

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118a.3
(328 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
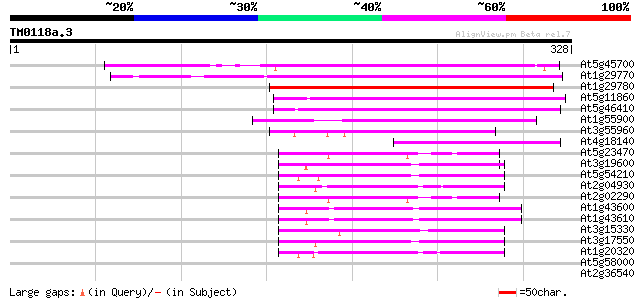
Score E
Sequences producing significant alignments: (bits) Value
At5g45700 unknown protein 199 1e-51
At1g29770 hypothetical protein 185 3e-47
At1g29780 hypothetical protein 173 1e-43
At5g11860 unknown protein 120 1e-27
At5g46410 unknown protein 103 1e-22
At1g55900 unknown protein 83 2e-16
At3g55960 unknown protein 78 8e-15
At4g18140 putative protein 62 6e-10
At5g23470 putative protein 54 9e-08
At3g19600 hypothetical protein 52 3e-07
At5g54210 putative protein 51 8e-07
At2g04930 hypothetical protein 51 1e-06
At2g02290 hypothetical protein 49 3e-06
At1g43600 hypothetical protein 49 3e-06
At1g43610 hypothetical protein 49 4e-06
At3g15330 hypothetical protein 46 2e-05
At3g17550 hypothetical protein 45 7e-05
At1g20320 hypothetical protein 44 9e-05
At5g58000 putative protein 35 0.056
At2g36540 hypothetical protein 34 0.12
>At5g45700 unknown protein
Length = 272
Score = 199 bits (507), Expect = 1e-51
Identities = 127/279 (45%), Positives = 165/279 (58%), Gaps = 35/279 (12%)
Query: 56 TKSIKDIRRNNNRRWRRKTPIKNAVAASSAVLASIHRRITKFFSKLARLSPDRKRASYKI 115
TKSI +RN RR R+ + A++ V+A+I++ I + L +KI
Sbjct: 11 TKSISRHQRNYLRRHRKSQIGCSPSPATTTVIATINKSIYRCHRTFLCLFSRHATKGFKI 70
Query: 116 LKKTSPQDDSICRTLVFDGGDNDPSPPLPLLPPSISHRR----------TVFLDLDETLV 165
LK P++D F P S+ H R T+ LDLDETLV
Sbjct: 71 LK---PRED-------FQNS-----------PYSLFHERSGKSFDETKKTIVLDLDETLV 109
Query: 166 HSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASL 225
HS+ P +DFVV P IDG + F+V KRPGVDEFL+ + K+++VVFTA LREYASL
Sbjct: 110 HSSMEKPEVPYDFVVNPKIDGQILTFFVIKRPGVDEFLKKIGEKYQIVVFTAGLREYASL 169
Query: 226 VLDRVD-RNRLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAI 284
VLD++D R++S YRD+C + DG+LVKDL RDL+RVVIVDDNPNS+A QPENA
Sbjct: 170 VLDKLDPERRVISRSFYRDACSEIDGRLVKDLGFVMRDLRRVVIVDDNPNSYALQPENAF 229
Query: 285 LIRPFVDDVFDRELWKLRMFFDGVDCC--DDMRDAVKQF 321
I+PF DD+ D EL KL F G DC +DMR A+K+F
Sbjct: 230 PIKPFSDDLEDVELKKLGEFLYG-DCVKFEDMRVALKEF 267
>At1g29770 hypothetical protein
Length = 278
Score = 185 bits (470), Expect = 3e-47
Identities = 110/266 (41%), Positives = 159/266 (59%), Gaps = 13/266 (4%)
Query: 60 KDIRRNNNRRWRRKTPIKNAVAASSAVLASIHRRITKFFSKLARLSPDRKRASYKILKKT 119
K R+N N + R + AVAA+ ++ S++ I F ++L R + L+
Sbjct: 20 KHARKNRNHQSRC---LAAAVAAAGSIFTSLNMSIFTFHNRLLRCVS-------RFLRLA 69
Query: 120 SPQDDSICRTLVFDGGDNDPSPPLPLLPPSISHRRTVFLDLDETLVHSNTAPPPE-SFDF 178
+ + R G PL+ + +RT+FLDLDETLVHS PP + DF
Sbjct: 70 TTSSATPTRRPTMKQGYKKLHKREPLIRRN-DKKRTIFLDLDETLVHSTMEPPIRVNVDF 128
Query: 179 VVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSH 238
+VR I+G + +V KRPGV EFLE ++ + V +FTA L EYAS VLD++D+NR++S
Sbjct: 129 MVRIKIEGAVIPMFVVKRPGVTEFLERISKNYRVAIFTAGLPEYASQVLDKLDKNRVISQ 188
Query: 239 RLYRDSCKQEDGKLVKDLSL-AGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRE 297
RLYRDSC + +G+ KDLSL A DL V++VDDNP S++ QP+N + I+PF+DD+ D+E
Sbjct: 189 RLYRDSCTEVNGRYAKDLSLVAKNDLGSVLLVDDNPFSYSLQPDNGVPIKPFMDDMEDQE 248
Query: 298 LWKLRMFFDGVDCCDDMRDAVKQFFT 323
L KL FFDG +D+RDA + +
Sbjct: 249 LMKLAEFFDGCYQYEDLRDAAAELLS 274
>At1g29780 hypothetical protein
Length = 247
Score = 173 bits (438), Expect = 1e-43
Identities = 90/167 (53%), Positives = 118/167 (69%), Gaps = 1/167 (0%)
Query: 153 RRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEV 212
+RT+ LDLDETLVH+ T P DF+V ++ M +V KRPGV EFLE L ++V
Sbjct: 75 KRTIILDLDETLVHATTHLPGVKHDFMVMVKMEREIMPIFVVKRPGVTEFLERLGENYKV 134
Query: 213 VVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSL-AGRDLKRVVIVDD 271
VVFTA L EYAS VLD++D+N ++S RLYRDSC + +GK VKDLSL G+DL+ +IVDD
Sbjct: 135 VVFTAGLEEYASQVLDKLDKNGVISQRLYRDSCTEVNGKYVKDLSLVVGKDLRSALIVDD 194
Query: 272 NPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAV 318
NP+S++ QPEN + I+ FVDD+ D+EL L F + +DMRDAV
Sbjct: 195 NPSSYSLQPENGVPIKAFVDDLKDQELLNLVEFLESCYAYEDMRDAV 241
>At5g11860 unknown protein
Length = 305
Score = 120 bits (301), Expect = 1e-27
Identities = 68/172 (39%), Positives = 104/172 (59%), Gaps = 2/172 (1%)
Query: 155 TVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVV 214
++ LDLDETLVHS P E DF + YV+ RP + EF+E ++ FE+++
Sbjct: 113 SLVLDLDETLVHSTLEPCGE-VDFTFPVNFNEEEHMVYVRCRPHLKEFMERVSRLFEIII 171
Query: 215 FTAALREYASLVLDRVD-RNRLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNP 273
FTA+ YA +L+ +D + +L HR+YRDSC DG +KDLS+ GRDL RV+IVD++P
Sbjct: 172 FTASQSIYAEQLLNVLDPKRKLFRHRVYRDSCVFFDGNYLKDLSVLGRDLSRVIIVDNSP 231
Query: 274 NSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQFFTVQ 325
+F Q EN + I + +D D+EL L F + + +D+R + + F ++
Sbjct: 232 QAFGFQVENGVPIESWFNDPSDKELLHLLPFLESLIGVEDVRPMIAKKFNLR 283
>At5g46410 unknown protein
Length = 453
Score = 103 bits (257), Expect = 1e-22
Identities = 66/169 (39%), Positives = 93/169 (54%), Gaps = 2/169 (1%)
Query: 155 TVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVV 214
T+ LDLDETLVHS T DF R + YV++RP + FLE + F VV+
Sbjct: 282 TLVLDLDETLVHS-TLESCNVADFSFRVFFNMQENTVYVRQRPHLYRFLERVGELFHVVI 340
Query: 215 FTAALREYASLVLDRVDRN-RLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNP 273
FTA+ YAS +LD +D + + +S R YRDSC DG KDL++ G DL +V I+D+ P
Sbjct: 341 FTASHSIYASQLLDILDPDGKFISQRFYRDSCILLDGIYTKDLTVLGLDLAKVAIIDNCP 400
Query: 274 NSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQFF 322
+ Q N I I+ + DD D L + F + + DD+R + + F
Sbjct: 401 QVYRLQINNGIPIKSWYDDPTDDGLITILPFLETLAVADDVRPIIGRRF 449
>At1g55900 unknown protein
Length = 376
Score = 83.2 bits (204), Expect = 2e-16
Identities = 55/166 (33%), Positives = 85/166 (51%), Gaps = 16/166 (9%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP L P+ H T+ LDL+ETL++++ F KRPGVD F
Sbjct: 180 LPDLHPAEQHVFTLVLDLNETLLYTDWKRERGWRTF----------------KRPGVDAF 223
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
LE L +E+VV++ + Y V +++D N + ++L R + K E+GK +DLS RD
Sbjct: 224 LEHLGKFYEIVVYSDQMEMYVLPVCEKLDPNGYIRYKLARGATKYENGKHYRDLSKLNRD 283
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGV 308
K+++ V N QPEN++ I+P+ + D L L F + V
Sbjct: 284 PKKILFVSANAFESTLQPENSVPIKPYKLEADDTALVDLIPFLEYV 329
>At3g55960 unknown protein
Length = 305
Score = 77.8 bits (190), Expect = 8e-15
Identities = 55/151 (36%), Positives = 81/151 (53%), Gaps = 19/151 (12%)
Query: 153 RRTVFLDLDETLV--HSNTAPPPESFDFVVRPVI--------------DGVPMEFYVK-- 194
R V LDLDETLV + ++ P + + + DG P YV
Sbjct: 97 RLKVVLDLDETLVCAYETSSLPAALRNQAIEAGLKWFELECLSTDKEYDGKPKINYVTVF 156
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGK-LV 253
+RPG+ EFLE L+ ++++FTA L YA ++DR+D +++++RLYR S + V
Sbjct: 157 ERPGLHEFLEQLSEFADLILFTAGLEGYARPLVDRIDTRKVLTNRLYRPSTVSTQYRDHV 216
Query: 254 KDLSLAGRDLKRVVIVDDNPNSFANQPENAI 284
KDL +++ R VIVD+NP SF QP N I
Sbjct: 217 KDLLSTSKNMCRTVIVDNNPFSFLLQPSNGI 247
>At4g18140 putative protein
Length = 307
Score = 61.6 bits (148), Expect = 6e-10
Identities = 37/99 (37%), Positives = 55/99 (55%), Gaps = 1/99 (1%)
Query: 225 LVLDRVDRN-RLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENA 283
++LD +D + + VS R YRDSC DG KDL++ G DL +V IVD+ P + Q N
Sbjct: 205 MLLDILDPDGKFVSQRFYRDSCILSDGIYTKDLTVLGLDLAKVAIVDNCPQVYRLQINNG 264
Query: 284 ILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQFF 322
I I+ + DD D L L F + + +D+R + + F
Sbjct: 265 IPIKSWYDDPTDDGLITLLPFLETLADANDVRPVIAKRF 303
>At5g23470 putative protein
Length = 302
Score = 54.3 bits (129), Expect = 9e-08
Identities = 45/145 (31%), Positives = 71/145 (48%), Gaps = 25/145 (17%)
Query: 158 LDLDETLVHS-NTAPPPESFDFVVRPVID---------GVPMEFYVKKRPGVDEFLEALA 207
LDLD+TL+H+ T+ ES +++ V G P E +K RP V +FL+
Sbjct: 91 LDLDQTLIHTIKTSLLYESEKYIIEEVESRKDIKRFNTGFPEESLIKLRPFVHQFLKECN 150
Query: 208 AKFEVVVFTAALREYASLVLDRVD------RNRLVSHRLYRDSCKQEDGKLVKDLSLAGR 261
F + V+T +YA LVL+ +D NR+++ R + G DL LA
Sbjct: 151 EMFSMYVYTKGGYDYARLVLEMIDPDKFYFGNRVITRR-------ESPGFKTLDLVLA-- 201
Query: 262 DLKRVVIVDDNPNSFANQPENAILI 286
D + +VIVDD + + + +N + I
Sbjct: 202 DERGIVIVDDTSSVWPHDKKNLLQI 226
>At3g19600 hypothetical protein
Length = 601
Score = 52.4 bits (124), Expect = 3e-07
Identities = 43/147 (29%), Positives = 63/147 (42%), Gaps = 22/147 (14%)
Query: 158 LDLDETLVHSNTAP-----------------PPESFDFVVRPVIDGVPMEFYVKKRPGVD 200
LDLD TL+HS P + + VR + +E VK RP +
Sbjct: 93 LDLDLTLIHSVRVPCLSEAEKYLIEEAGSTTREDLWKMKVRGDPISITIEHLVKLRPFLC 152
Query: 201 EFLEALAAKFEVVVFTAALREYASLVLDRVDRNRL-VSHRLYRDSCKQEDGKLVKDLSLA 259
EFL+ F + V+T R YA +L +D +L HR+ + + K L +
Sbjct: 153 EFLKEANEMFTMYVYTKGTRPYAEAILKLIDPKKLYFGHRV----ITRNESPHTKTLDMV 208
Query: 260 GRDLKRVVIVDDNPNSFANQPENAILI 286
D + VVIVDD ++ N N +LI
Sbjct: 209 LADERGVVIVDDTRKAWPNNKSNLVLI 235
Score = 47.4 bits (111), Expect = 1e-05
Identities = 41/146 (28%), Positives = 64/146 (43%), Gaps = 18/146 (12%)
Query: 158 LDLDETLVHSNTAPP-------------PESFDFVVRPVIDGVPMEFYVKKRPGVDEFLE 204
LDLD TL+H+ P + D + + G PMEF K RP + +FL+
Sbjct: 390 LDLDHTLLHTVMVPSLSQAEKYLIEEAGSATRDDLWKIKAVGDPMEFLTKLRPFLRDFLK 449
Query: 205 ALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDS-CKQEDGKLVKDLSLAGRDL 263
F + V+T R YA VL+ +D +L + D + + +K L +
Sbjct: 450 EANEFFTMYVYTKGSRVYAKQVLELIDPKKL----YFGDRVITKTESPHMKTLDFVLAEE 505
Query: 264 KRVVIVDDNPNSFANQPENAILIRPF 289
+ VVIVDD N + + N + I +
Sbjct: 506 RGVVIVDDTRNVWPDHKSNLVDISKY 531
>At5g54210 putative protein
Length = 306
Score = 51.2 bits (121), Expect = 8e-07
Identities = 42/143 (29%), Positives = 67/143 (46%), Gaps = 15/143 (10%)
Query: 158 LDLDETLVHS---NTAPPPESFDFV-------VRPVIDGVPMEFYVKKRPGVDEFLEALA 207
LDLD TL+H+ + E++ +R + G EF +K RP V EFL+
Sbjct: 93 LDLDHTLLHTVMISNLTKEETYLIEEEDSREDLRRLNGGYSSEFLIKLRPFVHEFLKEAN 152
Query: 208 AKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDS-CKQEDGKLVKDLSLAGRDLKRV 266
F + V+T R+YA VL+ +D ++ + D + + +K L L D V
Sbjct: 153 KMFSMYVYTMGDRDYAMNVLNLIDPEKV----YFGDRVITRNESPYIKTLDLVLADECGV 208
Query: 267 VIVDDNPNSFANQPENAILIRPF 289
VIVDD P+ + + N + I +
Sbjct: 209 VIVDDTPHVWPDHKRNLLEITKY 231
>At2g04930 hypothetical protein
Length = 277
Score = 50.8 bits (120), Expect = 1e-06
Identities = 43/147 (29%), Positives = 67/147 (45%), Gaps = 20/147 (13%)
Query: 158 LDLDETLVHSNTAPPPESFD---------------FVVRPVIDGVPMEFYVKKRPGVDEF 202
LDLD TL+HS + + RP+ G P++ +K RP V +F
Sbjct: 71 LDLDHTLLHSKLVSNLSQAERYLIQEASSRTREDLWKFRPI--GHPIDRLIKLRPFVRDF 128
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L+ F + V+T R YA +L+ +D +L R K E ++ K L+L +
Sbjct: 129 LKEANEMFTMFVYTMGSRIYAKAILEMIDPKKLYFGN--RVITKDESPRM-KTLNLVLAE 185
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPF 289
+ VVIVDD + + + N I IR +
Sbjct: 186 ERGVVIVDDTRDIWPHHKNNLIQIRKY 212
>At2g02290 hypothetical protein
Length = 302
Score = 49.3 bits (116), Expect = 3e-06
Identities = 43/145 (29%), Positives = 67/145 (45%), Gaps = 25/145 (17%)
Query: 158 LDLDETLVHS-NTAPPPESFDFVVRPVID---------GVPMEFYVKKRPGVDEFLEALA 207
LDLD TLVH+ + ES ++ V G P E +K R V +FL+
Sbjct: 91 LDLDHTLVHTIKVSQLSESEKYITEEVESRKDLRRFNTGFPEESLIKLRSFVHQFLKECN 150
Query: 208 AKFEVVVFTAALREYASLVLDRVD------RNRLVSHRLYRDSCKQEDGKLVKDLSLAGR 261
F + V+T +YA LVL+ +D NR+++ R + G DL LA
Sbjct: 151 EMFSLYVYTKGGYDYAQLVLEMIDPDKIYFGNRVITRR-------ESPGFKTLDLVLA-- 201
Query: 262 DLKRVVIVDDNPNSFANQPENAILI 286
D + +V+VDD + + + +N + I
Sbjct: 202 DERGIVVVDDKSSVWPHDKKNLLQI 226
>At1g43600 hypothetical protein
Length = 221
Score = 49.3 bits (116), Expect = 3e-06
Identities = 45/153 (29%), Positives = 64/153 (41%), Gaps = 16/153 (10%)
Query: 158 LDLDETLVHSNTAPP-----------PESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEAL 206
LDLD TL+HS +S + R +DG EF +K RP + EFL
Sbjct: 23 LDLDHTLLHSVLVSDLSKREKYLLEETDSRQDLWRRNVDGY--EFIIKLRPFLHEFLLEA 80
Query: 207 AAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRV 266
F + V+T YA VL +D +++ + +E K L L D +RV
Sbjct: 81 NKLFTMHVYTMGSSSYAKQVLKLIDPDKVY---FGKRVITREASPFNKSLDLLAADKRRV 137
Query: 267 VIVDDNPNSFANQPENAILIRPFVDDVFDRELW 299
VIVDD + + N + I +V D W
Sbjct: 138 VIVDDTVHVWPFHKRNLLQITKYVYFKVDGTKW 170
>At1g43610 hypothetical protein
Length = 255
Score = 48.9 bits (115), Expect = 4e-06
Identities = 44/153 (28%), Positives = 64/153 (41%), Gaps = 16/153 (10%)
Query: 158 LDLDETLVHSNTAPP-----------PESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEAL 206
LDLD TL+HS +S + R +DG EF +K RP + EFL
Sbjct: 57 LDLDHTLLHSVLVSDLSKREKYLLEETDSRQDLWRRNVDGY--EFIIKLRPFLHEFLLEA 114
Query: 207 AAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRV 266
F + V+T YA VL +D +++ + +E K L L D +RV
Sbjct: 115 NKLFTMHVYTMGSSSYAKQVLKLIDPDKVY---FGKRVITREASPFNKSLDLLAADKRRV 171
Query: 267 VIVDDNPNSFANQPENAILIRPFVDDVFDRELW 299
VIVDD + + N + I ++ D W
Sbjct: 172 VIVDDTVHVWPFHKRNLLQITKYIYFKVDGTKW 204
>At3g15330 hypothetical protein
Length = 305
Score = 46.2 bits (108), Expect = 2e-05
Identities = 37/140 (26%), Positives = 66/140 (46%), Gaps = 12/140 (8%)
Query: 158 LDLDETLVHS-NTAPPPESFDFVVRPVIDGVPMEF------YVKKRPGVDEFLEALAAKF 210
LDLD TL+HS T+ ++ ++++ G + VK RP V+EFL+ F
Sbjct: 81 LDLDHTLIHSMKTSNLSKAEKYLIKEEKSGSRKDLRKYNNRLVKFRPFVEEFLKEANKLF 140
Query: 211 EVVVFTAALREYASLVLDRVDRNRL-VSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIV 269
+ +T Y V+ +D N++ R+ +++ +K L L D + +VIV
Sbjct: 141 TMTAYTKGGSTYGQAVVRMIDPNKIYFGDRI----ITRKESPDLKTLDLVLADERGIVIV 196
Query: 270 DDNPNSFANQPENAILIRPF 289
D+ PN + + N + I +
Sbjct: 197 DNTPNVWPHHKRNLLEITSY 216
>At3g17550 hypothetical protein
Length = 296
Score = 44.7 bits (104), Expect = 7e-05
Identities = 38/141 (26%), Positives = 61/141 (42%), Gaps = 13/141 (9%)
Query: 158 LDLDETLVHSNTAPPPESFD--------FVVRPVIDGVPMEFYVKKRPGVDEFLEALAAK 209
LDLD TL+HS + R + + ++ K RP V EFL+
Sbjct: 90 LDLDHTLLHSIRVSLLSETEKCLIEEACSTTREDLWKLDSDYLTKLRPFVHEFLKEANEL 149
Query: 210 FEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDS-CKQEDGKLVKDLSLAGRDLKRVVI 268
F + V+T R YA +L +D R+ + D +++ VK L L + + VVI
Sbjct: 150 FTMYVYTMGTRVYAESLLKLIDPKRI----YFGDRVITRDESPYVKTLDLVLAEERGVVI 205
Query: 269 VDDNPNSFANQPENAILIRPF 289
VDD + + + N + I +
Sbjct: 206 VDDTSDVWTHHKSNLVEINEY 226
>At1g20320 hypothetical protein
Length = 342
Score = 44.3 bits (103), Expect = 9e-05
Identities = 44/140 (31%), Positives = 63/140 (44%), Gaps = 13/140 (9%)
Query: 158 LDLDETLVHS---NTAPPPESF-----DFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAK 209
LDLD TL+HS + E + DF R + + E +K RP V EFL+
Sbjct: 81 LDLDHTLLHSIMISRLSEGEKYLLGESDF--REDLWTLDREMLIKLRPFVHEFLKEANEI 138
Query: 210 FEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIV 269
F + V+T R+YA VL +D ++ R + E G K L L D VVIV
Sbjct: 139 FSMYVYTMGNRDYAQAVLKWIDPKKVYFGD--RVITRDESG-FSKTLDLVLADECGVVIV 195
Query: 270 DDNPNSFANQPENAILIRPF 289
DD + + + N + I +
Sbjct: 196 DDTRHVWPDHERNLLQITKY 215
>At5g58000 putative protein
Length = 1065
Score = 35.0 bits (79), Expect = 0.056
Identities = 36/150 (24%), Positives = 62/150 (41%), Gaps = 22/150 (14%)
Query: 158 LDLDETLVHS----NTAPPPESFDFVVRPVIDGVP-----------MEFYVKKRPGVDEF 202
LDLD TL+++ + P E + DG M+ K RP V F
Sbjct: 752 LDLDHTLLNTTILRDLKPEEEYLKSHTHSLQDGCNVSGGSLFLLEFMQMMTKLRPFVHSF 811
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVD-RNRLVSHRLYRDSCKQEDGKL--VKDLSLA 259
L+ + F + ++T R YA + +D + R+ ++DG + K L +
Sbjct: 812 LKEASEMFVMYIYTMGDRNYARQMAKLLDPKGEYFGDRV----ISRDDGTVRHEKSLDVV 867
Query: 260 GRDLKRVVIVDDNPNSFANQPENAILIRPF 289
V+I+DD N++ +N I+I +
Sbjct: 868 LGQESAVLILDDTENAWPKHKDNLIVIERY 897
>At2g36540 hypothetical protein
Length = 249
Score = 33.9 bits (76), Expect = 0.12
Identities = 25/95 (26%), Positives = 41/95 (42%), Gaps = 5/95 (5%)
Query: 193 VKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKL 252
V KRP +EF++ +FEV ++++A +SL + V R + Y+ +
Sbjct: 86 VYKRPFAEEFMKFCLERFEVGIWSSACELVSSLNILIVTGPRECTDSGYKTLENRYKPLF 145
Query: 253 VKDLS-----LAGRDLKRVVIVDDNPNSFANQPEN 282
KDLS G + +DD P P+N
Sbjct: 146 FKDLSKVFKCFKGFSASNTIFIDDEPYKALRNPDN 180
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,835,857
Number of Sequences: 26719
Number of extensions: 353840
Number of successful extensions: 973
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 917
Number of HSP's gapped (non-prelim): 40
length of query: 328
length of database: 11,318,596
effective HSP length: 100
effective length of query: 228
effective length of database: 8,646,696
effective search space: 1971446688
effective search space used: 1971446688
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0118a.3


