

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.4
(482 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
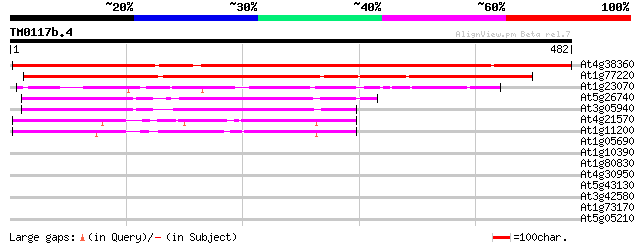
Score E
Sequences producing significant alignments: (bits) Value
At4g38360 unknown protein 760 0.0
At1g77220 unknown protei 476 e-134
At1g23070 hypothetical protein 269 2e-72
At5g26740 unknown protein 156 2e-38
At3g05940 unknown protein 152 3e-37
At4g21570 unknown protein 125 7e-29
At1g11200 unknown protein 122 3e-28
At1g05690 unknown protein 32 0.78
At1g10390 unknown protein 32 1.0
At1g80830 metal ion transporter (PMIT1) 30 2.3
At4g30950 chloroplast omega-6 fatty acid desaturase (fad6) 30 3.0
At5g43130 unkown protein 30 3.9
At3g42580 putative protein 29 5.0
At1g73170 unknown protein 29 5.0
At5g05210 unknown protein 29 6.6
>At4g38360 unknown protein
Length = 479
Score = 760 bits (1963), Expect = 0.0
Identities = 371/481 (77%), Positives = 421/481 (87%), Gaps = 11/481 (2%)
Query: 3 IIQLLHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIE 62
++ ++ P WA+ +A FL++TL+LS++L+F+HLS YKNPEEQKFLIGVILMVPCYSIE
Sbjct: 9 LLAAAYSAPAWASFMAGAFLVLTLSLSLFLVFDHLSTYKNPEEQKFLIGVILMVPCYSIE 68
Query: 63 SFVSLVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLL 122
SF SLV PSISVDCGILRDCYESFAMYCFGRYLVAC+GGEERTIEFMERQGR + KTPLL
Sbjct: 69 SFASLVKPSISVDCGILRDCYESFAMYCFGRYLVACIGGEERTIEFMERQGRKSFKTPLL 128
Query: 123 LHHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFG 182
H D KG + HPFP+ FLKPW L+ FYQ+VK MIIKS T++ A+ILEAFG
Sbjct: 129 DHK---DEKGIIKHPFPMNLFLKPWRLSPWFYQVVK------MIIKSLTALTALILEAFG 179
Query: 183 VYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFL 242
VYCEGEFK GCGYPY+AVVLNFSQSWALYCLVQFY TKDELAHI+PLAKFLTFKSIVFL
Sbjct: 180 VYCEGEFKWGCGYPYLAVVLNFSQSWALYCLVQFYGATKDELAHIQPLAKFLTFKSIVFL 239
Query: 243 TWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMG 302
TWWQGVAIALL + GLFKS IAQ LQ K+SVQDFIICIEMGIAS+VHLYVFPAKPY LMG
Sbjct: 240 TWWQGVAIALLSSLGLFKSSIAQSLQLKTSVQDFIICIEMGIASVVHLYVFPAKPYGLMG 299
Query: 303 DRHPGSISVLGDY-SADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGG 361
DR GS+SVLGDY S DCP+DPDEIRDSERPTK+RLP DVD +SGMTI+ES+RDV +GG
Sbjct: 300 DRFTGSVSVLGDYASVDCPIDPDEIRDSERPTKVRLPHPDVDIRSGMTIKESMRDVFVGG 359
Query: 362 SGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPAR 421
YIVKDV+FTV QAVEP+EK IT+FNEKLH+ISQNIKKHDK++RR+KDDSC+ SSSP+R
Sbjct: 360 GEYIVKDVRFTVTQAVEPMEKSITKFNEKLHKISQNIKKHDKEKRRVKDDSCM-SSSPSR 418
Query: 422 RVIRGIDDPLLNGSISDSGISRAKKHRRKSGYTSAESGGESSSDQSYGAYQVRGHRWVTK 481
RVIRGIDDPLLNGS SDSG++R KKHRRKSGYTSAESGGESSSDQ+YG ++VRG RW+TK
Sbjct: 419 RVIRGIDDPLLNGSFSDSGVTRTKKHRRKSGYTSAESGGESSSDQAYGGFEVRGRRWITK 478
Query: 482 E 482
+
Sbjct: 479 D 479
>At1g77220 unknown protei
Length = 484
Score = 476 bits (1224), Expect = e-134
Identities = 237/439 (53%), Positives = 317/439 (71%), Gaps = 9/439 (2%)
Query: 13 WATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSI 72
W + ASVF+++ + L MYL+FEHL++Y PEEQKFLIG+ILMVP Y++ESF+SLVN
Sbjct: 41 WPILSASVFVVIAILLPMYLIFEHLASYNQPEEQKFLIGLILMVPVYAVESFLSLVNSEA 100
Query: 73 SVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKG 132
+ +C ++RDCYE+FA+YCF RYL+ACL GEERTIEFME+Q TPLL S+ G
Sbjct: 101 AFNCEVIRDCYEAFALYCFERYLIACLDGEERTIEFMEQQTVITQSTPLLEGTCSY---G 157
Query: 133 TVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLG 192
V HPFP+ F+K W L +FY VK GIVQYMI+K +++A+ILEAFGVY EG+F
Sbjct: 158 VVEHPFPMNCFVKDWSLGPQFYHAVKIGIVQYMILKMICALLAMILEAFGVYGEGKFAWN 217
Query: 193 CGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIAL 252
GYPY+AVVLNFSQ+WALYCLVQFY V KD+LA IKPLAKFLTFKSIVFLTWWQG+ +A
Sbjct: 218 YGYPYLAVVLNFSQTWALYCLVQFYNVIKDKLAPIKPLAKFLTFKSIVFLTWWQGIIVAF 277
Query: 253 LYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHPGSISVL 312
L++ GL K +A+ L K+ +QD+IICIEMGIA++VHLYVFPA PY+ G+R +++V+
Sbjct: 278 LFSMGLVKGSLAKEL--KTRIQDYIICIEMGIAAVVHLYVFPAAPYK-RGERCVRNVAVM 334
Query: 313 GDY-SADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIVKDVKF 371
DY S D P DP+E++DSER T+ R D D + + +SVRDVV+G IV D++F
Sbjct: 335 SDYASIDVPPDPEEVKDSERTTRTRYGRHD-DREKRLNFPQSVRDVVLGSGEIIVDDMRF 393
Query: 372 TVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRGIDDPL 431
TV VEPVE+GI + N H+IS+N+K+ ++ ++ KDDS + + + + + L
Sbjct: 394 TVSHVVEPVERGIAKINRTFHQISENVKRFEQQKKTTKDDSYVIPLNQWAKEFSDVHENL 453
Query: 432 LN-GSISDSGISRAKKHRR 449
+ GS+SDSG+ +H +
Sbjct: 454 YDGGSVSDSGLGSTNRHHQ 472
>At1g23070 hypothetical protein
Length = 379
Score = 269 bits (688), Expect = 2e-72
Identities = 162/421 (38%), Positives = 240/421 (56%), Gaps = 60/421 (14%)
Query: 7 LHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
LH P + I+ F + + LS+Y + +HL Y NP + +S
Sbjct: 12 LHLPSL---IIGGSFATVAICLSLYSILQHLRFYTNP-------------------AIIS 49
Query: 67 LVNPSISVDCGILRDCYESFAMYCFGRYLVACLG---GEERTIEFMERQGRSASKTPLLL 123
L N S+ C ILR+CYE+FA+Y FG YLVACLG GE R +E++E + SK PLL
Sbjct: 50 LSNSKFSLPCDILRNCYEAFALYSFGSYLVACLGELCGERRVVEYLENE----SKKPLLE 105
Query: 124 HHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQY---MIIKSFTSIMAVILEA 180
+ K + F K+ P++L R + I KFG+VQY MI+K+F + + +LE
Sbjct: 106 EGANESKKKKKKNSF-WKFLCDPYVLGRELFVIEKFGLVQYVSQMILKTFCAFLTFLLEL 164
Query: 181 FGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIV 240
GVY +GEFK G Q WAL+CLVQFY VT + L IKPLAKF++FK+IV
Sbjct: 165 LGVYGDGEFKWYYG-----------QMWALFCLVQFYNVTHERLKEIKPLAKFISFKAIV 213
Query: 241 FLTWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYEL 300
F TWWQG IALL +G+ + + + ++ +QDF+ICIEM IA++ HL+VFPA+PY
Sbjct: 214 FATWWQGFGIALLCYYGI----LPKEGRFQNGLQDFLICIEMAIAAVAHLFVFPAEPY-- 267
Query: 301 MGDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIG 360
H +S G +A+ E++ E + T V+A SG +I+ESV+D+VI
Sbjct: 268 ----HYIPVSECGKITAE--TSKTEVK-LEEGGLVETTETQVEA-SGTSIKESVQDIVID 319
Query: 361 GSGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPA 420
G ++VKDV T++QA+ PVEKG+T+ + +H+ + + K+ + ++ + +S P
Sbjct: 320 GGQHVVKDVVLTINQAIGPVEKGVTKIQDTIHQ--KLLDSDGKEETEVTEEVTVETSVPP 377
Query: 421 R 421
+
Sbjct: 378 K 378
>At5g26740 unknown protein
Length = 422
Score = 156 bits (395), Expect = 2e-38
Identities = 102/306 (33%), Positives = 155/306 (50%), Gaps = 26/306 (8%)
Query: 11 PVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNP 70
P + IVA + + +AL+++ ++ HL Y P Q++++ +I MVP Y+ SF+SLV P
Sbjct: 6 PFYLNIVAFLCTVGAIALAIFHIYRHLLNYTEPTYQRYIVRIIFMVPVYAFMSFLSLVLP 65
Query: 71 SISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDY 130
S+ +R+ YE++ +Y F +A +GG + + GRS + L+
Sbjct: 66 KSSIYFDSIREVYEAWVIYNFLSLCLAWVGGPGSVV--LSLSGRSLKPSWSLM------- 116
Query: 131 KGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFK 190
FP P L RF + K G +Q++I+K + ++L A G Y +G F
Sbjct: 117 ----TCCFP------PLTLDGRFIRRCKQGCLQFVILKPILVAVTLVLYAKGKYKDGNFN 166
Query: 191 LGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAI 250
Y Y+ ++ S + ALY LV FY +D L P+ KF+ KS+VFLT+WQGV +
Sbjct: 167 PDQAYLYLTIIYTISYTVALYALVLFYMACRDLLQPFNPVPKFVIIKSVVFLTYWQGVLV 226
Query: 251 ALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHPGSIS 310
L G KS ++ + Q+FIIC+EM IA+ H Y FP K Y G GS S
Sbjct: 227 FLAAKSGFIKS-----AEAAAHFQNFIICVEMLIAAACHFYAFPYKEY--AGANVGGSGS 279
Query: 311 VLGDYS 316
G S
Sbjct: 280 FSGSLS 285
>At3g05940 unknown protein
Length = 422
Score = 152 bits (385), Expect = 3e-37
Identities = 90/288 (31%), Positives = 149/288 (51%), Gaps = 24/288 (8%)
Query: 11 PVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNP 70
P++ I+A + + +AL+++ +++HL Y P Q++++ ++ MVP Y++ SF++LV P
Sbjct: 6 PIYLIILAFLCTVGAIALALFHIYKHLLNYTEPIYQRYIVRIVFMVPVYALMSFLALVLP 65
Query: 71 SISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDY 130
S+ +R+ YE++ +Y F +A +GG + + GRS + L+
Sbjct: 66 KSSIYFNSIREVYEAWVIYNFLSLCLAWVGGPGSVV--ISLTGRSLKPSWHLM------- 116
Query: 131 KGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFK 190
+ P L RF + K G +Q++I+K + ++L A G Y +G F
Sbjct: 117 ----------TCCIPPLPLDGRFIRRCKQGCLQFVILKPILVAVTLVLYAKGKYKDGNFS 166
Query: 191 LGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAI 250
Y Y+ ++ S + ALY LV FY KD L P+ KF+ KS+VFLT+WQGV +
Sbjct: 167 PDQSYLYLTIIYTISYTVALYALVLFYVACKDLLQPFNPVPKFVIIKSVVFLTYWQGVLV 226
Query: 251 ALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPY 298
L G + + + Q+FIIC+EM IA+ H Y FP K Y
Sbjct: 227 FLFAKSGFIRDE-----EEAALFQNFIICVEMLIAAAAHFYAFPYKEY 269
>At4g21570 unknown protein
Length = 294
Score = 125 bits (313), Expect = 7e-29
Identities = 93/306 (30%), Positives = 155/306 (50%), Gaps = 35/306 (11%)
Query: 3 IIQLLHTPPVWATIVASVF-LLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSI 61
++ L P T S F +L+TL ++ L+ +HL +KNP+EQK ++ ++LM P Y++
Sbjct: 1 MMDLTKLKPPQITFYCSAFSVLLTLHFTIQLVSQHLFHWKNPKEQKAILIIVLMAPIYAV 60
Query: 62 ESFVSLVNPSISVDCGI----LRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSAS 117
SF+ L+ S + +++CYE+ + F + + L S S
Sbjct: 61 VSFIGLLEVKGSETFFLFLESIKECYEALVIAKFLALMYSYLN-------------ISMS 107
Query: 118 KTPLLLHHHSHDYKGT-VNHPFPLKYFLKPWI--LTRRFYQIVKFGIVQYMIIKSFTSIM 174
K L KG ++H FP+ F +P + L R +++K+ Q+++I+ S +
Sbjct: 108 KNIL-----PDGIKGREIHHSFPMTLF-QPHVVRLDRHTLKLLKYWTWQFVVIRPVCSTL 161
Query: 175 AVILEAFGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFL 234
+ L+ G Y + + +++NFS S ALY LV FY V ELA PLAKFL
Sbjct: 162 MIALQLIGFYPSW-----LSWTF-TIIVNFSVSLALYSLVIFYHVFAKELAPHNPLAKFL 215
Query: 235 TFKSIVFLTWWQGVAIALLYTFGLFKSP--IAQGLQSKSSVQDFIICIEMGIASIVHLYV 292
K IVF +WQG+A+ +L G KS + Q + ++Q+ ++C+EM I + V +
Sbjct: 216 CIKGIVFFVFWQGIALDILVAMGFIKSHHFWLEVEQIQEAIQNVLVCLEMVIFAAVQKHA 275
Query: 293 FPAKPY 298
+ A PY
Sbjct: 276 YHAGPY 281
>At1g11200 unknown protein
Length = 295
Score = 122 bits (307), Expect = 3e-28
Identities = 92/305 (30%), Positives = 155/305 (50%), Gaps = 33/305 (10%)
Query: 3 IIQLLHTPPVWATIVASVF-LLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSI 61
+I L P T++ SVF +L+++ +M L+ +HL +K P EQ+ ++ ++LM P Y+I
Sbjct: 1 MIDLSTLSPAEITVMGSVFCVLLSMHFTMQLVSQHLFYWKKPNEQRAILIIVLMAPVYAI 60
Query: 62 ESFVSLVNPSIS----VDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSAS 117
SFV L++ S + +++CYE+ + F + + + SA
Sbjct: 61 NSFVGLLDAKGSKPFFMFLDAVKECYEALVIAKFLALMYSYVN-----------ISMSAR 109
Query: 118 KTPLLLHHHSHDYKGT-VNHPFPLKYFL-KPWILTRRFYQIVKFGIVQYMIIKSFTSIMA 175
P ++KG ++H FP+ F+ + L + +K Q+ II+ SI+
Sbjct: 110 IIP-------DEFKGREIHHSFPMTLFVPRTTHLDYLTLKQLKQWTWQFCIIRPVCSILM 162
Query: 176 VILEAFGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLT 235
+ L+ G+Y + + + A+ LN S S ALY LV+FY V EL KPL KF+
Sbjct: 163 ITLQILGIY-----PVWLSWIFTAI-LNVSVSLALYSLVKFYHVFAKELEPHKPLTKFMC 216
Query: 236 FKSIVFLTWWQGVAIALLYTFGLFKSP--IAQGLQSKSSVQDFIICIEMGIASIVHLYVF 293
K IVF +WQG+ + +L GL KS + Q + ++Q+ ++C+EM + SI+ Y F
Sbjct: 217 VKGIVFFCFWQGIVLKILVGLGLIKSHHFWLEVDQLEEALQNVLVCLEMIVFSIIQQYAF 276
Query: 294 PAKPY 298
PY
Sbjct: 277 HVAPY 281
>At1g05690 unknown protein
Length = 364
Score = 32.0 bits (71), Expect = 0.78
Identities = 19/78 (24%), Positives = 37/78 (47%), Gaps = 16/78 (20%)
Query: 27 ALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSL---------VNPSISVD-- 75
A+ M++ F + S Y+ E +KF++ ++++ CYS+ S L +N +D
Sbjct: 105 AVYMFIRFLYSSCYEEEEMKKFVLHLLVLSHCYSVPSLKRLCVEILDQGWINKENVIDVL 164
Query: 76 -----CGILRDCYESFAM 88
C + R C+ +M
Sbjct: 165 QLARNCDVTRICFVCLSM 182
>At1g10390 unknown protein
Length = 1041
Score = 31.6 bits (70), Expect = 1.0
Identities = 21/96 (21%), Positives = 44/96 (44%), Gaps = 3/96 (3%)
Query: 296 KPYELMGDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVR 355
+PY+ + G +++ ++ AD ++R S+ T+ R+ ++ VR
Sbjct: 853 RPYKTLSGHRAGEAAIVYEHGADIEALMPKLRQSDYFTEPRIQELAAKERADPGYCRRVR 912
Query: 356 DVVIGGSGYIVKDVKFTVHQAVEPVE-KGITRFNEK 390
D V+G GY +KF V ++ + + +FN +
Sbjct: 913 DFVVGRHGY--GSIKFMGETDVRRLDLESLVQFNTR 946
>At1g80830 metal ion transporter (PMIT1)
Length = 532
Score = 30.4 bits (67), Expect = 2.3
Identities = 36/145 (24%), Positives = 62/145 (41%), Gaps = 10/145 (6%)
Query: 199 AVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYTFGL 258
+++L+F +AL L++F + +H+ P+A I LTW G I + + L
Sbjct: 398 SMILSFELPFALVPLLKFTSCKTKMGSHVNPMA-------ITALTWVIGGLIMGINIYYL 450
Query: 259 FKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHPGSISVLGDYSAD 318
S I + S + + C +G A I LY+ +R S+ + D
Sbjct: 451 VSSFIKLLIHSHMKLILVVFCGILGFAGIA-LYLAAIAYLVFRKNRVATSLLISRDSQNV 509
Query: 319 CPLDPDEIRDSERPTKLRLPTTDVD 343
L +I + + P R+ T+DVD
Sbjct: 510 ETLPRQDIVNMQLP--CRVSTSDVD 532
>At4g30950 chloroplast omega-6 fatty acid desaturase (fad6)
Length = 448
Score = 30.0 bits (66), Expect = 3.0
Identities = 29/104 (27%), Positives = 47/104 (44%), Gaps = 12/104 (11%)
Query: 230 LAKFLTFKS---IVFLTW-WQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIA 285
L F+ KS ++ L W W G AI + G A SK+ + + I+ G
Sbjct: 139 LGLFMIAKSPWYLLPLAWAWTGTAITGFFVIG---HDCAHKSFSKNKLVEDIV----GTL 191
Query: 286 SIVHLYVFPAKPYELMGDRHPGSISVLGDYSADCPLDPDEIRDS 329
+ + L V+P +P+ DRH ++L +A P+ P+E S
Sbjct: 192 AFLPL-VYPYEPWRFKHDRHHAKTNMLVHDTAWQPVPPEEFESS 234
>At5g43130 unkown protein
Length = 689
Score = 29.6 bits (65), Expect = 3.9
Identities = 23/113 (20%), Positives = 47/113 (41%), Gaps = 5/113 (4%)
Query: 364 YIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRV 423
+I D++ +++ + V++ EK ++ +KK + + D +++ A R
Sbjct: 533 FITSDIRLQINEMNQKVKEEW----EKKQAEAEKLKKPSEGNKEDDDKMRTTAANVAARA 588
Query: 424 IRGIDDPLLNGSISDSGISRAKKHRRKSGYTSAES-GGESSSDQSYGAYQVRG 475
G DD L + ++ K G S GG++S D+ G + G
Sbjct: 589 AVGGDDAFLKWQLMAEARQKSVSEAGKDGNQKTTSGGGKNSKDRQDGGRRFSG 641
>At3g42580 putative protein
Length = 903
Score = 29.3 bits (64), Expect = 5.0
Identities = 30/116 (25%), Positives = 46/116 (38%), Gaps = 30/116 (25%)
Query: 375 QAVEPVEKGITRFNEKLHRISQ-----------------NIKKHDKDRRRIKDDSC--IA 415
+ VE V K + + N+K+H++S+ N++ + DD IA
Sbjct: 337 EKVEMVNKKVEKGNKKVHQVSEQAETISLPIEELSSSDDNVEDPSNQKGSFSDDEMKDIA 396
Query: 416 SSSPARRVIRGIDDPLLNGSISDSGISRAKKHRRKSGYT--SAESGGESSSDQSYG 469
+ P R + G D +G +S AKKH S Y + GG S Q G
Sbjct: 397 PTQPVPRNVPGNDG---DGEVS------AKKHHTVSDYVMENIYLGGASPPHQEVG 443
>At1g73170 unknown protein
Length = 666
Score = 29.3 bits (64), Expect = 5.0
Identities = 25/77 (32%), Positives = 31/77 (39%), Gaps = 2/77 (2%)
Query: 321 LDPDEIRDS--ERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIVKDVKFTVHQAVE 378
L P+EIR + E P L +D R D VI VKD++F V Q E
Sbjct: 87 LVPEEIRQTLKEHPEISELIEIVLDLGRKPLARFPSGDFVISDDAVRVKDLEFAVSQVGE 146
Query: 379 PVEKGITRFNEKLHRIS 395
+ LHRIS
Sbjct: 147 FTNDNRAGISRTLHRIS 163
>At5g05210 unknown protein
Length = 386
Score = 28.9 bits (63), Expect = 6.6
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Query: 392 HRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRGIDDPLLNGSISDSGISRAKKHRRKS 451
+R++ + K + D R KDD + R+ R ID+ L G S R+ + R+K+
Sbjct: 142 NRLNDDDSKEETDNNRQKDDRSVTYEELRERLHRKIDE--LKGGRGGSDRPRSNERRKKN 199
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,888,909
Number of Sequences: 26719
Number of extensions: 466490
Number of successful extensions: 1264
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1229
Number of HSP's gapped (non-prelim): 17
length of query: 482
length of database: 11,318,596
effective HSP length: 103
effective length of query: 379
effective length of database: 8,566,539
effective search space: 3246718281
effective search space used: 3246718281
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0117b.4


