

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.11
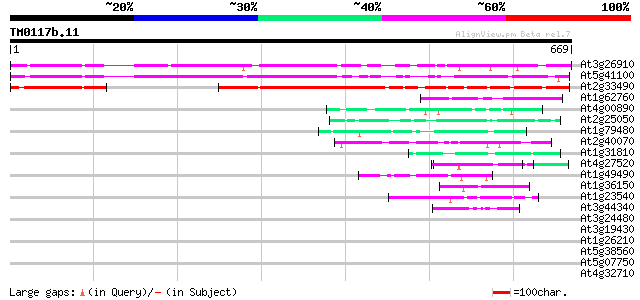
(669 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g26910 unknown protein 431 e-121
At5g41100 unknown protein 405 e-113
At2g33490 unknown protein 310 2e-84
At1g62760 hypothetical protein 49 1e-05
At4g00890 48 2e-05
At2g25050 hypothetical protein 47 3e-05
At1g79480 hypothetical protein 47 3e-05
At2g40070 En/Spm-like transposon protein 46 8e-05
At1g31810 hypothetical protein 45 1e-04
At4g27520 early nodulin-like 2 predicted GPI-anchored protein 45 2e-04
At1g49490 hypothetical protein 44 2e-04
At1g36150 hypothetical protein 44 4e-04
At1g23540 putative serine/threonine protein kinase 43 5e-04
At3g44340 putative protein 42 8e-04
At3g24480 disease resistance protein, putative 42 0.001
At3g19430 putative late embryogenesis abundant protein 42 0.001
At1g26210 hypothetical protein 42 0.001
At5g38560 putative protein 42 0.001
At5g07750 putative protein 41 0.002
At4g32710 putative protein kinase 41 0.002
>At3g26910 unknown protein
Length = 621
Score = 431 bits (1108), Expect = e-121
Identities = 289/690 (41%), Positives = 386/690 (55%), Gaps = 113/690 (16%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MK+S++KLR L H+H + K + ++ Q++EL +A +DMQDMR+CYD LL+AAAA
Sbjct: 1 MKASIEKLRRLTSHSHKVDVK--EKGDVMATTQIDELDRAGKDMQDMRECYDRLLAAAAA 58
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
TA+SAYEF+ESL +MGSCL + ND EE+ ++L MLGK+Q +LQ+L+D Y
Sbjct: 59 TANSAYEFSESLGEMGSCLEQIAPHND--EESSRILFMLGKVQSELQRLLDTY------- 109
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
RSHI +TIT PSE+LL +LR VE+MK+QCD KR
Sbjct: 110 ---------------------------RSHIFETITSPSEALLKDLRYVEDMKQQCDGKR 142
Query: 181 DVYEYMATRFRERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLLT 240
+VYE + E+GR K K E + + A E+ +EAT+ +FRLKSLK+GQ+RSLL
Sbjct: 143 NVYEMSLVK--EKGRPKSSKGERHIPPESRPAYSEFHDEATMCIFRLKSLKEGQARSLLI 200
Query: 241 QAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHID----YHFNGLEEEDGDEGDEGYE 296
QA RHH AQ+ F +KSLE VE HVK E+QHID H N +E + D
Sbjct: 201 QAVRHHTAQMRLFHTGLKSLEAVERHVKVAVEKQHIDCDLSVHGNEMEASEDD------- 253
Query: 297 DDDGYDENDDGELSFDYGQIEQEQDVSTSRN--SMELDQVELTLPRGSTAEAAKENLDKL 354
DDDG N +GELSFDY EQ+ + S+ + ++D +L+ PR ST A N D
Sbjct: 254 DDDGRYMNREGELSFDYRTNEQKVEASSLSTPWATKMDDTDLSFPRPSTTRPAAVNADHR 313
Query: 355 QRNLFSFRVR-TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAK-SSISS 412
+ S R + S SAPLF + KPD SE+LRQ PS F++YVLPTP D++ S S
Sbjct: 314 EEYPVSTRDKYLSSHSAPLFPEKKPDVSERLRQANPS----FNAYVLPTPNDSRYSKPVS 369
Query: 413 GSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTAT 472
+ NP+P +N + N+WHSSPLE K + K++ SN+
Sbjct: 370 QALNPRP------TNHSAGNIWHSSPLEPIK---------------SGKDGKDAESNSFY 408
Query: 473 TRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPT 532
RLP P S D + RHAFSGPL P T+P+++ +SG P
Sbjct: 409 GRLPRP-------STTDTHHHQQQAAGRHAFSGPL--RPSSTKPITM--ADSYSGAFCPL 457
Query: 533 PIP------QPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNF---PSNSRLLGLVGY 583
P P SSSP+VSP+ASP SSP+++ELHELPRPP +F P ++ GLVG+
Sbjct: 458 PTPPVLQSHPHSSSSPRVSPTASPPPASSPRLNELHELPRPPGHFAPPPRRAKSPGLVGH 517
Query: 584 SGPLVPRGQKVSAPNNLVISS---AASPLPIPPQAMARSFSIPSSGARVTALHSSSALES 640
S PL Q+ S V S+ ASPLP+PP + RS+SIPS RV S
Sbjct: 518 SAPLTAWNQERSTVTVAVPSATNIVASPLPVPPLVVPRSYSIPSRNQRVV---------S 568
Query: 641 SHISSISEDI-ASPPLTPIALSNSRPSSDG 669
+ +DI ASPPLTP++LS P + G
Sbjct: 569 QRLVERRDDIVASPPLTPMSLSRPLPQATG 598
>At5g41100 unknown protein
Length = 582
Score = 405 bits (1040), Expect = e-113
Identities = 289/680 (42%), Positives = 377/680 (54%), Gaps = 133/680 (19%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MK+S +LR L + D + P Q+E LA+A +DMQDMR+ YD LL AAA
Sbjct: 2 MKASFGRLRRFALPKADAI----DIGELFPTAQIEGLARAAKDMQDMREGYDRLLEVAAA 57
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
A+SAYEF+ESL +MGSCL + ND +E+G +LLMLGK+QF+L+KL+D Y
Sbjct: 58 MANSAYEFSESLGEMGSCLEQIAPHND--QESGGILLMLGKVQFELKKLVDTY------- 108
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
RS I +TIT PSESLL++LR VE+MK+QC+EKR
Sbjct: 109 ---------------------------RSQIFKTITRPSESLLSDLRTVEDMKQQCEEKR 141
Query: 181 DVYEYMATRF-RERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLL 239
DV ++M +++ + KG K E +QL+TARDE +EATL +FRLKSLK+GQ+RSLL
Sbjct: 142 DVVKHMLMEHVKDKVQVKGTKGERLIRRQLETARDELQDEATLCIFRLKSLKEGQARSLL 201
Query: 240 TQAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYE-DD 298
TQAARHH AQ+ F +KSLE VE HV+ ++QHID + + G+E D + DD
Sbjct: 202 TQAARHHTAQMHMFFAGLKSLEAVEQHVRIAADRQHIDC----VLSDPGNEMDCSEDNDD 257
Query: 299 DGYDENDDGELSFDYGQIEQEQDV-STSRNSMELDQVELTLPRGSTAEAAKENLDKLQRN 357
D N DGELSFDY EQ +V ST SM++D +L+ R S A +A N D + +
Sbjct: 258 DDRLVNRDGELSFDYITSEQRVEVISTPHGSMKMDDTDLSFQRPSPAGSATVNADPREEH 317
Query: 358 LFSFR-VRTGSQSAPLFADNKPDSSEK-LRQMRPSLSRKFSSYVLPTPVDAKSSISSGSN 415
S R RT S SAPLF D K D +++ +RQM PS ++Y+LPTPVD+KSS
Sbjct: 318 SVSNRDRRTSSHSAPLFPDKKADLADRSMRQMTPSA----NAYILPTPVDSKSS------ 367
Query: 416 NPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRL 475
P +K T +N + NLWHSSPLE K + K++ SN +RL
Sbjct: 368 -PIFTKPVTQTNH-SANLWHSSPLEPIK-----------------TAHKDAESN-LYSRL 407
Query: 476 PPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIP 535
P P HAFSGPL P TR LP P+
Sbjct: 408 PRP--------------------SEHAFSGPL--KPSSTR--------------LPVPVA 431
Query: 536 -QPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNF--PSNSRLLGLVGYSGPLVPRGQ 592
Q SSSP++SP+ASP + SSP+I+ELHELPRPP F P S+ GLVG+S PL Q
Sbjct: 432 VQAQSSSPRISPTASPPLASSPRINELHELPRPPGQFAPPRRSKSPGLVGHSAPLTAWNQ 491
Query: 593 KVSAPNNLVISS--AASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDI 650
+ S N+V+S+ ASPLP+PP + RS+SIPS R A + +
Sbjct: 492 ERS---NVVVSTNIVASPLPVPPLVVPRSYSIPSRNQRAMAQQPLPERNQNR-------V 541
Query: 651 ASP---PLTPIALSNSRPSS 667
ASP PLTP +L N R S
Sbjct: 542 ASPPPLPLTPASLMNLRSLS 561
>At2g33490 unknown protein
Length = 521
Score = 310 bits (794), Expect = 2e-84
Identities = 195/423 (46%), Positives = 267/423 (63%), Gaps = 31/423 (7%)
Query: 250 LCFFKKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYEDDDGYDENDDGEL 309
LCFFKKA+ SLE V+PHV+ VTE QHIDYHF+GLE++DGD+ E E +DG + +DDGEL
Sbjct: 108 LCFFKKALSSLEEVDPHVQMVTESQHIDYHFSGLEDDDGDDEIENNE-NDGSEVHDDGEL 166
Query: 310 SFDYGQIEQEQDV-STSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFR--VRTG 366
SF+Y +++QD S++ S EL ++T P+ A+EN + R SFR VR
Sbjct: 167 SFEYRVNDKDQDADSSAGGSSELGNSDITFPQIGGPYTAQENEEGNYRKSHSFRRDVRAV 226
Query: 367 SQSAPLFADNK-PDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTN 425
SQSAPLF +N+ SEKL +MR +L+RKF++Y LPTPV+ S SS ++ + +N
Sbjct: 227 SQSAPLFPENRTTPPSEKLLRMRSTLTRKFNTYALPTPVETTRSPSSTTSPGHKNVGSSN 286
Query: 426 SNEA-TTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLL 484
+A T +W+SSPLE + + S V + VL+ESN N T+RLPPPL DGLL
Sbjct: 287 PTKAITKQIWYSSPLETRGPAKVS---SRSMVALKEQVLRESNKN--TSRLPPPLADGLL 341
Query: 485 SSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKV 544
S +KR +FSGPLTS P+P +P+S S ++SGP+ P+ + P S
Sbjct: 342 FSR-------LGTLKRRSFSGPLTSKPLPNKPLSTTS-HLYSGPIPRNPVSKLPKVSS-- 391
Query: 545 SPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISS 604
SP+ASPT VS+PKISELHELPRPP S+++ +GYS PLV R Q +S P +I++
Sbjct: 392 SPTASPTFVSTPKISELHELPRPPPR--SSTKSSRELGYSAPLVSRSQLLSKP---LITN 446
Query: 605 AASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSR 664
+ASPLPIPP A+ RSFSIP+S R + L S + + + SPPLTP++L +
Sbjct: 447 SASPLPIPP-AITRSFSIPTSNLRASDLDMS----KTSLGTKKLGTPSPPLTPMSLIHPP 501
Query: 665 PSS 667
P +
Sbjct: 502 PQA 504
Score = 146 bits (369), Expect = 3e-35
Identities = 72/115 (62%), Positives = 99/115 (85%), Gaps = 6/115 (5%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MK+SL++LRG+ H H+ D + + L Q +ELAQA++D++DMRDCYD+LL+AAAA
Sbjct: 1 MKTSLRRLRGV-----LHKHESKDRRDLRALVQKDELAQASQDVEDMRDCYDSLLNAAAA 55
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVC 115
TA+SAYEF+ESLR++G+CLLEKTALND +EE+G+VL+MLGK+QF+LQKL+DKY+C
Sbjct: 56 TANSAYEFSESLRELGACLLEKTALND-DEESGRVLIMLGKLQFELQKLVDKYLC 109
>At1g62760 hypothetical protein
Length = 312
Score = 48.5 bits (114), Expect = 1e-05
Identities = 42/170 (24%), Positives = 73/170 (42%), Gaps = 19/170 (11%)
Query: 490 YVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSAS 549
+++ S + + S L+ +P + P S + P P P+ PSS P PS+S
Sbjct: 19 FITTSSSSLSPSSSSPSLSPSPPSSSPSSAPPSSL--SPSSPPPLSLSPSSPPPPPPSSS 76
Query: 550 PTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPL 609
P SP +S P PP++ PS++ P S+P L +S ++ P
Sbjct: 77 PLSSLSPSLS-----PSPPSSSPSSA------------PPSSLSPSSPPPLSLSPSSPPP 119
Query: 610 PIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIA 659
P P + S S SS + + + +++S ++ + I L+P A
Sbjct: 120 PPPSSSPLSSLSPSSSSSTYSNQTNLDYIKTSCNITLYKTICYNSLSPYA 169
Score = 34.7 bits (78), Expect = 0.18
Identities = 40/127 (31%), Positives = 61/127 (47%), Gaps = 31/127 (24%)
Query: 539 SSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPN 598
+SS +SPS+S SP +S P PP++ PS++ P S+P
Sbjct: 22 TSSSSLSPSSS-----SPSLS-----PSPPSSSPSSAP------------PSSLSPSSPP 59
Query: 599 NLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPI 658
L +S ++ P P PP + S PS ++ SS+ S+ SS+S +SPP P+
Sbjct: 60 PLSLSPSSPP-PPPPSSSPLSSLSPS----LSPSPPSSSPSSAPPSSLSP--SSPP--PL 110
Query: 659 ALSNSRP 665
+LS S P
Sbjct: 111 SLSPSSP 117
>At4g00890
Length = 431
Score = 47.8 bits (112), Expect = 2e-05
Identities = 65/275 (23%), Positives = 103/275 (36%), Gaps = 40/275 (14%)
Query: 378 PDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSS 437
P+S+E +Q S+ + P + K+ +S S + Q+ S
Sbjct: 102 PESTESQQQFLASVPLR------PLTTEPKTPLSPSSTLKATEESQSQP---------SP 146
Query: 438 PLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAY--- 494
PLE R + T ++ L A T PP +L++ ++ S
Sbjct: 147 PLESIVETWFRPSPPTSTETGDETQLPIPPPQEAKT--PPSSPSMMLNATEEFESQPKPP 204
Query: 495 ---SKKIKRHAFSGPLTSN-----PMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSP 546
SK I PL S P+P++ + + + S P+ P + S SP
Sbjct: 205 LLPSKSIDETRLRSPLMSQASSPPPLPSKSIDENETRSQSPPISPPKSDKQARSQTHSSP 264
Query: 547 SASPTIVSSPKISELHE--LPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAP----NNL 600
S P + SPK SE H+ P PP PS + + L P +P ++AP ++
Sbjct: 265 SPPPLL--SPKASENHQSKSPMPP---PSPTAQISLSSLKSP-IPSPATITAPPPPFSSP 318
Query: 601 VISSAASPLPIPPQAMARSFSIPSSGARVTALHSS 635
+ + SP P PQ PS T H+S
Sbjct: 319 LSQTTPSPKPSLPQIEPNQIKSPSPKLTNTESHAS 353
>At2g25050 hypothetical protein
Length = 742
Score = 47.4 bits (111), Expect = 3e-05
Identities = 65/276 (23%), Positives = 107/276 (38%), Gaps = 41/276 (14%)
Query: 382 EKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQ 441
++ R +R S+ R S + +P +S ++S P P++ T + A+ + +HSSP
Sbjct: 433 DESRGLRVSVQRNVHSKIF-SPRMVQSPVTS----PLPNRSPTQGSPASISRFHSSPSSL 487
Query: 442 KKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRH 501
+ D S + +T+++ P + L + H S+ KK
Sbjct: 488 GITSILHDHGSC-----------KDEESTSSSPASPSI--SFLPTLHPLTSSQPKK---- 530
Query: 502 AFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISEL 561
S P P V S S T P PP ++ S P P IS L
Sbjct: 531 ------ASPQCPQSPTPVHSNGPPSAEAAVTSSPLPPLKPLRIL-SRPPPPPPPPPISSL 583
Query: 562 HELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFS 621
P P + SNS + GP P + +SS+ P P+PP+ + + +
Sbjct: 584 RSTPSPSST--SNS-----IATQGPPPPPPPPPLQSHRSALSSSPLPPPLPPKKLLATTN 636
Query: 622 IPSSGARVTALHSSSALESSHISSISEDIASPPLTP 657
P LHS+S + + S + + SPP+ P
Sbjct: 637 PPPPPP--PPLHSNSRMGAPTSSLV---LKSPPVPP 667
Score = 32.3 bits (72), Expect = 0.88
Identities = 51/216 (23%), Positives = 74/216 (33%), Gaps = 22/216 (10%)
Query: 377 KPDSSEKLRQMRPSLSRKFSSYVLPT--PVDAKSSISSGSNNPK-PSKMQTNSNEATTNL 433
K + S PS+S LPT P+ + + P+ P+ + +N +
Sbjct: 500 KDEESTSSSPASPSIS------FLPTLHPLTSSQPKKASPQCPQSPTPVHSNGPPSAEAA 553
Query: 434 WHSSPLEQKKHETIRDEFSSP-------TVRNAQSVLKESNSNTATTRLPPPLVDGLLSS 486
SSPL K I P ++R+ S SNS AT PPP L S
Sbjct: 554 VTSSPLPPLKPLRILSRPPPPPPPPPISSLRSTPSPSSTSNS-IATQGPPPPPPPPPLQS 612
Query: 487 NHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSP 546
+ +S+ T+NP P P + S P + PP V P
Sbjct: 613 HRSALSSSPLPPPLPPKKLLATTNPPPPPPPPLHSNSRMGAPTSSLVLKSPP-----VPP 667
Query: 547 SASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVG 582
+P +S + +P PP L L G
Sbjct: 668 PPAPAPLSRSHNGNIPPVPGPPLGLKGRGILQNLKG 703
>At1g79480 hypothetical protein
Length = 356
Score = 47.0 bits (110), Expect = 3e-05
Identities = 61/256 (23%), Positives = 89/256 (33%), Gaps = 37/256 (14%)
Query: 369 SAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSN------NPKPSKM 422
+AP F N P++ P LS LP P D+ S+ +S N NP P
Sbjct: 57 NAPPFCINPPNTPPS--SSYPGLSPPPGPITLPNPPDSSSNPNSNPNPPESSSNPNPPDS 114
Query: 423 QTNSNE-ATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVD 481
+N N + +P E + D S+P N SN N T PP
Sbjct: 115 SSNPNSNPNPPVTVPNPPESSSNPNPPDSSSNPN-SNPNPPESSSNPNPPVTVPNPPES- 172
Query: 482 GLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSS 541
SSN + P S+ P P+++ S P P +P PP S
Sbjct: 173 ---SSNPN----------------PPESSSNPNPPITIPYPPESSSPNPPEIVPSPPESG 213
Query: 542 PKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNL- 600
P P + +P P + F L + Y P+ P V+ +
Sbjct: 214 YTPGPVLGPPYSEPGPSTPTGSIPSPSSGF------LPPIVYPPPMAPPSPSVTPTSAYW 267
Query: 601 VISSAASPLPIPPQAM 616
++ + P PI +AM
Sbjct: 268 CVAKPSVPDPIIQEAM 283
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 45.8 bits (107), Expect = 8e-05
Identities = 70/276 (25%), Positives = 112/276 (40%), Gaps = 50/276 (18%)
Query: 388 RPSLSRK---FSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKH 444
RPSL+ S+ PTP+ +S+SS P SK T++ + ++ S+P K
Sbjct: 192 RPSLTNSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPTTSTARSAGSVTRSTPSTTTKS 251
Query: 445 ETIRDEFSSPTVRNAQSVLKESNSNTATTR--LPPPLVDGLLSSNHDYVSAYSKKIKRHA 502
+ P+ + S+T T+R LPP S S +++ A
Sbjct: 252 -------AGPSRSTTPLSRSTARSSTPTSRPTLPP-------SKTISRSSTPTRRPIASA 297
Query: 503 FSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSP-KISEL 561
+ T+NP ++ ++ S P P+P PS +P +S +ASPT+ S P K S++
Sbjct: 298 SAATTTANP------TISQIKP-SSPAPAKPMP-TPSKNPALSRAASPTVRSRPWKPSDM 349
Query: 562 --HELPRPP---TNFPSN--SRLLGLVG----YSGPLVPRGQKVSAPNNLVISSAASPLP 610
L PP T P S G G SG + P G P S + P
Sbjct: 350 PGFSLETPPNLRTTLPERPLSATRGRPGAPSSRSGSVEPGGPPGGRPRRQSCSPSRGRAP 409
Query: 611 IPPQAMARSFSIPSSGARVTALHSSSALESSHISSI 646
+ SSG+ V A++ + S ++S +
Sbjct: 410 -----------MYSSGSSVPAVNRGYSKASDNVSPV 434
>At1g31810 hypothetical protein
Length = 1201
Score = 45.4 bits (106), Expect = 1e-04
Identities = 47/182 (25%), Positives = 66/182 (35%), Gaps = 24/182 (13%)
Query: 476 PPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIP 535
PPPL SN D ++ + I + P P+P+R + P P+
Sbjct: 549 PPPLPS---FSNRDPLTTLHQPINKTPPPPPPPPPPLPSRSI-------------PPPLA 592
Query: 536 QPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVS 595
QPP P P P+ S P S P PP +F G G P
Sbjct: 593 QPPPPRPPPPPPPPPSSRSIPSPSAPPPPPPPPPSF-------GSTGNKRQAQPPPPPPP 645
Query: 596 APNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPL 655
P + ++ +P P PP + S SI G T ++IS+ + A PPL
Sbjct: 646 PPPTRIPAAKCAPPPPPPPPTSHSGSI-RVGPPSTPPPPPPPPPKANISNAPKPPAPPPL 704
Query: 656 TP 657
P
Sbjct: 705 PP 706
Score = 36.2 bits (82), Expect = 0.061
Identities = 40/140 (28%), Positives = 56/140 (39%), Gaps = 29/140 (20%)
Query: 505 GPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHEL 564
GP ++ P P P ++ + P P P P PPSS+ +P P P +S+
Sbjct: 675 GPPSTPPPPPPPPPKANIS--NAPKPPAPPPLPPSSTRLGAPPPPP----PPPLSKTPAP 728
Query: 565 PRPP---TNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARS-- 619
P PP T P LG SGP P G K + +P P PP R+
Sbjct: 729 PPPPLSKTPVPPPPPGLGRGTSSGP-PPLGAK----------GSNAPPPPPPAGRGRASL 777
Query: 620 -------FSIPSSGARVTAL 632
S+P++ + TAL
Sbjct: 778 GLGRGRGVSVPTAAPKKTAL 797
Score = 33.1 bits (74), Expect = 0.51
Identities = 36/124 (29%), Positives = 45/124 (36%), Gaps = 17/124 (13%)
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPP--SSSPKVSPSASPTIVSSPKISE--- 560
P P P P S FS P P P PP +S+ SPS P P S
Sbjct: 501 PSQPPPPPPPPPLFMSTTSFSPSQPPPPPPPPPLFTSTTSFSPSQPPPPPPLPSFSNRDP 560
Query: 561 ---LHE--------LPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPL 609
LH+ P PP PS S + + P P P++ I S ++P
Sbjct: 561 LTTLHQPINKTPPPPPPPPPPLPSRS-IPPPLAQPPPPRPPPPPPPPPSSRSIPSPSAPP 619
Query: 610 PIPP 613
P PP
Sbjct: 620 PPPP 623
Score = 32.3 bits (72), Expect = 0.88
Identities = 36/120 (30%), Positives = 46/120 (38%), Gaps = 22/120 (18%)
Query: 511 PMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTN 570
P P P S P P P P PP PK + S +P + P PP
Sbjct: 660 PPPPPPTSHSGSIRVGPPSTPPPPPPPP---PKANISNAP------------KPPAPPPL 704
Query: 571 FPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSI--PSSGAR 628
PS++R L P P K AP +S +P+P PP + R S P GA+
Sbjct: 705 PPSSTR---LGAPPPPPPPPLSKTPAPPPPPLSK--TPVPPPPPGLGRGTSSGPPPLGAK 759
Score = 31.2 bits (69), Expect = 2.0
Identities = 39/133 (29%), Positives = 44/133 (32%), Gaps = 16/133 (12%)
Query: 511 PMPTRPVSVDSVQMFSGPLLPTPIPQPP--SSSPKVSPSASPTIVSSPKI-----SELHE 563
P P P S FS P P P PP S+ SPS P P + S
Sbjct: 485 PPPPPPPLFTSTTSFSPSQPPPPPPPPPLFMSTTSFSPSQPPPPPPPPPLFTSTTSFSPS 544
Query: 564 LPRPPTNFPSNSRLLGLVGYSGPL-----VPRGQKVSAPNNLVISSAASP----LPIPPQ 614
P PP PS S L P+ P P+ + A P P PP
Sbjct: 545 QPPPPPPLPSFSNRDPLTTLHQPINKTPPPPPPPPPPLPSRSIPPPLAQPPPPRPPPPPP 604
Query: 615 AMARSFSIPSSGA 627
S SIPS A
Sbjct: 605 PPPSSRSIPSPSA 617
>At4g27520 early nodulin-like 2 predicted GPI-anchored protein
Length = 349
Score = 44.7 bits (104), Expect = 2e-04
Identities = 38/119 (31%), Positives = 51/119 (41%), Gaps = 14/119 (11%)
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELP 565
P TS P P P S V S P+ P P P SS + PS++P ++SP S +
Sbjct: 213 PTTSPPAP--PKSTSPVSPSSAPMTSPPAPMAPKSSSTIPPSSAP--MTSPPGSMAPKSS 268
Query: 566 RPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPS 624
P +N P+ S L P G S+P++ SA P P A A S P+
Sbjct: 269 SPVSNSPT---------VSPSLAPGGSTSSSPSDSPSGSAMGPSGDGPSA-AGDISTPA 317
Score = 44.3 bits (103), Expect = 2e-04
Identities = 51/175 (29%), Positives = 70/175 (39%), Gaps = 37/175 (21%)
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIP----------QPPSSSPKVSPSASPTIV 553
+ P +S P P S P+ PT P P SS VSP+ SP
Sbjct: 143 AAPGSSTPGSMTPPGGAHSPKSSSPVSPTTSPPGSTTPPGGAHSPKSSSAVSPATSPPGS 202
Query: 554 SSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPP 613
+PK P PT P P K ++P +S +++P+ PP
Sbjct: 203 MAPKSGS----PVSPTTSP----------------PAPPKSTSP----VSPSSAPMTSPP 238
Query: 614 QAMA--RSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPS 666
MA S +IP S A +T+ S A +SS S S + SP L P ++S PS
Sbjct: 239 APMAPKSSSTIPPSSAPMTSPPGSMAPKSSSPVSNSPTV-SPSLAPGGSTSSSPS 292
Score = 43.1 bits (100), Expect = 5e-04
Identities = 36/116 (31%), Positives = 50/116 (43%), Gaps = 13/116 (11%)
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLPTP------IPQPPSSSPKVSPSASP--TIVSS 555
S P+TS P P P S ++ S P+ P P S+SP VSPS +P + SS
Sbjct: 231 SAPMTSPPAPMAPKSSSTIPPSSAPMTSPPGSMAPKSSSPVSNSPTVSPSLAPGGSTSSS 290
Query: 556 PKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPI 611
P S P + PS + S P GQK S+ N + + S + L +
Sbjct: 291 PSDSPSGSAMGPSGDGPSAAG-----DISTPAGAPGQKKSSANGMTVMSITTVLSL 341
>At1g49490 hypothetical protein
Length = 847
Score = 44.3 bits (103), Expect = 2e-04
Identities = 51/170 (30%), Positives = 76/170 (44%), Gaps = 28/170 (16%)
Query: 417 PKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLP 476
P P++ T S+E T SS +Q + SP A + ++ S ++ T++P
Sbjct: 653 PTPTQAPTPSSETTQVPTPSSESDQS-------QILSPV--QAPTPVQSSTPSSEPTQVP 703
Query: 477 PPLVDGLLSSNHDYVSAYSKKIKRHA-FSGPLTSNPMPTRPV-SVDSVQMFSGPLLPTPI 534
P SS+ Y + ++ P TS+ P S +S Q S PTPI
Sbjct: 704 TP------SSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQ--SPSQAPTPI 755
Query: 535 PQP-----PSSSPKVSPSASPTIVSSPKISELHELPRP----PTNFPSNS 575
+P P+S P SP+ S VSSP+ SE E P P P++ PS+S
Sbjct: 756 LEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPSSVPSSS 805
Score = 41.2 bits (95), Expect = 0.002
Identities = 73/303 (24%), Positives = 105/303 (34%), Gaps = 64/303 (21%)
Query: 368 QSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSN 427
+ +P +KP+ S K Q +P S K +P P KP
Sbjct: 458 KESPKPQPSKPEDSPKPEQPKPEESPKPEQPQIPEPT-------------KP-------- 496
Query: 428 EATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSN 487
SP + + T D + + V+N +S TR+PPP + S
Sbjct: 497 --------VSPPNEAQGPTPDDPYDASPVKNRRS---PPPPKVEDTRVPPP--QPPMPSP 543
Query: 488 HDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSP- 546
YS H+ P+ S+P P P + P P P P PP SP P
Sbjct: 544 SPPSPIYSPPPPVHSPPPPVYSSPPP--PHVYSPPPPVASP--PPPSPPPPVHSPPPPPV 599
Query: 547 -SASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSA 605
S P + S P S ++ P PP++ P YS P S P +
Sbjct: 600 FSPPPPVFSPPPPSPVYS-PPPPSHSPPPP------VYS----PPPPTFSPPPTHNTNQP 648
Query: 606 ASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRP 665
P P QA P+ + T + + S+ S I SP P + +S P
Sbjct: 649 PMGAPTPTQA-------PTPSSETTQVPTPSS------ESDQSQILSPVQAPTPVQSSTP 695
Query: 666 SSD 668
SS+
Sbjct: 696 SSE 698
Score = 39.7 bits (91), Expect = 0.005
Identities = 70/301 (23%), Positives = 106/301 (34%), Gaps = 47/301 (15%)
Query: 401 PTPVDAKSSISSGSNNPKPSKM----QTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTV 456
P P +K S PKP + Q E T + SP + + T D + + V
Sbjct: 461 PKPQPSKPEDSPKPEQPKPEESPKPEQPQIPEPTKPV---SPPNEAQGPTPDDPYDASPV 517
Query: 457 RNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSA------------YSKKIKRHAFS 504
+N +S TR+PPP S + + YS H +S
Sbjct: 518 KNRRS---PPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYS 574
Query: 505 GP--LTSNPMPTRPVSVDSVQ---MFSGP--LLPTPIPQPPSSSPKVSPSASPTIVSSPK 557
P + S P P+ P V S +FS P + P P P S P S S P + S P
Sbjct: 575 PPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPP 634
Query: 558 ISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAP-------NNLVISSAASPLP 610
+ PP +N +G + P + P + ++S +P P
Sbjct: 635 PTF-----SPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPTP 689
Query: 611 I---PPQAMARSFSIPSSGARVTALHSSSALESSHIS--SISEDIASPPLTPIALSNSRP 665
+ P + PSS A + S + + + S + + P TP + SN P
Sbjct: 690 VQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVP-TPSSESNQSP 748
Query: 666 S 666
S
Sbjct: 749 S 749
Score = 38.9 bits (89), Expect = 0.009
Identities = 40/177 (22%), Positives = 74/177 (41%), Gaps = 25/177 (14%)
Query: 378 PDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSS 437
P S + Q+ S S +L +PV A + + S + + +P+++ T S+ + + S
Sbjct: 659 PTPSSETTQVPTPSSESDQSQIL-SPVQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLS 717
Query: 438 PLEQKKHETIRDEFSSPTVRNAQSVLK--ESNSNTATTRLPPPLVDGLLSSNHDYVSAYS 495
P++ +PT + S + S SN + ++ P P+++ + + + S
Sbjct: 718 PVQAPT------PVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPN-----S 766
Query: 496 KKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTI 552
K ++ S S+P + V P P P PSS P SPS +I
Sbjct: 767 KPVQSPTPSSEPVSSPEQSEEVEA-----------PEPTPVNPSSVPSSSPSTDTSI 812
Score = 35.4 bits (80), Expect = 0.10
Identities = 33/112 (29%), Positives = 46/112 (40%), Gaps = 23/112 (20%)
Query: 531 PTPIPQPPSS----SPKVSPSASPTIVSSPKIS-ELHELPRPPTNF---PSNSRLLGLVG 582
PT +P P SS +P +SP +PT V +P S E ++P P + PS + L
Sbjct: 699 PTQVPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEP 758
Query: 583 YSGPL---------VPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSS 625
P P + VS+P A P P+ P S+PSS
Sbjct: 759 VHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPS------SVPSS 804
>At1g36150 hypothetical protein
Length = 256
Score = 43.5 bits (101), Expect = 4e-04
Identities = 38/117 (32%), Positives = 50/117 (42%), Gaps = 12/117 (10%)
Query: 513 PTRPVSVDSVQMFSGPLLPTPIPQPPS--------SSPKVSPSASPTIVSSPKISELHEL 564
P VS + P + +P P S SSP VS S P SSP +S H
Sbjct: 119 PNCDVSTPAASTPVSPPVESPTTSPSSAKSPAITPSSPAVSHSPPPVRHSSPPVS--HSS 176
Query: 565 PRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISS--AASPLPIPPQAMARS 619
P + P SR V +S P+V V A ++ SS AASP P P +++ S
Sbjct: 177 PPVSHSSPPTSRSSPAVSHSSPVVAASSPVKAVSSSTASSPRAASPSPSPSPSISSS 233
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 43.1 bits (100), Expect = 5e-04
Identities = 52/189 (27%), Positives = 78/189 (40%), Gaps = 24/189 (12%)
Query: 452 SSPTVRNAQSVLKESNSNTATTRLPP-PLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSN 510
S P ++ L E ++ ++LPP P + L+ + S S + P TSN
Sbjct: 41 SPPADSSSTPPLSEPSTPPPDSQLPPLPSILPPLTDSPPPPSDSSPPVDSTPSPPPPTSN 100
Query: 511 PMPTRPVSVDSVQM------FSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHEL 564
P+ P ++ + P + PP SSP SP+ PT SP L
Sbjct: 101 ESPSPPEDSETPPAPPNESNDNNPPPSQDLQSPPPSSP--SPNVGPTNPESPP---LQSP 155
Query: 565 PRPPTNFPSNSRLLGLVGYSG--PLVPRGQKVSAPNNLVISSAASPLP-IPPQAMARSFS 621
P PP + P+NS + + P+ P G S P N ++ SP P +PP+
Sbjct: 156 PAPPASDPTNSPPASPLDPTNPPPIQPSGPATSPPAN--PNAPPSPFPTVPPKT------ 207
Query: 622 IPSSGARVT 630
PSSG V+
Sbjct: 208 -PSSGPVVS 215
Score = 35.4 bits (80), Expect = 0.10
Identities = 46/163 (28%), Positives = 60/163 (36%), Gaps = 20/163 (12%)
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPTP---IPQPPSSSPKVSPSASPTIVSSPKISELH 562
P+ S+P P+ P S S P P P +P PS P ++ S P SSP +
Sbjct: 34 PVDSSP-PSPPADSSSTPPLSEPSTPPPDSQLPPLPSILPPLTDSPPPPSDSSPPVDSTP 92
Query: 563 ELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSI 622
P PPT+ S S P S NN + L PP S
Sbjct: 93 S-PPPPTSNESPS-----PPEDSETPPAPPNESNDNN---PPPSQDLQSPPP------SS 137
Query: 623 PSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRP 665
PS T S L+S S+ SPP +P+ +N P
Sbjct: 138 PSPNVGPTN-PESPPLQSPPAPPASDPTNSPPASPLDPTNPPP 179
Score = 32.0 bits (71), Expect = 1.1
Identities = 59/209 (28%), Positives = 78/209 (37%), Gaps = 46/209 (22%)
Query: 367 SQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNS 426
S + PL + P +L + PS+ + P P D+ + S + P P+ S
Sbjct: 47 SSTPPLSEPSTPPPDSQLPPL-PSILPPLTDSP-PPPSDSSPPVDSTPSPPPPT-----S 99
Query: 427 NEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSS 486
NE+ SP E D + P N ESN N PPP D L S
Sbjct: 100 NESP------SPPE--------DSETPPAPPN------ESNDNN-----PPPSQD--LQS 132
Query: 487 NHDYVSAYSKKI-KRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVS 545
S+ S + + S PL S P P P S + + PL PT P S P S
Sbjct: 133 PPP--SSPSPNVGPTNPESPPLQSPPAP--PASDPTNSPPASPLDPTNPPPIQPSGPATS 188
Query: 546 PSASPTIVSSPKISELHELPRPPTNFPSN 574
P A+P SP P P PS+
Sbjct: 189 PPANPNAPPSP-------FPTVPPKTPSS 210
>At3g44340 putative protein
Length = 1122
Score = 42.4 bits (98), Expect = 8e-04
Identities = 37/109 (33%), Positives = 50/109 (44%), Gaps = 20/109 (18%)
Query: 505 GPLTSNPMPTR--PVSVDSVQMF-SGPLL--PTPIPQPPSSSPKVSPSASPTIVSSPKIS 559
GPL++ P P P + F SGP++ P P QPP++ P S +SSP
Sbjct: 218 GPLSNGPPPPMMGPGAFPRGSQFTSGPMMAPPPPYGQPPNAGPFTGNSP----LSSPPA- 272
Query: 560 ELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASP 608
H +P PPTNFP V Y P +P G AP + S+ +P
Sbjct: 273 --HSIP-PPTNFPG-------VPYGRPPMPGGFPYGAPPQQLPSAPGTP 311
Score = 36.2 bits (82), Expect = 0.061
Identities = 41/161 (25%), Positives = 59/161 (36%), Gaps = 5/161 (3%)
Query: 467 NSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLT-SNPMPTRPVSVDSVQMF 525
N+N + PP V G N + ++A + + + P+ S P P+ P S Q F
Sbjct: 13 NNNQQNSGGPPNFVPGS-QGNPNSLAANMQNLNINRPPPPMPGSGPRPSPPFG-QSPQSF 70
Query: 526 SGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRP--PTNFPSNSRLLGLVGY 583
P P P + P+A P++S+ P P PSN G
Sbjct: 71 PQQQQQQPRPSPMARPGPPPPAAMARPGGPPQVSQPGGFPPVGRPVAPPSNQPPFGGRPS 130
Query: 584 SGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPS 624
+GPLV G P S +P P + AR S
Sbjct: 131 TGPLVGGGSSFPQPGGFPASGPPGGVPSGPPSGARPIGFGS 171
>At3g24480 disease resistance protein, putative
Length = 494
Score = 42.0 bits (97), Expect = 0.001
Identities = 33/109 (30%), Positives = 42/109 (38%), Gaps = 20/109 (18%)
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHE 563
S P+ + P P P P LP P+ PP S P SP SP + S P +H
Sbjct: 404 SPPIVALPPPPPP----------SPPLPPPVYSPPPSPPVFSPPPSPPVYSPPPPPSIHY 453
Query: 564 LPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIP 612
PP +S + GPL P ++ S ASP P P
Sbjct: 454 SSPPPPPVHHSSPPPPSPEFEGPLPP----------VIGVSYASPPPPP 492
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 42.0 bits (97), Expect = 0.001
Identities = 52/166 (31%), Positives = 65/166 (38%), Gaps = 32/166 (19%)
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVS---PSASPTIVS-SPKISEL 561
P S P PT PVS P PTP P PS +P VS P+ +P++ S +P +S
Sbjct: 86 PTPSVPSPTPPVS---------PPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVS-- 134
Query: 562 HELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFS 621
P PPT PS P P VS P S SP P P S
Sbjct: 135 ---PPPPTPTPS---------VPSPTPP----VSPPPPTPTPSVPSPTPPVPTDPMPSPP 178
Query: 622 IPSSGARVTALHSSSALESSHISSISEDIASPP-LTPIALSNSRPS 666
P S T S + + + + SPP +TP + S PS
Sbjct: 179 PPVSPPPPTPTPSVPSPPDVTPTPPTPSVPSPPDVTPTPPTPSVPS 224
Score = 32.7 bits (73), Expect = 0.67
Identities = 21/67 (31%), Positives = 32/67 (47%), Gaps = 9/67 (13%)
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLPTP-IPQPPSSSPKVSPSASPTIVSSPKISELH 562
S P + P+PT P+ + P PTP +P PP +P +P++ S P ++
Sbjct: 161 SVPSPTPPVPTDPMPSPPPPVSPPPPTPTPSVPSPPDVTP---TPPTPSVPSPPDVT--- 214
Query: 563 ELPRPPT 569
P PPT
Sbjct: 215 --PTPPT 219
Score = 31.6 bits (70), Expect = 1.5
Identities = 22/67 (32%), Positives = 30/67 (43%), Gaps = 8/67 (11%)
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPS--SSPKVSPS-ASPTIVSSPKISELH 562
P+ S P P P P TP P PS S P V+P+ +P++ S P ++
Sbjct: 173 PMPSPPPPVSPPPPTPTPSVPSPPDVTPTPPTPSVPSPPDVTPTPPTPSVPSPPDVT--- 229
Query: 563 ELPRPPT 569
P PPT
Sbjct: 230 --PTPPT 234
Score = 29.6 bits (65), Expect = 5.7
Identities = 32/140 (22%), Positives = 49/140 (34%), Gaps = 5/140 (3%)
Query: 528 PLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPL 587
P P P PP +P V P P + P PP + P + + + P+
Sbjct: 74 PPAPVPPVSPPPPTPSVPSPTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPV 133
Query: 588 VPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSIS 647
P P+ + S P+ PP S P+ + S S + +
Sbjct: 134 SP-PPPTPTPS---VPSPTPPVSPPPPTPTPSVPSPTPPVPTDPMPSPPPPVSPPPPTPT 189
Query: 648 EDIASPP-LTPIALSNSRPS 666
+ SPP +TP + S PS
Sbjct: 190 PSVPSPPDVTPTPPTPSVPS 209
>At1g26210 hypothetical protein
Length = 168
Score = 42.0 bits (97), Expect = 0.001
Identities = 40/149 (26%), Positives = 68/149 (44%), Gaps = 23/149 (15%)
Query: 275 HIDYHFNGLEE-----EDGDEGDEGYEDDDGYDENDDGELSFDYGQIEQEQDVSTSRNSM 329
+I+ F+G ++ +D D+ + E DDGY ENDDG+ S D G E + +++ +S
Sbjct: 25 YIEDAFHGNDQSSVVVDDDDDDTQVKEADDGY-ENDDGDTSDDGGDEESDDSMASDASSG 83
Query: 330 ELDQVELTLPRGSTAEAAKENLDK---LQRNLFSFRVRTGSQSAPLFADNKPDSSEKLRQ 386
+Q LP+ AA++N K LQ+ Q N+ + S+ +
Sbjct: 84 PSNQ----LPKHINKHAARKNGSKQVYLQKR----------QHTEKTISNEGEKSDLKAR 129
Query: 387 MRPSLSRKFSSYVLPTPVDAKSSISSGSN 415
R S + + S VD K+ +S +N
Sbjct: 130 TRTSAASRVQSRGKDKVVDDKTGVSKVAN 158
>At5g38560 putative protein
Length = 681
Score = 41.6 bits (96), Expect = 0.001
Identities = 37/123 (30%), Positives = 48/123 (38%), Gaps = 2/123 (1%)
Query: 504 SGPLTSNPMPTRPVSVDSVQ-MFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELH 562
S P +S+P P+ PV + S P P I PP S+P +P A P VS P +
Sbjct: 63 SPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDAS 122
Query: 563 ELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSI 622
P PT + P P G+ S P + + PP A S S
Sbjct: 123 PSPPAPTTTNPPPKPSPSPPGETP-SPPGETPSPPKPSPSTPTPTTTTSPPPPPATSASP 181
Query: 623 PSS 625
PSS
Sbjct: 182 PSS 184
Score = 39.3 bits (90), Expect = 0.007
Identities = 51/225 (22%), Positives = 82/225 (35%), Gaps = 13/225 (5%)
Query: 397 SYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTV 456
S V P P+ + S +S + P P + Q + A + Q + P V
Sbjct: 2 SLVPPLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVV 61
Query: 457 RNAQSVLKESNSNTATTRLPPPLVDG-----LLSSNHDYVSAYSKKIKRHAFSGPLT--S 509
+ S T PP + +++S A + S P +
Sbjct: 62 SSPPPSSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDA 121
Query: 510 NPMPTRPVSVDSVQMFSGPLLPTPIPQPPS---SSPKVSPSA-SPTIVSSPKISELHELP 565
+P P P + + S P P P PP S PK SPS +PT +SP
Sbjct: 122 SPSPPAPTTTNPPPKPS-PSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPPATSAS 180
Query: 566 RPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLP 610
PP++ P++ L P+VPR + ++ P ++ + LP
Sbjct: 181 -PPSSNPTDPSTLAPPPTPLPVVPREKPIAKPTGPASNNGNNTLP 224
Score = 34.3 bits (77), Expect = 0.23
Identities = 38/114 (33%), Positives = 48/114 (41%), Gaps = 13/114 (11%)
Query: 506 PLTSNPMPTRPVSVDSVQMF-SGPLLPTPIPQPPSSSPK-VSPSASPTIVSSPKISELHE 563
P +SN T P + + S P TP P PP S P VS S P +VSSP S
Sbjct: 12 PPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSS--- 68
Query: 564 LPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMA 617
PP + P + V S P P S P S+ A+ P PPQ ++
Sbjct: 69 --SPPPSPPVITSPPPTVASSPP--PPVVIASPPP----STPATTPPAPPQTVS 114
>At5g07750 putative protein
Length = 1289
Score = 41.2 bits (95), Expect = 0.002
Identities = 96/421 (22%), Positives = 148/421 (34%), Gaps = 83/421 (19%)
Query: 292 DEGYEDDDGYDEN---------DDGELSFDYGQIE-QEQDVSTSRNSMELDQVELTLPRG 341
D Y D D N DDG+ +E +E D ST + + D+ L
Sbjct: 410 DVQYRSDGKADSNIDSVKDIGIDDGDEQRKRRTVEAKENDSSTVQTQSKGDEESNDLESM 469
Query: 342 STAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADN--KPDSSEK------LRQMRPSL-- 391
S N ++ + R + G+ + P A + KP S ++ +R +P+
Sbjct: 470 SQKTNTSLNKPISEKPQATLRKQVGANAKPAAAGDSLKPKSKQQETQGPNVRMAKPNAVS 529
Query: 392 --------SRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKK 443
S K S +V P S+ +S + + K K T+ + + K
Sbjct: 530 RWIPSNKGSYKDSMHVAYPPTRINSAPASITTSLKDGKRATSPDGVIP--------KDAK 581
Query: 444 HETIRDEFSSPTVRNAQSVLKESNSNTATT----------RLPPPLVDGLLSSNHDYVSA 493
+ +R SSP +R+ + +S+ T + PPPL L+S V
Sbjct: 582 TKYLRASVSSPDMRSRAPICSSPDSSPKETPSSLPPASPHQAPPPLPS--LTSEAKTVLH 639
Query: 494 YSKKIKRH----------AFSGPLTSN--PMPTRPVSVDSVQMFSGPLLPTPIPQPPSSS 541
S+ + +S TS P P P S + SG +LP P P PP S
Sbjct: 640 SSQAVASPPPPPPPPPLPTYSHYQTSQLPPPPPPPPPFSSERPNSGTVLPPPPPPPPPFS 699
Query: 542 PKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVP----RGQKVSAP 597
+ P+ S T++ P P PP F S G V P P ++ P
Sbjct: 700 SE-RPN-SGTVLPPP--------PPPPLPFSSERPNSGTVLPPPPSPPWKSVYASALAIP 749
Query: 598 NNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTP 657
S A + P PP P A + SS L++S + S PP
Sbjct: 750 AICSTSQAPTSSPTPP---------PPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFAS 800
Query: 658 I 658
+
Sbjct: 801 V 801
Score = 37.7 bits (86), Expect = 0.021
Identities = 39/157 (24%), Positives = 57/157 (35%), Gaps = 13/157 (8%)
Query: 507 LTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPR 566
L S P P P SV+ S LLP P P PP V ++ + P P
Sbjct: 786 LPSPPPPPPPPPFASVRRNSETLLPPPPPPPPPPFASVRRNSETLLPPPP--------PP 837
Query: 567 PPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSG 626
PP S S P P + + ++ A+ +PP + + PS
Sbjct: 838 PPWKSLYASTFETHEACSTSSSPPPPPPPPPFSPLNTTKANDYILPPPPLPYTSIAPSPS 897
Query: 627 ARVTALHSSSALESSHISSISEDIASPPLTPIALSNS 663
++ LH S+ S + + A PP P SN+
Sbjct: 898 VKILPLHGISSAPSPPVKT-----APPPPPPPPFSNA 929
Score = 33.5 bits (75), Expect = 0.39
Identities = 41/142 (28%), Positives = 53/142 (36%), Gaps = 33/142 (23%)
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPP-----------------SSSPKVSP 546
S L P P P SV+ S LLP P P PP SSSP P
Sbjct: 805 SETLLPPPPPPPPPPFASVRRNSETLLPPPPPPPPWKSLYASTFETHEACSTSSSPPPPP 864
Query: 547 SASPTIVSSPKISELHELPRPPTNFPS-----NSRLLGLVGYSGPLVPRGQKVSAPNNLV 601
P + + + LP PP + S + ++L L G S SAP+
Sbjct: 865 PPPPFSPLNTTKANDYILPPPPLPYTSIAPSPSVKILPLHGIS----------SAPSP-P 913
Query: 602 ISSAASPLPIPPQAMARSFSIP 623
+ +A P P PP + A S P
Sbjct: 914 VKTAPPPPPPPPFSNAHSVLSP 935
>At4g32710 putative protein kinase
Length = 731
Score = 41.2 bits (95), Expect = 0.002
Identities = 49/172 (28%), Positives = 66/172 (37%), Gaps = 22/172 (12%)
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLP--TPIPQPPSSSPKVSPSASP--TIVSSPKISEL 561
P TS P + P + DSV S P P +P+P PP SSP PS P +SP +
Sbjct: 11 PATSPPAMSLPPA-DSVPDTSSPPAPPLSPLP-PPLSSPPPLPSPPPLSAPTASPPPLPV 68
Query: 562 HELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASP-------LPIPPQ 614
P PP P L P P + S P+ V + + SP P PP
Sbjct: 69 ESPPSPPIESPPPPLL------ESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPP 122
Query: 615 AMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPS 666
+ S ++P T +L S ++ PP P S +PS
Sbjct: 123 SSPPSETVPPGN---TISPPPRSLPSESTPPVNTASPPPPSPPRRRSGPKPS 171
Score = 35.0 bits (79), Expect = 0.14
Identities = 62/209 (29%), Positives = 79/209 (37%), Gaps = 56/209 (26%)
Query: 476 PPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLT-----SNPMPTRPVSVD---------- 520
PPPL++ + S S + + S PL +P P+ P S
Sbjct: 80 PPPLLESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISPP 139
Query: 521 --SVQMFSGPLLPTPIPQPPS-----SSPKVSPSASPTIVSSP--------KISELHELP 565
S+ S P + T P PPS S PK PS P I SSP + E P
Sbjct: 140 PRSLPSESTPPVNTASPPPPSPPRRRSGPK--PSFPPPINSSPPNPSPNTPSLPETSPPP 197
Query: 566 RPP---TNFPSNSR----------LLGLVGYSGPLVPRGQKVSAPNNLVISSAASPL--- 609
+PP T FPS+S L G +G L P G V+ P VI SP
Sbjct: 198 KPPLSTTPFPSSSTPPPKKSPAAVTLPFFGPAGQL-PDG-TVAPPIGPVIEPKTSPAESI 255
Query: 610 -PIPPQAMARSFSIPSSGARVTALHSSSA 637
P PQ + +P S T+ H SSA
Sbjct: 256 SPGTPQPL-----VPKSLPVTTSYHRSSA 279
Score = 34.3 bits (77), Expect = 0.23
Identities = 42/143 (29%), Positives = 58/143 (40%), Gaps = 26/143 (18%)
Query: 529 LLPTPIPQPPSSSPKVS--PSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGP 586
L P+ P P +S P +S P+ S SSP L LP PP + P P
Sbjct: 3 LSPSSSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLP-PPLSSP-------------P 48
Query: 587 LVPRGQKVSAPNNLVISSAASPLPI--PPQAMARSFSIPSSGARVTALHSSSALESSHIS 644
+P +SAP +++ PLP+ PP S P + S + S H+S
Sbjct: 49 PLPSPPPLSAP-----TASPPPLPVESPPSPPIESPPPPLLESPPPPPLESPSPPSPHVS 103
Query: 645 SISEDIASPPLTPIALSNSRPSS 667
+ S SPPL + S P S
Sbjct: 104 APS---GSPPLPFLPAKPSPPPS 123
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.129 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,335,191
Number of Sequences: 26719
Number of extensions: 743695
Number of successful extensions: 6663
Number of sequences better than 10.0: 382
Number of HSP's better than 10.0 without gapping: 123
Number of HSP's successfully gapped in prelim test: 265
Number of HSP's that attempted gapping in prelim test: 4555
Number of HSP's gapped (non-prelim): 1055
length of query: 669
length of database: 11,318,596
effective HSP length: 106
effective length of query: 563
effective length of database: 8,486,382
effective search space: 4777833066
effective search space used: 4777833066
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0117b.11


