

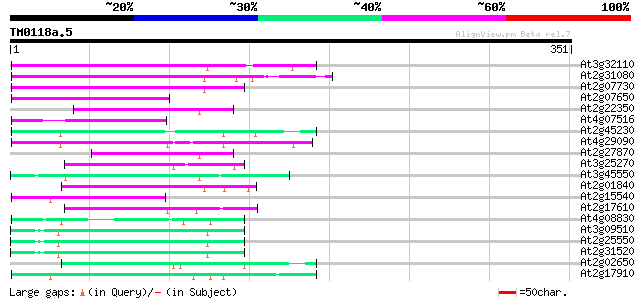
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118a.5
(351 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g32110 non-LTR reverse transcriptase, putative 95 7e-20
At2g31080 putative non-LTR retroelement reverse transcriptase 87 1e-17
At2g07730 putative non-LTR retroelement reverse transcriptase 86 3e-17
At2g07650 putative non-LTR retrolelement reverse transcriptase 75 4e-14
At2g22350 putative non-LTR retroelement reverse transcriptase 70 2e-12
At4g07516 putative protein 67 2e-11
At2g45230 putative non-LTR retroelement reverse transcriptase 66 2e-11
At4g29090 putative protein 62 4e-10
At2g27870 putative non-LTR retroelement reverse transcriptase 60 1e-09
At3g25270 hypothetical protein 60 2e-09
At3g45550 putative protein 57 2e-08
At2g01840 putative non-LTR retroelement reverse transcriptase 56 3e-08
At2g15540 putative non-LTR retroelement reverse transcriptase 55 7e-08
At2g17610 putative non-LTR retroelement reverse transcriptase 54 1e-07
At4g08830 putative protein 52 6e-07
At3g09510 putative non-LTR reverse transcriptase 51 8e-07
At2g25550 putative non-LTR retroelement reverse transcriptase 51 8e-07
At2g31520 putative non-LTR retroelement reverse transcriptase 49 4e-06
At2g02650 putative reverse transcriptase 49 4e-06
At2g17910 putative non-LTR retroelement reverse transcriptase 47 2e-05
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 94.7 bits (234), Expect = 7e-20
Identities = 63/206 (30%), Positives = 95/206 (45%), Gaps = 19/206 (9%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDHEGQN-GRCWQWVWKLKVPEKM*LFVWLILNKALQTNG 60
WG + G +T + A+ L N D Q+ + VWK++ PE++ +F+WL++N+A+ TN
Sbjct: 1548 WGMTSDGRFTVKSAFAMLTNDDSPRQDMSSLYGRVWKVQAPERVRVFLWLVVNQAIMTNS 1607
Query: 61 NRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRL-GATTWRSFATMDVEAWITS 119
R R HL S C E+ILH LRDCP +W R+ +SF TM + W+ S
Sbjct: 1608 ERKRRHLCDSDVCQVCRGGIESILHVLRDCPAMSGIWDRIVPRRLQQSFFTMSLLEWLYS 1667
Query: 120 LAR-------SNHAISFLSGIWSVWLWRNNMCFEETPWNLAEAWRRLSHVHDEMLQTSQD 172
R S+ + F +W W WR + F E R+ + D ++ S
Sbjct: 1668 NLRQGLMTEGSDWSTMFAMAVWWGWKWRCSNIFGEN----KTCRDRVRFIKDLAVEVSIA 1723
Query: 173 WSP------GDLNSLLCVRWHPPARG 192
+S L +RW PP G
Sbjct: 1724 YSREVELRLSGLRVNKPIRWTPPMEG 1749
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 87.4 bits (215), Expect = 1e-17
Identities = 64/215 (29%), Positives = 98/215 (44%), Gaps = 26/215 (12%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDHEGQN-GRCWQWVWKLKVPEKM*LFVWLILNKALQTNG 60
W + G +T R AY L + N G + +WKL PE++ +F+WL+ + TN
Sbjct: 872 WKGTQDGAFTVRSAYSLLQGDVGDRPNMGSFFNRIWKLITPERVRVFIWLVSQNVIMTNV 931
Query: 61 NRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLR-LGATTWRSFATMDVEAWI-T 118
R R HL+++ S C+ AEETILH LRDCP +W R L F + + W+ T
Sbjct: 932 ERVRRHLSENAICSVCNGAEETILHVLRDCPAMEPIWRRLLPLRRHHEFFSQSLLEWLFT 991
Query: 119 SL--ARSNHAISFLSGIWSVWLWR-------NNMCFEETPW--NLAEAWRRLSHVHDEML 167
++ + F GIW W WR +C + + ++AE RR+ HV
Sbjct: 992 NMDPVKGIWPTLFGMGIWWAWKWRCCDVFGERKICRDRLKFIKDMAEEVRRV-HV----- 1045
Query: 168 QTSQDWSPGDLNSLLCVRWHPPARGGSN*MWMTVT 202
+ P + +RW P+ G W+ +T
Sbjct: 1046 -GAVGNRPNGVRVERMIRWQVPSDG-----WVKIT 1074
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 85.9 bits (211), Expect = 3e-17
Identities = 50/149 (33%), Positives = 73/149 (48%), Gaps = 3/149 (2%)
Query: 2 WGPSPTGCYTAREAYGWLN-NLDHEGQNGRCWQWVWKLKVPEKM*LFVWLILNKALQTNG 60
W + G +T R AY L + G + +WKL PE++ +F+WL+ + + TN
Sbjct: 613 WKGTQNGDFTVRSAYELLKPEAEERPLIGSFLKQIWKLVAPERVRVFIWLVSHMVIMTNV 672
Query: 61 NRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWRSFATMDVEAWITSL 120
R R HL+ + S C+ A+E+ILH LRDCP +W RL ++ E T+L
Sbjct: 673 ERVRRHLSDIATCSVCNGADESILHVLRDCPAMTPIWQRLLPQRRQNEFFSQFEWLFTNL 732
Query: 121 --ARSNHAISFLSGIWSVWLWRNNMCFEE 147
A+ + F GIW W WR F E
Sbjct: 733 DPAKGDWPTLFSMGIWWAWKWRCGDVFGE 761
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 75.5 bits (184), Expect = 4e-14
Identities = 37/100 (37%), Positives = 55/100 (55%), Gaps = 1/100 (1%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDHEGQN-GRCWQWVWKLKVPEKM*LFVWLILNKALQTNG 60
WG S G + Y +L + Q+ R + VW + PE++ LF+WL+ +A+ TN
Sbjct: 534 WGESQDGRFKVSTTYSFLTRDEAPRQDMKRFFDRVWSVTAPERVRLFLWLVAQQAIMTNQ 593
Query: 61 NRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRL 100
R R HL+ + C A+E+I+H LRDCP +WLRL
Sbjct: 594 ERHRRHLSTTTICQVCKGAKESIIHVLRDCPAMEGIWLRL 633
>At2g22350 putative non-LTR retroelement reverse transcriptase
Length = 321
Score = 70.1 bits (170), Expect = 2e-12
Identities = 37/103 (35%), Positives = 53/103 (50%), Gaps = 3/103 (2%)
Query: 41 PEKM*LFVWLILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRL 100
PE++ +F+WL++ + + TN R+R HL+ + C EETILH LRDCP +W RL
Sbjct: 3 PERVRVFLWLVVQQVIITNVERYRRHLSDTRVCQICQGGEETILHVLRDCPAMAGIWSRL 62
Query: 101 -GATTWRSFATMDVEAWI--TSLARSNHAISFLSGIWSVWLWR 140
R F T + WI R + F+ +W W WR
Sbjct: 63 VPRDQIRQFFTASLLEWIYKNLRERGSWPTVFVMAVWWGWKWR 105
>At4g07516 putative protein
Length = 740
Score = 66.6 bits (161), Expect = 2e-11
Identities = 35/97 (36%), Positives = 50/97 (51%), Gaps = 13/97 (13%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDHEGQNGRCWQWVWKLKVPEKM*LFVWLILNKALQTNGN 61
WG S +T + AY +L VW + VPE++ F+WL + + + TN
Sbjct: 473 WGGSINRDFTVKLAYMFLTR-------------VWSIVVPERVRAFIWLPVIQVVMTNVE 519
Query: 62 RFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWL 98
R R HL+ + S C EETI+H LRDCP R +W+
Sbjct: 520 RKRRHLSDTDLYSVCKQGEETIIHILRDCPAMRGIWI 556
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 66.2 bits (160), Expect = 2e-11
Identities = 51/225 (22%), Positives = 85/225 (37%), Gaps = 49/225 (21%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDHEGQNGR---------CWQWVWKLKVPEKM*LFVWLIL 52
W S +G Y+ + Y + + ++ N + +Q +WKL VP K+ F+W +
Sbjct: 1012 WEYSRSGHYSVKSGYWVMTEIINQRNNPQEVLQPSLDPIFQQIWKLDVPPKIHHFLWRCV 1071
Query: 53 NKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWRSFATMD 112
N L N HLA+ S RC + ET+ H L CP +R W
Sbjct: 1072 NNCLSVASNLAYRHLAREKSCVRCPSHGETVNHLLFKCPFARLTW------AISPLPAPP 1125
Query: 113 VEAWITSLARSNHAISFLSG---------------IWSVWLWRNNMCFEETPWNL----- 152
W SL R+ H + + +W +W RN++ F+ +
Sbjct: 1126 GGEWAESLFRNMHHVLSVHKSQPEESDHHALIPWILWRLWKNRNDLVFKGREFTAPQVIL 1185
Query: 153 -----AEAWRRLSHVHDEMLQTSQDWSPGDLNSLLCVRWHPPARG 192
+AW ++ +++D CV+W PP+ G
Sbjct: 1186 KATEDMDAWNNRKEPQPQVTSSTRD---------RCVKWQPPSHG 1221
>At4g29090 putative protein
Length = 575
Score = 62.4 bits (150), Expect = 4e-10
Identities = 51/211 (24%), Positives = 87/211 (41%), Gaps = 25/211 (11%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDHEGQNGR---------CWQWVWKLKVPEKM*LFVWLIL 52
W + +G YT + Y L + ++ + + +Q +WK + K+ F+W L
Sbjct: 215 WDYTSSGDYTVKSGYWVLTQIINKRSSPQEVSEPSLNPIYQKIWKSQTSPKIQHFLWKCL 274
Query: 53 NKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELW------LRLGATTWR 106
+ +L G HL++ + RC + +ET+ H L C +R W + LG W
Sbjct: 275 SNSLPVAGALAYRHLSKESACIRCPSCKETVNHLLFKCTFARLTWAISSIPIPLGG-EWA 333
Query: 107 SFATMDVEAWITSLARSNHAISFLSG-----IWSVWLWRNNMCFEETPWNLAEAWRRLSH 161
+++ W+ +L N S +W +W RN + F +N E RR
Sbjct: 334 DSIYVNL-YWVFNLGNGNPQWEKASQLVPWLLWRLWKNRNELVFRGREFNAQEVLRRAED 392
Query: 162 VHDEMLQTSQDWSPG---DLNSLLCVRWHPP 189
+E ++ S G +N C RW PP
Sbjct: 393 DLEEWRIRTEAESCGTKPQVNRSSCGRWRPP 423
>At2g27870 putative non-LTR retroelement reverse transcriptase
Length = 314
Score = 60.5 bits (145), Expect = 1e-09
Identities = 34/97 (35%), Positives = 44/97 (45%), Gaps = 8/97 (8%)
Query: 52 LNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRL-GATTWRSFAT 110
+ + L TN R R HL+ S C AE+TI+H LRDCP +W+RL A R F T
Sbjct: 1 MQQVLMTNAERRRRHLSDSDICQICKGAEKTIIHILRDCPAMEGIWIRLVPAGKRREFFT 60
Query: 111 MDVEAWI-------TSLARSNHAISFLSGIWSVWLWR 140
+ W+ S + F IW W WR
Sbjct: 61 QSLLEWLFANLGDRRKTCESTWSTLFALSIWWAWKWR 97
>At3g25270 hypothetical protein
Length = 343
Score = 59.7 bits (143), Expect = 2e-09
Identities = 38/122 (31%), Positives = 59/122 (48%), Gaps = 10/122 (8%)
Query: 35 VWKLKVPEKM*LFVWLILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSR 94
+WKLK K+ F+W +L+ AL T N R H+ P RC +ET H DC +++
Sbjct: 18 IWKLKTAPKIKHFLWKLLSGALATGDNLKRRHIRNHPQCHRCCQEDETSQHLFFDCFYAQ 77
Query: 95 ELWLRLG------ATTWRSFATMDVEAWITSLARSNHAISFLSGIWSVW-LW--RNNMCF 145
++W G TT + T +E ++S + F IW +W LW RN + F
Sbjct: 78 QVWRASGIPHQELRTTGITMET-KMELLLSSCLANRQPQLFNLAIWILWRLWKSRNQLVF 136
Query: 146 EE 147
++
Sbjct: 137 QQ 138
>At3g45550 putative protein
Length = 851
Score = 56.6 bits (135), Expect = 2e-08
Identities = 51/194 (26%), Positives = 75/194 (38%), Gaps = 21/194 (10%)
Query: 1 IWGPSPTGCYTAREAYGWLNNLDHEGQNGRCWQ---------WVWKLKVPEKM*LFVWLI 51
IW + TG YT R Y WL+ D + +W L + K+ F+W I
Sbjct: 625 IWSYNSTGDYTVRSGY-WLSTHDPSNTIPTMAKPHGSVDLKTKIWNLPIMPKLKHFLWRI 683
Query: 52 LNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWR-SFAT 110
L+KAL T + P RC E+I H L CP + W +R S +
Sbjct: 684 LSKALPTTDRLTTRGMRIDPGCPRCRRENESINHALFTCPFATMAWRLSDTPLYRSSILS 743
Query: 111 MDVEAWI---------TSLARSNHAISFLSGIWSVWLWRNNMCFEETPWNLAEAWRRLSH 161
++E I T++ S I F +W +W RNN+ F + + R
Sbjct: 744 NNIEDNISNILLLLQNTTITDSQKLIPFWL-LWRIWKARNNVVFNNLRESPSITVVRAKA 802
Query: 162 VHDEMLQTSQDWSP 175
+E L +Q P
Sbjct: 803 ETNEWLNATQTQGP 816
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 55.8 bits (133), Expect = 3e-08
Identities = 36/134 (26%), Positives = 65/134 (47%), Gaps = 12/134 (8%)
Query: 33 QWVWKLKVPEKM*LFVWLILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPH 92
Q +W+LK+ K+ F+W L+ AL T ++ P+ RC A+ETI H + C +
Sbjct: 1386 QEIWRLKITPKIKHFIWRCLSGALSTTTQLRNRNIPADPTCQRCCNADETINHIIFTCSY 1445
Query: 93 SRELWLRLG-ATTWRSFATMDVEAWITSL--ARSNHAISFLSGI------WSVWLWRNNM 143
++ +W + + R T ++E I + + N + L+G+ W +W RN
Sbjct: 1446 AQVVWRSANFSGSNRLCFTDNLEENIRLILQGKKNQNLPILNGLMPFWIMWRLWKSRNEY 1505
Query: 144 CFEET---PWNLAE 154
F++ PW +A+
Sbjct: 1506 LFQQLDRFPWKVAQ 1519
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 54.7 bits (130), Expect = 7e-08
Identities = 34/104 (32%), Positives = 46/104 (43%), Gaps = 8/104 (7%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDH-------EGQNGRCWQ-WVWKLKVPEKM*LFVWLILN 53
W + +G YT + Y +L +G + Q VWK+K K F W L+
Sbjct: 885 WSFTKSGNYTVKSGYWAARDLSRPTCDLPFQGPSVSALQAQVWKIKTTRKFKHFEWQCLS 944
Query: 54 KALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELW 97
L TN F H+ RC A EE+I H L CP SR++W
Sbjct: 945 GCLATNQRLFSRHIGTEKVCPRCGAEEESINHLLFLCPPSRQIW 988
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 53.9 bits (128), Expect = 1e-07
Identities = 35/129 (27%), Positives = 58/129 (44%), Gaps = 9/129 (6%)
Query: 35 VWKLKVPEKM*LFVWLILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSR 94
+WK P K+ F W + AL T GN R L + RC A E + H L C S+
Sbjct: 501 IWKQNAPPKIKHFWWRSAHNALPTAGNLKRRRLITDDTCQRCGEASEDVNHLLFQCRVSK 560
Query: 95 ELW------LRLGATTWRSFATMDVEA--WITSLARSNHAISFLSGIWSVWLWRNNMCFE 146
E+W L G + + ++E+ + AR + ++ G W +W RN++ F
Sbjct: 561 EIWEQAHIKLCPGDSLMSNSFNQNLESIQKLNQSARKDVSLFPFIG-WRIWKMRNDLIFN 619
Query: 147 ETPWNLAEA 155
W++ ++
Sbjct: 620 NKRWSIPDS 628
>At4g08830 putative protein
Length = 947
Score = 51.6 bits (122), Expect = 6e-07
Identities = 42/157 (26%), Positives = 62/157 (38%), Gaps = 29/157 (18%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDHEGQNGRC--WQWVWKLKVPEKM*LFVWLILNKALQTN 59
WG S G +T + AY L DH+ + + +W++ E++ F+W
Sbjct: 662 WGYSADGVFTVKSAYRLLTE-DHDPRPNMAAFFDRLWRVVALERVKTFLW---------- 710
Query: 60 GNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWRS--FATMDVEAWI 117
H+ + C +ETILH L+DCP +W RL RS F + W+
Sbjct: 711 ------HIGDTSVCQVCKGGDETILHVLKDCPSIAGIWRRL-VQVQRSYDFFNGSLFGWL 763
Query: 118 -TSLARSN------HAISFLSGIWSVWLWRNNMCFEE 147
+L N A F +W W WR F E
Sbjct: 764 YVNLGMKNAETGYAWATLFAIVVWWSWKWRCGYVFGE 800
>At3g09510 putative non-LTR reverse transcriptase
Length = 484
Score = 51.2 bits (121), Expect = 8e-07
Identities = 44/167 (26%), Positives = 67/167 (39%), Gaps = 23/167 (13%)
Query: 1 IWGPSPTGCYTAREAYGWLNNLDHEGQNG-----------RCWQWVWKLKVPEKM*LFVW 49
IW + TG YT R Y WL L H+ +W L + K+ F+W
Sbjct: 120 IWNYNTTGEYTVRSGY-WL--LTHDPSTNIPAINPPHGSIDLKTRIWNLPIMPKLKHFLW 176
Query: 50 LILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWRS-F 108
L++AL T + PS RC E+I H L CP + W ++ R+
Sbjct: 177 RALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFTCPFATMAWRLSDSSLIRNQL 236
Query: 109 ATMDVEAWITSLAR--------SNHAISFLSGIWSVWLWRNNMCFEE 147
+ D E I+++ H + + IW +W RNN+ F +
Sbjct: 237 MSNDFEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARNNVVFNK 283
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 51.2 bits (121), Expect = 8e-07
Identities = 44/167 (26%), Positives = 67/167 (39%), Gaps = 23/167 (13%)
Query: 1 IWGPSPTGCYTAREAYGWLNNLDHEGQNG-----------RCWQWVWKLKVPEKM*LFVW 49
IW + TG YT R Y WL L H+ +W L + K+ F+W
Sbjct: 1386 IWNYNTTGEYTVRSGY-WL--LTHDPSTNIPAINPPHGSIDLKTRIWNLPIMPKLKHFLW 1442
Query: 50 LILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWRS-F 108
L++AL T + PS RC E+I H L CP + W ++ R+
Sbjct: 1443 RALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFTCPFATMAWRLSDSSLIRNQL 1502
Query: 109 ATMDVEAWITSLAR--------SNHAISFLSGIWSVWLWRNNMCFEE 147
+ D E I+++ H + + IW +W RNN+ F +
Sbjct: 1503 MSNDFEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARNNVVFNK 1549
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 48.9 bits (115), Expect = 4e-06
Identities = 43/167 (25%), Positives = 66/167 (38%), Gaps = 23/167 (13%)
Query: 1 IWGPSPTGCYTAREAYGWLNNLDHEGQNG-----------RCWQWVWKLKVPEKM*LFVW 49
IW + TG YT R Y WL L H+ +W L + K+ F+W
Sbjct: 1160 IWNYNTTGEYTVRSGY-WL--LTHDPSTNIPAINPPHGSIDLKTRIWNLPIMPKLKHFLW 1216
Query: 50 LILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWRS-F 108
L++AL T + P RC E+I H L CP + W ++ R+
Sbjct: 1217 RALSQALATTERLTTRGMRIDPICPRCHRENESINHALFTCPFATMAWWLSDSSLIRNQL 1276
Query: 109 ATMDVEAWITSLAR--------SNHAISFLSGIWSVWLWRNNMCFEE 147
+ D E I+++ H + + IW +W RNN+ F +
Sbjct: 1277 MSNDFEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARNNVVFNK 1323
>At2g02650 putative reverse transcriptase
Length = 365
Score = 48.9 bits (115), Expect = 4e-06
Identities = 46/187 (24%), Positives = 72/187 (37%), Gaps = 36/187 (19%)
Query: 33 QWVWKLKVPEKM*LFVWLILNKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPH 92
Q +WKL V K+ F+W + AL TN ++ P RC EETI H + +CP+
Sbjct: 35 QAIWKLHVAPKIKHFLWRCVTGALATNTRLRSRNIDADPICQRCCIEEETIHHIMFNCPY 94
Query: 93 SRELWLRLG---ATTW---RSFATMDVEAWITSLARSNHAISFLSGIWSVW-LWRNNMCF 145
++ +W W SF S ++ +++ W +W LW++ F
Sbjct: 95 TQSVWRSANIIIGNQWGPPSSFEDNLNRLIQLSKTQTTNSLDRFLPFWIMWRLWKSRNVF 154
Query: 146 ------------------EETPW-NLAEAWRRLS-HVHDEMLQTSQDWSPGDLNSLLCVR 185
+ T W N E + HV +QTS+ S +
Sbjct: 155 LFQQKCQSPDYEARKGIQDATEWLNANETTENTNVHVATNPIQTSRRDSS---------Q 205
Query: 186 WHPPARG 192
W+PP G
Sbjct: 206 WNPPPEG 212
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 47.0 bits (110), Expect = 2e-05
Identities = 45/208 (21%), Positives = 78/208 (36%), Gaps = 18/208 (8%)
Query: 2 WGPSPTGCYTAREAYGWLNNLDH---------EGQNGRCWQWVWKLKVPEKM*LFVWLIL 52
W S G Y+ + Y +L+ H + + +W L K+ +F+W L
Sbjct: 982 WLHSHNGLYSVKTGYEFLSKQVHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWKAL 1041
Query: 53 NKALQTNGNRFRCHLAQSPSRSRCSAAEETILHCLRDCPHSRELWLRLGATTWRSFATMD 112
+ A+ + C ETI H L +CP +R++W ++ S +
Sbjct: 1042 HGAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQVWAITHLSSAGSEFSNS 1101
Query: 113 V---EAWITSLARSN---HAISFLSG--IWSVWLWRNNMCFEETPWNLAEAWRRLSHVHD 164
V + + L + N H + F+S +W +W RN + FE + +
Sbjct: 1102 VYTNMSRLIDLTQQNDLPHHLRFVSPWILWFLWKNRNALLFEGKGSITTTLVDKAYEAYH 1161
Query: 165 EMLQTSQDWSPGDLNSLLCVRWHPPARG 192
E ++Q D L +W PP G
Sbjct: 1162 EWF-SAQTHMQNDEKHLKITKWCPPLPG 1188
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.342 0.147 0.572
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,203,301
Number of Sequences: 26719
Number of extensions: 326587
Number of successful extensions: 1148
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 1065
Number of HSP's gapped (non-prelim): 65
length of query: 351
length of database: 11,318,596
effective HSP length: 100
effective length of query: 251
effective length of database: 8,646,696
effective search space: 2170320696
effective search space used: 2170320696
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.5 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0118a.5


