPNFEEAPWPSLSP 356
Query: 291 AAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQF 350
A DFVK+LL KD R RLTAAQAL HPW+ G +IP D+ + ++ ++ + ++
Sbjct: 357 EAVDFVKRLLNKDYRKRLTAAQALCHPWL-VGSHELKIPSDMIIYKLVKVYIMSTSLRKS 415
Query: 351 ALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQA 410
AL ALA L +LA L++QF + KNG IS++ + A+ K + +K+SRV + +
Sbjct: 416 ALAALAKTLTVPQLAYLREQFTLLGPSKNGYISMQNYKTAILKSSTDAMKDSRVFDFVHM 475
Query: 411 IDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHT 470
I +DF EF A+ L V+QLE ++ W+ ++ A+E F+ D + I EEL
Sbjct: 476 ISCLQYKKLDFEEFCASALSVYQLEAMET--WEQHARRAYELFEKDGNRPIMIEELASEL 533
Query: 471 GLRGS--IDPLLEEADIDKDGKISLSEFRRLLRTAS 504
GL S + +L++ DGK+S F RLL S
Sbjct: 534 GLGPSVPVHVVLQDWIRHSDGKLSFLGFVRLLHGVS 569
>At3g56760 calcium-dependent protein kinase-like
Length = 577
Score = 408 bits (1049), Expect = e-114
Identities = 230/516 (44%), Positives = 321/516 (61%), Gaps = 20/516 (3%)
Query: 6 STNTATKPQQSTAAQKKQPPPNRHQRQR-AQRHVA---GSRHVPSGMRAD------FGYD 55
S++ ++ P + P P +H R A+RH + +P G + FG+
Sbjct: 58 SSSVSSTPLRIFKRPFPPPSPAKHIRALLARRHGSVKPNEASIPEGSECEVGLDKKFGFS 117
Query: 56 KDFDERYSLGKLLGHGQFGYTYVGVDKAS---GDRVAVKRLEKSKMVLPAAVEDVKQEVN 112
K F Y + +G G FGYT K G VAVK + KSKM A+EDV++EV
Sbjct: 118 KQFASHYEIDGEVGRGHFGYTCSAKGKKGSLKGQDVAVKVIPKSKMTTAIAIEDVRREVK 177
Query: 113 ILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRIL--ANRYTEKDAAVVVRQML 170
IL+A+ GH+N+VQFY+AF+DD VYIVMELC+GGELLD+IL +Y+E DA V+ Q+L
Sbjct: 178 ILRALTGHKNLVQFYDAFEDDENVYIVMELCQGGELLDKILQRGGKYSEVDAKKVMIQIL 237
Query: 171 KVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVA 230
V A CHL+G+VHRD+KPENFLF + E SPLKA DFGLSD+++P ++ +DIVGSAYYVA
Sbjct: 238 SVVAYCHLQGVVHRDLKPENFLFTTKDESSPLKAIDFGLSDYVRPDERLNDIVGSAYYVA 297
Query: 231 PEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISN 290
PEVL R G ++D+WSIGVI YILLCG RPFW ++E GIF+ VL+ +P+F PWP++S
Sbjct: 298 PEVLHRTYGTEADMWSIGVIAYILLCGSRPFWARSESGIFRAVLKAEPNFEEAPWPSLSP 357
Query: 291 AAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQF 350
A DFVK+LL KD R RLTAAQAL HPW+ G +IP D+ + ++ ++ S ++
Sbjct: 358 DAVDFVKRLLNKDYRKRLTAAQALCHPWL-VGSHELKIPSDMIIYKLVKVYIMSSSLRKS 416
Query: 351 ALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQA 410
AL ALA L +L L++QF+ + KNG IS++ + A+ K + K+SRVL+ +
Sbjct: 417 ALAALAKTLTVPQLTYLQEQFNLLGPSKNGYISMQNYKTAILKSSTEATKDSRVLDFVHM 476
Query: 411 IDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHT 470
I +DF EF A+ L V+QLE ++ W+ ++ A+E ++ D + I EEL
Sbjct: 477 ISCLQYKKLDFEEFCASALSVYQLEAMET--WEQHARRAYELYEKDGNRVIMIEELATEL 534
Query: 471 GLRGS--IDPLLEEADIDKDGKISLSEFRRLLRTAS 504
GL S + +L++ DGK+S F RLL S
Sbjct: 535 GLGPSVPVHVVLQDWIRHSDGKLSFLGFVRLLHGVS 570
>At2g38910 putative calcium-dependent protein kinase
Length = 583
Score = 408 bits (1048), Expect = e-114
Identities = 212/503 (42%), Positives = 316/503 (62%), Gaps = 19/503 (3%)
Query: 12 KPQQSTAAQKKQPPPNRHQRQRAQRHVAGSRHVPSGMRADFGYDK---DFDERYSLGKLL 68
+P + K+ P ++ Q++ A R +G++ D + + + YS+G+ L
Sbjct: 85 EPPPKPITENKEDPNSKPQKKEAHM----KRMASAGLQIDSVLGRKTENLKDIYSVGRKL 140
Query: 69 GHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVVQFYN 128
G GQFG T++ VDK +G A K + K K+ P VEDV++E+ I+ ++GH NV+Q
Sbjct: 141 GQGQFGTTFLCVDKKTGKEFACKTIAKRKLTTPEDVEDVRREIQIMHHLSGHPNVIQIVG 200
Query: 129 AFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRGLVHRDMK 187
A++D V++VME+C GGEL DRI+ YTEK AA + R ++ V CH G++HRD+K
Sbjct: 201 AYEDAVAVHVVMEICAGGELFDRIIQRGHYTEKKAAELARIIVGVIEACHSLGVMHRDLK 260
Query: 188 PENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDVWSI 247
PENFLF S E++ LK DFGLS F KPG+ F D+VGS YYVAPEVL++ + DVWS
Sbjct: 261 PENFLFVSGDEEAALKTIDFGLSVFFKPGETFTDVVGSPYYVAPEVLRKHYSHECDVWSA 320
Query: 248 GVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRAR 307
GVI YILL G PFWD+TE GIF++VL+ DF +PWP++S +AKD V+++LI+DP+ R
Sbjct: 321 GVIIYILLSGVPPFWDETEQGIFEQVLKGDLDFISEPWPSVSESAKDLVRRMLIRDPKKR 380
Query: 308 LTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADL 367
+T + L HPW R G A + P+D +VL+ L+QF ++ K+ A++ +A L+EEE+A L
Sbjct: 381 MTTHEVLCHPWARVDGVALDKPLDSAVLSRLQQFSAMNKLKKIAIKVIAESLSEEEIAGL 440
Query: 368 KDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAA 427
K+ F ID D +G I+LEE+++ L + + LK+S +L ++QA D + G +D+ EF+AA
Sbjct: 441 KEMFKMIDTDNSGHITLEELKKGLDR-VGADLKDSEILGLMQAADIDNSGTIDYGEFIAA 499
Query: 428 TLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRG----SIDPLLEEA 483
+H++++E+ D AF FD D GYIT +EL+ G +D +L E
Sbjct: 500 MVHLNKIEKED------HLFTAFSYFDQDGSGYITRDELQQACKQFGLADVHLDDILREV 553
Query: 484 DIDKDGKISLSEFRRLLRTASMG 506
D D DG+I SEF +++ G
Sbjct: 554 DKDNDGRIDYSEFVDMMQDTGFG 576
>At4g04720 putative calcium dependent protein kinase
Length = 531
Score = 405 bits (1040), Expect = e-113
Identities = 216/504 (42%), Positives = 318/504 (62%), Gaps = 18/504 (3%)
Query: 20 QKKQPPPNRHQRQRAQRHVAGSRHVPSGMR-ADFGYDKDFDER---YSLGKLLGHGQFGY 75
+K Q P + Q + ++ P +R D K F++ YSLGK LG GQFG
Sbjct: 34 RKPQTPTPKPMTQPIHQQISTPSSNPVSVRDPDTILGKPFEDIRKFYSLGKELGRGQFGI 93
Query: 76 TYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVVQFYNAFDDDSY 135
TY+ + +G+ A K + K K++ EDVK+E+ I++ ++G N+V+ A++D
Sbjct: 94 TYMCKEIGTGNTYACKSILKRKLISKQDKEDVKREIQIMQYLSGQPNIVEIKGAYEDRQS 153
Query: 136 VYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRGLVHRDMKPENFLFK 194
+++VMELC GGEL DRI+A Y+E+ AA ++R ++ V CH G+VHRD+KPENFL
Sbjct: 154 IHLVMELCAGGELFDRIIAQGHYSERAAAGIIRSIVNVVQICHFMGVVHRDLKPENFLLS 213
Query: 195 STREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYIL 254
S E++ LKATDFGLS FI+ GK + DIVGSAYYVAPEVL+R G + D+WS GVI YIL
Sbjct: 214 SKEENAMLKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLRRSYGKEIDIWSAGVILYIL 273
Query: 255 LCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQAL 314
L G PFW + E GIF EV++ + DF +PWP+IS +AKD V+K+L KDP+ R+TAAQ L
Sbjct: 274 LSGVPPFWAENEKGIFDEVIKGEIDFVSEPWPSISESAKDLVRKMLTKDPKRRITAAQVL 333
Query: 315 SHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAI 374
HPW++ GGEA + PID +VL+ ++QF ++ K+ AL+ +A L+EEE+ LK F I
Sbjct: 334 EHPWIK-GGEAPDKPIDSAVLSRMKQFRAMNKLKKLALKVIAESLSEEEIKGLKTMFANI 392
Query: 375 DVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQL 434
D DK+G+I+ EE++ L + L +L E+ V ++++A D + +G +D+ EF++AT+H ++L
Sbjct: 393 DTDKSGTITYEELKTGLTR-LGSRLSETEVKQLMEAADVDGNGTIDYYEFISATMHRYKL 451
Query: 435 EEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL-----RMHTGLRGSIDPLLEEADIDKDG 489
+ + AF+ FD D G+IT +EL G SI ++ E D D DG
Sbjct: 452 DRDE------HVYKAFQHFDKDNSGHITRDELESAMKEYGMGDEASIKEVISEVDTDNDG 505
Query: 490 KISLSEFRRLLRTASMGSQTVTTP 513
+I+ EF ++R+ S Q P
Sbjct: 506 RINFEEFCAMMRSGSTQPQGKLLP 529
>At3g51850 calcium-dependent like protein kinase
Length = 528
Score = 402 bits (1034), Expect = e-112
Identities = 214/511 (41%), Positives = 319/511 (61%), Gaps = 22/511 (4%)
Query: 1 MGSCFSTNTATKPQQSTAAQKKQPPPNRHQRQRAQRHVAGSRHVPSGMRADFGYDKDFDE 60
MG+C + A + K + H R+ A G + P + +D + + ++
Sbjct: 1 MGNCCRSPAAVAREDV----KSNYSGHDHARKDA---AGGKKSAPIRVLSDVPKE-NIED 52
Query: 61 RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
RY L + LG G+FG TY+ ++++S D +A K + K K+ +EDVK+EV I+K +
Sbjct: 53 RYLLDRELGRGEFGVTYLCIERSSRDLLACKSISKRKLRTAVDIEDVKREVAIMKHLPKS 112
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLR 179
++V A +DD+ V++VMELCEGGEL DRI+A YTE+ AA V + +++V CH
Sbjct: 113 SSIVTLKEACEDDNAVHLVMELCEGGELFDRIVARGHYTERAAAGVTKTIVEVVQLCHKH 172
Query: 180 GLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSG 239
G++HRD+KPENFLF + +E+SPLKA DFGLS F KPG+KF +IVGS YY+APEVLKR G
Sbjct: 173 GVIHRDLKPENFLFANKKENSPLKAIDFGLSIFFKPGEKFSEIVGSPYYMAPEVLKRNYG 232
Query: 240 PQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKL 299
P+ D+WS GVI YILLCG PFW ++E G+ + +LR DF+R+PWP IS AK+ V+++
Sbjct: 233 PEIDIWSAGVILYILLCGVPPFWAESEQGVAQAILRGVIDFKREPWPNISETAKNLVRQM 292
Query: 300 LIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASIL 359
L DP+ RLTA Q L HPW++ +A +P+ V + L+QF +RFK+ ALR +A L
Sbjct: 293 LEPDPKRRLTAKQVLEHPWIQNAKKAPNVPLGDVVKSRLKQFSVMNRFKRKALRVIAEFL 352
Query: 360 NEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLV 419
+ EE+ D+K F+ +D D +G +S+EE++ L +D S +L ES V +++A+D+ G +
Sbjct: 353 STEEVEDIKVMFNKMDTDNDGIVSIEELKAGL-RDFSTQLAESEVQMLIEAVDTKGKGTL 411
Query: 420 DFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRGSID-- 477
D+ EFVA +LH+ ++ + + AF FD D +GYI P+EL G D
Sbjct: 412 DYGEFVAVSLHLQKVANDE------HLRKAFSYFDKDGNGYILPQELCDALKEDGGDDCV 465
Query: 478 ----PLLEEADIDKDGKISLSEFRRLLRTAS 504
+ +E D DKDG+IS EF +++T +
Sbjct: 466 DVANDIFQEVDTDKDGRISYEEFAAMMKTGT 496
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,759,791
Number of Sequences: 26719
Number of extensions: 522922
Number of successful extensions: 4121
Number of sequences better than 10.0: 977
Number of HSP's better than 10.0 without gapping: 359
Number of HSP's successfully gapped in prelim test: 618
Number of HSP's that attempted gapping in prelim test: 2106
Number of HSP's gapped (non-prelim): 1251
length of query: 520
length of database: 11,318,596
effective HSP length: 104
effective length of query: 416
effective length of database: 8,539,820
effective search space: 3552565120
effective search space used: 3552565120
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0119.8


