

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
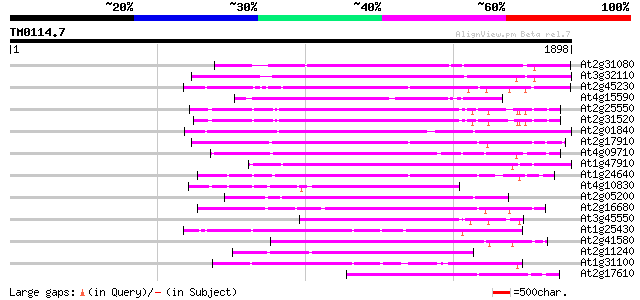
Query= TM0114.7
(1898 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g31080 putative non-LTR retroelement reverse transcriptase 684 0.0
At3g32110 non-LTR reverse transcriptase, putative 662 0.0
At2g45230 putative non-LTR retroelement reverse transcriptase 587 e-167
At4g15590 reverse transcriptase like protein 569 e-162
At2g25550 putative non-LTR retroelement reverse transcriptase 546 e-155
At2g31520 putative non-LTR retroelement reverse transcriptase 538 e-152
At2g01840 putative non-LTR retroelement reverse transcriptase 527 e-149
At2g17910 putative non-LTR retroelement reverse transcriptase 526 e-149
At4g09710 RNA-directed DNA polymerase -like protein 509 e-144
At1g47910 reverse transcriptase, putative 482 e-135
At1g24640 hypothetical protein 480 e-135
At4g10830 putative protein 468 e-131
At2g05200 putative non-LTR retroelement reverse transcriptase 462 e-130
At2g16680 putative non-LTR retroelement reverse transcriptase 443 e-124
At3g45550 putative protein 409 e-113
At1g25430 hypothetical protein 399 e-110
At2g41580 putative non-LTR retroelement reverse transcriptase 385 e-106
At2g11240 pseudogene 372 e-102
At1g31100 hypothetical protein 362 1e-99
At2g17610 putative non-LTR retroelement reverse transcriptase 353 7e-97
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 684 bits (1766), Expect = 0.0
Identities = 427/1239 (34%), Positives = 641/1239 (51%), Gaps = 94/1239 (7%)
Query: 692 VYASPIPANREILWRHMTLLRRRFLLPWLLLGDFNEILFPSEVRGGDF-LPNRAAMFASV 750
VYA+P + R LW + + P L+ GDFN IL+ E GG+ L + F
Sbjct: 6 VYAAPSVSRRSGLWGELKDVVNGLEGPLLIGGDFNTILWVDERMGGNGRLSPDSLAFGDW 65
Query: 751 LDTCQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDH 810
++ L+DLG G +FTW + + +++KRLDR ++ + +AVV L + SDH
Sbjct: 66 INELSLIDLGFKGNKFTWRRGRQESTVVAKRLDRVFVCAHARLKWQEAVVSHLPFMASDH 125
Query: 811 CPILLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRSGEECITSKLERVREASTVF 870
P+ +Q Q R L W
Sbjct: 126 APLYVQLEPLQQ-------RKLRKW----------------------------------- 143
Query: 871 NKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAELQREYRNILRQEELL*YQKA 930
N+EVFG+I RK + A ++ VQ L ++ D++ E L +E +L QEE L +QK+
Sbjct: 144 NREVFGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEVLLKEMDLVLEQEETLWFQKS 203
Query: 931 RENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFT-- 988
RE + LGDRNT +FHT T+IRRR+NR+ LK +D W D+ L+ + ++ L++
Sbjct: 204 REKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMALTYYKRLYSLE 263
Query: 989 GITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKK 1048
++ + + T F S+S K+ LLQ K EV A+ SM + APG DG+QP FY++
Sbjct: 264 DVSEVRNMLPTGG-FASISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPDGYQPVFYQQ 322
Query: 1049 YWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVIT 1108
W VG S+ V E F+ G + + D L+VLI KV +P ++ RP+SLCNV FK+IT
Sbjct: 323 CWETVGPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQFRPVSLCNVLFKIIT 382
Query: 1109 KVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEK 1168
K++V RL+ + KLIGP Q+SF+PGR ++DN L QE +H M + ++G + K+DLEK
Sbjct: 383 KMMVTRLKNVISKLIGPAQASFIPGRLSIDNIVLVQEAVHSMRRKKGRKGWMLLKLDLEK 442
Query: 1169 AYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPM 1228
AYD V W FLQETLE G E + IM+ VT +S+LWNG R +F P RGLRQGDP+
Sbjct: 443 AYDRVRWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSVLWNGERTDSFVPARGLRQGDPL 502
Query: 1229 SPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLL 1288
SPYLFVLC+ERL LI++ V K +WKPI +S G +SH+ FADD++LF EASVAQ+ ++
Sbjct: 503 SPYLFVLCLERLCHLIEASVGKREWKPIAVSCGGSKLSHVCFADDLILFAEASVAQIRII 562
Query: 1289 ASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGR 1348
++ FC++SG K++L KSK S VS ++ IS + I +LGKYLG P+ R
Sbjct: 563 RRVLERFCEASGQKVSLEKSKIFFSHNVSREMEQLISEESGIGCTKELGKYLGMPILQKR 622
Query: 1349 IHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKIN 1408
+++ F +LE + +L W+ LSLAGR+ L K+V++++P + M +P ++
Sbjct: 623 MNKETFGEVLERVSARLAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPVSTLDTLD 682
Query: 1409 QLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNK 1468
+ R+F+W HL++W KI +PK GG+G+R N AL+ K W L+
Sbjct: 683 RYSRTFLWGSTMEKKKQHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKES 742
Query: 1469 LWVRVLSHKY----LSNSSVLQVQAKPQDSQVWKGL-LKARDRLHTGFVFRLGNGET-SL 1522
LW RV+ KY + ++S L+ Q P+ S W+ + + R+ + G + G+G T
Sbjct: 743 LWARVVRKKYKVGGVQDTSWLKPQ--PRWSSTWRSVAVGLREVVVKGVGWVPGDGCTIRF 800
Query: 1523 WHDDWSGMGNIAPAVPY-VDIHDIDRRLCDLVD----NGTWNMQVLYTSLPVDVLERLQK 1577
W D W P V D+ R+ D WN+++L LP V RL
Sbjct: 801 WLDRWLLQ---EPLVELGTDMIPEGERIKVAADYWLPGSGWNLEILGLYLPETVKRRLLS 857
Query: 1578 IKPTIVPNRQDVWTWETNSMGIYTVRGAYRWLQDQQSHLPVVDD-WNWIWKLKVPEKIRT 1636
+ + D +W+ G +TVR AY LQ P + +N IWKL PE++R
Sbjct: 858 VVVQVFLGNGDEISWKGTQDGAFTVRSAYSLLQGDVGDRPNMGSFFNRIWKLITPERVRV 917
Query: 1637 FVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELWLKFGAF-AW 1695
F+WL QN + N+ R R ++ + CS C+ EE ILH LR CP + +W +
Sbjct: 918 FIWLVSQNVIMTNVERVRRHLSENAICSVCNGAEETILHVLRDCPAMEPIWRRLLPLRRH 977
Query: 1696 PNFTAGDHSSWIRSQARSNNGV---KFIVGLWGVWKWRNNMIFEDSPWSLEEAWRRVCHE 1752
F + W+ + G+ F +G+W WKWR +F + R++C +
Sbjct: 978 HEFFSQSLLEWLFTNMDPVKGIWPTLFGMGIWWAWKWRCCDVFGE---------RKICRD 1028
Query: 1753 HDEIVAVLGEEAGSMDCW-LGS-----------RWQPPMAGSIKLNVDGSYRDVDDSSGV 1800
+ + + EE + +G+ RWQ P G +K+ DG+ R +
Sbjct: 1029 RLKFIKDMAEEVRRVHVGAVGNRPNGVRVERMIRWQVPSDGWVKITTDGASRGNHGLAAA 1088
Query: 1801 GGLARDPSGNWLFGFLAHRRGGNAFLAEAQALLLGLELVWARGYRDIVVEVDC----ADL 1856
GG R+ G WL GF + A LAE GL + W +G+R + +++DC L
Sbjct: 1089 GGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYYGLLIAWDKGFRRVELDLDCKLVVGFL 1148
Query: 1857 LQSLDDEDRRRFLPILGDIRNMKDRGWRISLERVRRDCN 1895
+ + FL L + R W + + V R+ N
Sbjct: 1149 STGVSNAHPLSFLVRL--CQGFFTRDWLVRVSHVYREAN 1185
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 662 bits (1707), Expect = 0.0
Identities = 435/1318 (33%), Positives = 657/1318 (49%), Gaps = 79/1318 (5%)
Query: 616 DVFILMETRCQFRRVSAFWNSLGFFPIHIEEARGFSGGIWVLANTSVPYNFRVIDTHQQV 675
DV + ET + S LGF +A G SGG+W+L T + V T Q +
Sbjct: 595 DVLAIFETHAGGDQASRICQGLGFENSFRVDAVGHSGGLWLLWRTGIGEVSVVDSTDQFI 654
Query: 676 VTFEV-WKDNLSWVCSAVYASPIPANREILWRHMTLLRRRFLLPWLLLGDFNEILFPSEV 734
+V KDN++ V VYA+P + R LW + + R P ++ GDFN I+ E
Sbjct: 655 HAKDVNGKDNVNLV--VVYAAPTASRRSGLWDRLGDVIRSMDGPVVIGGDFNTIVRLDER 712
Query: 735 RGGDF-LPNRAAMFASVLDTCQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKV 793
GG+ L + + F ++ L+DLG G +FTW + +R ++KRLDR L ++
Sbjct: 713 SGGNGRLSSDSLAFGEWINDHSLIDLGFKGNKFTWKRGREERFFVAKRLDRVLCCAHARL 772
Query: 794 AFPDAVVDVLNRVHSDHCPILLQCGSPQTLAHN--RPFRFLAAWADHPDFAAVVDNAWRS 851
+ +A V L + SDH P+ +Q +P+ + RPFRF AAW HP F ++ +W
Sbjct: 773 KWQEASVLHLPFLASDHAPLYVQL-TPEVSGNRGRRPFRFEAAWLSHPGFKELLLTSW-- 829
Query: 852 GEECITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAEL 911
NK++ VQ LD + D++ E EL
Sbjct: 830 -------------------NKDIS----------TPEALKVQELLDLHQSDDLLKKEEEL 860
Query: 912 QREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGD 971
+++ +L QEE++ QK+RE GDRNT +FHT T+IRRR+N++ L+ DG W +
Sbjct: 861 LKDFDVVLEQEEVVWMQKSREKWFVHGDRNTKFFHTSTIIRRRRNQIEMLQDNDGRWLSN 920
Query: 972 EEVLKRVVHGFFVNLFT-GITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSM 1030
+ L+ ++ L++ ++ + F +LS S L +P EV A+ SM
Sbjct: 921 AQELETHAIDYYKRLYSLDDLDAVVEQLPQEGFTALSEADFSSLTKPFSPLEVEGAIRSM 980
Query: 1031 NSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPST 1090
Y APG DGFQP FY++ W VG+S+ V + F G + D+LVVLI KV +P
Sbjct: 981 GKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFFSSGSFPQETNDVLVVLIAKVLKPEK 1040
Query: 1091 VRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHM 1150
+ RPISLCNV FK ITKV+V RL+ + KLIGP Q+SF+PGR + DN + QEV+H M
Sbjct: 1041 ITQFRPISLCNVLFKTITKVMVGRLKGVINKLIGPAQTSFIPGRLSTDNIVVVQEVVHSM 1100
Query: 1151 SKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNG 1210
+ +G + K+DLEKAYD + W L++TL+ G P + IM V + +LWNG
Sbjct: 1101 RRKKGVKGWMLLKLDLEKAYDRIRWDLLEDTLKAAGLPGTWVQWIMKCVEGPSMRLLWNG 1160
Query: 1211 CRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFF 1270
+ AFKP RGLRQGDP+SPYLFVLC+ERL LI+S + WKPI++S++GP +SH+ F
Sbjct: 1161 EKTDAFKPLRGLRQGDPLSPYLFVLCIERLCHLIESSIAAKKWKPIKISQSGPRLSHICF 1220
Query: 1271 ADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPI 1330
ADD++LF EAS+ Q+ +L ++ FC +SG K++L KSK SK V + IS + +
Sbjct: 1221 ADDLILFAEASIDQIRVLRGVLEKFCGASGQKVSLEKSKIYFSKNVLRDLGKRISEESGM 1280
Query: 1331 PFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMP 1390
DLGKYLG P+ RI++ F +++ +L W+ MLS AGR+ L K+V+ ++
Sbjct: 1281 KATKDLGKYLGVPILQKRINKETFGEVIKRFSSRLAGWKGRMLSFAGRLTLTKAVLTSIL 1340
Query: 1391 TYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELA 1450
+TM +P+ ++++ R+F+W HLV W ++ P+ GGLG+R
Sbjct: 1341 VHTMSTIKLPQSTLDGLDKVSRAFLWGSTLEKKKQHLVAWTRVCLPRREGGLGIRSATAM 1400
Query: 1451 NTALMGKAIWCLMHKPNKLWVRVLSHKYLSNS--SVLQVQAKPQDSQVWKGL-LKARDRL 1507
N AL+ K W +++ + LW +V+ KY AK S W+ + + RD +
Sbjct: 1401 NKALIAKVGWRVLNDGSSLWAQVVRSKYKVGDVHDRNWTVAKSNWSSTWRSVGVGLRDVI 1460
Query: 1508 HTGFVFRLGNG-ETSLWHDDWSGMGNIA--PAVPYVDIHDIDRRLC---DLVDNGT-WNM 1560
+ +G+G + W D W IA VP + + LC DL +GT W+M
Sbjct: 1461 WREQHWVIGDGRQIRFWTDRWLSETPIADDSIVPL----SLAQMLCTARDLWRDGTGWDM 1516
Query: 1561 QVLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWL-QDQQSHLPVV 1619
+ + + L + V D W S G +TV+ A+ L D +
Sbjct: 1517 SQIAPFVTDNKRLDLLAVIVDSVTGAHDRLAWGMTSDGRFTVKSAFAMLTNDDSPRQDMS 1576
Query: 1620 DDWNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRT 1679
+ +WK++ PE++R F+WL + ++ N R R + S C C E ILH LR
Sbjct: 1577 SLYGRVWKVQAPERVRVFLWLVVNQAIMTNSERKRRHLCDSDVCQVCRGGIESILHVLRD 1636
Query: 1680 CPHSQELWLKF-GAFAWPNFTAGDHSSWIRSQAR-------SNNGVKFIVGLWGVWKWRN 1731
CP +W + +F W+ S R S+ F + +W WKWR
Sbjct: 1637 CPAMSGIWDRIVPRRLQQSFFTMSLLEWLYSNLRQGLMTEGSDWSTMFAMAVWWGWKWRC 1696
Query: 1732 NMIFEDSPWSLEEAWRRVCHEHDEIVAVLGEEAGSMDCWLGS-------RWQPPMAGSIK 1784
+ IF ++ + RV D V V + ++ L RW PPM G K
Sbjct: 1697 SNIFGENKTCRD----RVRFIKDLAVEVSIAYSREVELRLSGLRVNKPIRWTPPMEGWYK 1752
Query: 1785 LNVDGSYRDVDDSSGVGGLARDPSGNWLFGFLAHRRGGNAFLAEAQALLLGLELVWARGY 1844
+N DG+ R + GG+ R+ +G W GF + +A LAE + GL + WA+
Sbjct: 1753 INTDGASRGNPGLASAGGVLRNSAGAWCGGFAVNIGRCSAPLAELWGVYYGLYMAWAKQL 1812
Query: 1845 RDIVVEVD----CADLLQSLDDEDRRRFLPILGDIRNMKDRGWRISLERVRRDCNAPA 1898
+ +EVD L + + FL L N + W + + V R+ N+ A
Sbjct: 1813 THLELEVDSEVVVGFLKTGIGETHPLSFLVRL--CHNFLSKDWTVRISHVYREANSLA 1868
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 587 bits (1512), Expect = e-167
Identities = 405/1361 (29%), Positives = 651/1361 (47%), Gaps = 78/1361 (5%)
Query: 588 KLISWNVRGALYANGKLAIRELVRSKKPDVFILMETRCQFRRVSAFWNSLGFFPIHIEEA 647
+++SWN +G +RE+ P+V L ET+ + + LGFF +H E
Sbjct: 2 RILSWNCQGVGNTPTVRHLREIRGLYFPEVIFLCETKKRRNYLENVVGHLGFFDLHTVEP 61
Query: 648 RGFSGGIWVLANTSVPYNFRVIDTHQQVV-TFEVWKDNLSWVCSAVYASPIPANREILWR 706
G SGG+ ++ SV +V+ + ++++ +W+D + + +Y P+ A R LW
Sbjct: 62 IGKSGGLALMWKDSV--QIKVLQSDKRLIDALLIWQDK-EFYLTCIYGEPVQAERGELWE 118
Query: 707 HMTLLRRRFLLPWLLLGDFNEILFPSEVRGGDFLPNRAAM-FASVLDTCQLVDLGAVGRR 765
+T L PW+L GDFNE++ PSE GG + + F +L++C L ++ G +
Sbjct: 119 RLTRLGLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCLEFRQMLNSCGLWEVNHSGYQ 178
Query: 766 FTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCPILLQCGSPQTLAH 825
F+W+ ND L+ RLDR + + W FP A L ++ SDH P++
Sbjct: 179 FSWYGNRNDELVQC-RLDRTVANQAWMELFPQAKATYLQKICSDHSPLINNLVGDNWRKW 237
Query: 826 NRPFRFLAAWADHPDFAAVVDNAWRSGEECITSKLERVREASTVFNKEVFGNIFRRKRHV 885
F++ W F ++ N W S + T+ L + AS +E+ K
Sbjct: 238 -AGFKYDKRWVQREGFKDLLCNFW-SQQSTKTNALMMEKIASC--RREISKWKRVSKPSS 293
Query: 886 EARLRGVQRELDRRVTS------DMVLFEAELQREYRNILRQEELL*YQKARENRVHLGD 939
R++ +Q +LD ++ + EL +EY N EE +K+R + GD
Sbjct: 294 AVRIQELQFKLDAATKQIPFDRRELARLKKELSQEYNN----EEQFWQEKSRIMWMRNGD 349
Query: 940 RNTAYFHTQTLIRRRKNRVHRLKLEDGT-WCGDEEVLKRVVHGFFVNLFTGITPLASPRV 998
RNT YFH T RR +NR+ +L E+G W DE+ L RV +F LF +
Sbjct: 350 RNTKYFHAATKNRRAQNRIQKLIDEEGREWTSDED-LGRVAEAYFKKLFASEDVGYTVEE 408
Query: 999 THDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLW 1058
+L P +S + + LL P+ K+EV+ A S+N + PG DG F Y+++W +GD +
Sbjct: 409 LENLTPLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQIT 468
Query: 1059 HMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPF 1118
MV+ F+ G + E + + LIPK+ + + D RPISLCNV +KVI K++ NRL+
Sbjct: 469 EMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANRLKKI 528
Query: 1119 LRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKST-AKRGMVAFKIDLEKAYDSVSWSF 1177
L LI Q++F+ GR DN +A E++H +S + +A K D+ KAYD V W F
Sbjct: 529 LPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPF 588
Query: 1178 LQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCM 1237
L++ + GF + I LIM V S + +L NG P RGLRQGDP+SPYLFV+C
Sbjct: 589 LEKAMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICT 648
Query: 1238 ERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCD 1297
E L ++QS K +++++ PPISHL FADD + +C+ + + + ++ +
Sbjct: 649 EMLVKMLQSAEQKNQITGLKVARGAPPISHLLFADDSMFYCKVNDEALGQIIRIIEEYSL 708
Query: 1298 SSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFP--LKGGRIHRNRFN 1355
+SG ++N KS K +SE + + I G YLG P +G ++ +
Sbjct: 709 ASGQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKV--ATLS 766
Query: 1356 FLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFI 1415
+L + + +K+ W++N LS G+ L K+V A+PTYTM F +P+ + +I ++ F
Sbjct: 767 YLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESVMAEFW 826
Query: 1416 WSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLS 1475
W K G G H W +++PK GGLG ++ E N AL+GK +W ++ + + L +V
Sbjct: 827 WKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKVFK 886
Query: 1476 HKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGET-SLWHDDWSGMGNIA 1534
+Y S S L + S WK + +A+ + G +GNGET ++W D W G
Sbjct: 887 SRYFSKSDPLNAPLGSRPSFAWKSIYEAQVLIKQGIRAVIGNGETINVWTDPWIGAKPAK 946
Query: 1535 PAVPYVDIHDIDRRLCD---------LVDNGTWNMQVLYTSLPVDVLERLQKIKPTIVPN 1585
A H + + + L D WN ++ P + E + ++P
Sbjct: 947 AAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLVSLLFPDNTQENILALRPGGKET 1006
Query: 1586 RQDVWTWETNSMGIYTVRGAYRWL---------QDQQSHLPVVDD-WNWIWKLKVPEKIR 1635
R D +TWE + G Y+V+ Y W+ Q+ P +D + IWKL VP KI
Sbjct: 1007 R-DRFTWEYSRSGHYSVKSGY-WVMTEIINQRNNPQEVLQPSLDPIFQQIWKLDVPPKIH 1064
Query: 1636 TFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELWL------- 1688
F+W + N L V + +A SC RC + E + H L CP ++ W
Sbjct: 1065 HFLWRCVNNCLSVASNLAYRHLAREKSCVRCPSHGETVNHLLFKCPFARLTWAISPLPAP 1124
Query: 1689 KFGAFAWPNFTAGDH-SSWIRSQARSNNGVKFIVG-LWGVWKWRNNMIFEDSPWSLE--- 1743
G +A F H S +SQ ++ I LW +WK RN+++F+ ++
Sbjct: 1125 PGGEWAESLFRNMHHVLSVHKSQPEESDHHALIPWILWRLWKNRNDLVFKGREFTAPQVI 1184
Query: 1744 -------EAWRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPMAGSIKLNVDGSYRDVDD 1796
+AW ++ + + +WQPP G +K N DG++
Sbjct: 1185 LKATEDMDAWNNRKEPQPQVTSSTRDRC--------VKWQPPSHGWVKCNTDGAWSKDLG 1236
Query: 1797 SSGVGGLARDPSGNWLF-GFLAHRRGGNAFLAEAQALLLGLELVWARGYRDIVVEVDCAD 1855
+ GVG + R+ +G L+ G A + E +AL + + YR ++ E D
Sbjct: 1237 NCGVGWVLRNHTGRLLWLGLRALPSQQSVLETEVEALRWAVLSLSRFNYRRVIFESDSQY 1296
Query: 1856 LLQSLDDE-DRRRFLPILGDIRNMKDRGWRISLERVRRDCN 1895
L+ + +E D P + DIRN+ + + RR+ N
Sbjct: 1297 LVSLIQNEMDIPSLAPRIQDIRNLLRHFEEVKFQFTRREGN 1337
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 569 bits (1466), Expect = e-162
Identities = 334/915 (36%), Positives = 509/915 (55%), Gaps = 59/915 (6%)
Query: 759 LGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCPILLQCG 818
+G G RFTW + + ++KRLDR L ++ + +A++ C
Sbjct: 1 MGFKGNRFTWRRGLVESTFVAKRLDRVLFCAHARLKWQEALL----------------CP 44
Query: 819 SPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRSGEECITSKLERVREASTVFNKEVFGNI 878
+ A RPFRF AAW H F ++ +W +G + L R+R +NKEVFGNI
Sbjct: 45 AQNVDARRRPFRFEAAWLSHEGFKELLTASWDTGLSTPVA-LNRLRWQLKKWNKEVFGNI 103
Query: 879 FRRKRHVEARLRGVQRELDRRVTSDMVLFEAELQREYRNILRQEELL*YQKARENRVHLG 938
RK V + L+ VQ L+ T D+++ E L +E+ +L QEE L +QK+RE + LG
Sbjct: 104 HVRKEKVVSDLKAVQDLLEVVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREKLLALG 163
Query: 939 DRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTGITPLASPRV 998
DRNT +FHT T+IRRR+NR+ LK + W ++E L+++ ++ L++ + ++ R
Sbjct: 164 DRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYS-LEDVSVVRG 222
Query: 999 T--HDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDS 1056
T + FP L+ E K+ L +P +DEV A+ SM + APG DG+QP FY++ W VG+S
Sbjct: 223 TLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGES 282
Query: 1057 LWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLR 1116
+ V E F+ G + ++ D+L+VL+ KV +P + RP+SLCNV FK+ITK++V RL+
Sbjct: 283 VSKFVMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRLK 342
Query: 1117 PFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWS 1176
+ KLIGP Q+SF+PGR + DN + QE +H M + ++G + K+DLEKAYD + W
Sbjct: 343 NVISKLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRWD 402
Query: 1177 FLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLC 1236
FL ETLE G E I IM V ++S+LWNG + +F P RGLRQGDP+SPYLFVLC
Sbjct: 403 FLAETLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLRQGDPISPYLFVLC 462
Query: 1237 MERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFC 1296
+ERL I++ V +GDWK I +S+ GP +SH+ FADD++LF EASVAQ
Sbjct: 463 IERLCHQIETAVGRGDWKSISISQGGPKVSHVCFADDLILFAEASVAQ------------ 510
Query: 1297 DSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNF 1356
K++L KSK S VS ++ I+ I +LGKYLG P+ RI+++ F
Sbjct: 511 -----KVSLEKSKIFFSNNVSRDLEGLITAETGIGSTRELGKYLGMPVLQKRINKDTFGE 565
Query: 1357 LLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIW 1416
+LE + +L W++ LSLAGR+ L K+V+ ++P +TM +P + +++++ R+F+W
Sbjct: 566 VLERVSSRLSGWKSRSLSLAGRITLTKAVLMSIPIHTMSSILLPASLLEQLDKVSRNFLW 625
Query: 1417 SGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSH 1476
HL++W+K+ +PK GGLG+R ++ N AL+ K W L++ LW RVL
Sbjct: 626 GSTVEKRKQHLLSWKKVCRPKAAGGLGLRASKDMNRALLAKVGWRLLNDKVSLWARVLRR 685
Query: 1477 KY----LSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGETSLWHDDWSGMGN 1532
KY + +SS L K S W+ + G R G + + H+
Sbjct: 686 KYKVTDVHDSSWL--VPKATWSSTWRSI---------GVGLREGVAKGWILHEPL----- 729
Query: 1533 IAPAVPYVDIHDIDRRLCDLVDNGT-WNMQVLYTSLPVDVLERLQKIKPTIVPNRQDVWT 1591
A + +++ R+ + G W+M L LP V +RL + V +D +
Sbjct: 730 CTRATCLLSPEELNARVEEFWTEGVGWDMVKLGQCLPRSVTDRLHAVVIKGVLGLRDRIS 789
Query: 1592 WETNSMGIYTVRGAYRWLQDQQSHLPVVDD-WNWIWKLKVPEKIRTFVWLTLQNSLQVNL 1650
W+ S G +TV AY L ++ P ++ + IW + PE++R F+WL Q + N+
Sbjct: 790 WQGTSDGDFTVGSAYVLLTQEEESKPCMESFFKRIWGVIAPERVRVFLWLVGQQVIMTNV 849
Query: 1651 HRFRCKMAASPSCSR 1665
R R + C R
Sbjct: 850 ERVRRHIGDIEVCQR 864
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 546 bits (1407), Expect = e-155
Identities = 400/1321 (30%), Positives = 603/1321 (45%), Gaps = 113/1321 (8%)
Query: 609 LVRSKKPDVFILMETRCQFRRVSAFWNSLGFFPIHIEEARGFSGGIWVLANTSVPYNF-- 666
L R D+ L+ET Q +V LGF + + G SGG+ ++ +V +
Sbjct: 404 LFRMYNYDILFLVETLNQCDKVCKLAYDLGFPNVITQPPNGRSGGLALMWKNNVSLSLIS 463
Query: 667 ---RVIDTHQQVVTFEVWKDNLSWVCSAVYASPIPANREILWRHMTLLRRRFLLPWLLLG 723
R+ID+H VTF +N S+ S VY P + R LW+ + + WLL+G
Sbjct: 464 QDERLIDSH---VTF----NNKSFYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVG 516
Query: 724 DFNEILFPSEVRGGDFLPNRAAM-FASVLDTCQLVDLGAVGRRFTWFQKANDRLILSKRL 782
DFNEIL +E GG F +++ C + D+ + G RF+W + + + L
Sbjct: 517 DFNEILSNAEKIGGPMREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGERHTHTVKCC-L 575
Query: 783 DRALGDMEWKVAFPDAVVDVLNRVHSDHCPILLQCGSPQTLAHNRPFRFLAAWADHPDFA 842
DR + W FP A ++ L+ SDH P+L+ ++ FRF D P F
Sbjct: 576 DRVFINSAWTATFPYAEIEFLDFTGSDHKPVLVHFNESFP-RRSKLFRFDNRLIDIPTFK 634
Query: 843 AVVDNAWRSGEEC----ITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDR 898
+V +WR+ IT ++ R+A N +R + +++ L E R
Sbjct: 635 RIVQTSWRTNRNSRSTPITERISSCRQAMARLKHASNLNSEQRIKKLQSSLNRAM-ESTR 693
Query: 899 RVTSDMVLFEAELQREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRV 958
RV ++ +LQ EE+ QK+R + GD+NT YFH T R +NRV
Sbjct: 694 RVDRQLI---PQLQESLAKAFSDEEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRV 750
Query: 959 HRLKLEDGT-WCGDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQP 1017
+ + + G + GD+E+ FF N+F+ SP D +++ L +
Sbjct: 751 NTIMDDQGRMFTGDKEIGNHA-QDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKE 809
Query: 1018 VRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDI 1077
E+ A+ + APG DG FYK W+ VG + VK+ F+ + ++
Sbjct: 810 FSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHT 869
Query: 1078 LVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTM 1137
+ +IPK+ P+T+ D RPI+LCNV +KVI+K LVNRL+ L ++ Q++F+PGR
Sbjct: 870 NICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIIN 929
Query: 1138 DNAFLAQEVIHHMS-KSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIM 1196
DN +A EV+H + + + +A K D+ KAYD V W FL+ T+ L+GF I IM
Sbjct: 930 DNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIM 989
Query: 1197 SSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPI 1256
++V S S+L NG P RG+RQGDP+SPYLF+LC + LS LI GD + +
Sbjct: 990 AAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGV 1049
Query: 1257 RLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGV 1316
R+ P I+HL FADD L FC+A+V L ++ SG KIN+ KS V
Sbjct: 1050 RIGNGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRV 1109
Query: 1317 SEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLA 1376
+ + + IP GKYLG P + GR + F ++++ ++++ +W A LS A
Sbjct: 1110 YGSTQSRLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPA 1169
Query: 1377 GRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQP 1436
G+ + KSV AMP Y M F +P+G+ +I L+ +F W G V W+++
Sbjct: 1170 GKEIMLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYS 1229
Query: 1437 KDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQV 1496
K GGLG RD N AL+ K W L+ PN L+ RV+ +Y + S+L + + Q S
Sbjct: 1230 KKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYG 1289
Query: 1497 WKGLLKARDRLHTGFVFRLGNGETSLWHDDWSGMGNIAPAVPYVDIHDIDRRLCDLVDNG 1556
W LL L G +G+G+ G+ NI + P + + + ++ N
Sbjct: 1290 WASLLDGIALLKKGTRHLIGDGQNIR-----IGLDNIVDSHPPRPL-NTEETYKEMTINN 1343
Query: 1557 TWNMQVLY-------TSLPVD-----VLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRG 1604
+ + Y S VD + R+ K + D W N+ G YTVR
Sbjct: 1344 LFERKGSYYFWDDSKISQFVDQSDHGFIHRIYLAK----SKKPDKIIWNYNTTGEYTVRS 1399
Query: 1605 AYRWL--QDQQSHLPV-------VDDWNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRC 1655
Y WL D +++P +D IW L + K++ F+W L +L
Sbjct: 1400 GY-WLLTHDPSTNIPAINPPHGSIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTR 1458
Query: 1656 KMAASPSCSRCSAPEEDILHCLRTCPHSQELWLKFGAFAWPNFTAGDHSSWIRSQARSN- 1714
M PSC RC E I H L TCP F AW SS IR+Q SN
Sbjct: 1459 GMRIDPSCPRCHRENESINHALFTCP--------FATMAW----RLSDSSLIRNQLMSND 1506
Query: 1715 ------NGVKFIVG--------------LWGVWKWRNNMI---FEDSPWSL-------EE 1744
N + F+ +W +WK RNN++ F +SP
Sbjct: 1507 FEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARNNVVFNKFRESPSKTVLSAKAETH 1566
Query: 1745 AWRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPMAGSIKLNVDGSYRDVDDSSGVGG-L 1803
W H + + + A + W+ P A +K N D + DV GG +
Sbjct: 1567 DWLNATQSHKKTPSPTRQIAENK-----IEWRNPPATYVKCNFDAGF-DVQKLEATGGWI 1620
Query: 1804 ARDPSG---NWLFGFLAHRRGGNAFLAEAQALLLGLELVWARGYRDIVVEVDCADLLQSL 1860
R+ G +W LAH N AE +ALL L+ W RGY + +E DC L+ +
Sbjct: 1621 IRNHYGTPISWGSMKLAHT--SNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLINLI 1678
Query: 1861 D 1861
+
Sbjct: 1679 N 1679
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 538 bits (1385), Expect = e-152
Identities = 394/1309 (30%), Positives = 597/1309 (45%), Gaps = 113/1309 (8%)
Query: 621 METRCQFRRVSAFWNSLGFFPIHIEEARGFSGGIWVLANTSVPYNF-----RVIDTHQQV 675
+ET Q +V LGF + + G SGG+ ++ +V + R+ID+H
Sbjct: 190 LETLNQCDKVCKLAYDLGFPNVITQPPNGRSGGLALMWKNNVSLSLISQDERLIDSH--- 246
Query: 676 VTFEVWKDNLSWVCSAVYASPIPANREILWRHMTLLRRRFLLPWLLLGDFNEILFPSEVR 735
VTF +N S+ S VY P + R LW+ + + WLL+GDFNEIL +E
Sbjct: 247 VTF----NNKSFYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKI 302
Query: 736 GGDFLPNRAAM-FASVLDTCQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVA 794
GG F +++ C + D+ + G RF+W + + + LDR + W
Sbjct: 303 GGPMREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGERHTHTVKCC-LDRVFINSAWTAT 361
Query: 795 FPDAVVDVLNRVHSDHCPILLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRSGEE 854
FP A + L+ SDH P+L+ ++ FRF D P F +V +WR+
Sbjct: 362 FPYAETEFLDFTGSDHKPVLVHFNESFP-RRSKLFRFDNRLIDIPTFKRIVQTSWRTNRN 420
Query: 855 C----ITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAE 910
IT ++ R+A N +R + +++ L E RRV ++ +
Sbjct: 421 SRSTPITERISSCRQAMARLKHASNLNSEQRIKKLQSSLNRAM-ESTRRVDRQLI---PQ 476
Query: 911 LQREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGT-WC 969
LQ EE+ QK+R + GD+NT YFH T R +NRV+ + + G +
Sbjct: 477 LQESLAKAFSDEEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFT 536
Query: 970 GDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMS 1029
GD+E+ FF N+F+ SP D +++ L + E+ A+
Sbjct: 537 GDKEIGNHA-QDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQ 595
Query: 1030 MNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPS 1089
+ APG DG FYK W+ VG + VK+ F+ + ++ + +IPK+ P+
Sbjct: 596 IGDDKAPGPDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPT 655
Query: 1090 TVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHH 1149
T+ D RPI+LCNV +KVI+K LVNRL+ L ++ Q++F+PGR DN +A EV+H
Sbjct: 656 TLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHS 715
Query: 1150 MS-KSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILW 1208
+ + + +A K D+ KAYD V W FL+ T+ L+GF I IM++V S S+L
Sbjct: 716 LKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLI 775
Query: 1209 NGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHL 1268
NG P RG+RQGDP+SPYLF+LC + LS LI GD + +R+ P I+HL
Sbjct: 776 NGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHL 835
Query: 1269 FFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVA 1328
FADD L FC+A+V L ++ SG KIN+ KS V + + +
Sbjct: 836 QFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSKLKQIL 895
Query: 1329 PIPFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAA 1388
IP GKYLG P + GR + F ++++ ++++ +W A LS AG+ + KSV A
Sbjct: 896 EIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALA 955
Query: 1389 MPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTE 1448
MP Y M F +P+G+ +I L+ +F W G V W+++ K GGLG RD
Sbjct: 956 MPVYAMSCFKLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLA 1015
Query: 1449 LANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLH 1508
N AL+ K W L+ PN L+ RV+ +Y + S+L + + Q S W LL L
Sbjct: 1016 KFNDALLAKQAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLLDGIALLK 1075
Query: 1509 TGFVFRLGNGETSLWHDDWSGMGNIAPAVPYVDIHDIDRRLCDLVDNGTWNMQVLY---- 1564
G +G+G+ G+ NI + P + + + ++ N + + Y
Sbjct: 1076 KGTRHLIGDGQNIR-----IGLDNIVDSHPPRPL-NTEETYKEMTINNLFERKGSYYFWD 1129
Query: 1565 ---TSLPVD-----VLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWL--QDQQS 1614
S VD + R+ K + D W N+ G YTVR Y WL D +
Sbjct: 1130 DSKISQFVDQSDHGFIHRIYLAK----SKKPDKIIWNYNTTGEYTVRSGY-WLLTHDPST 1184
Query: 1615 HLPV-------VDDWNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCS 1667
++P +D IW L + K++ F+W L +L M P C RC
Sbjct: 1185 NIPAINPPHGSIDLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPICPRCH 1244
Query: 1668 APEEDILHCLRTCPHSQELWLKFGAFAWPNFTAGDHSSWIRSQARSN-------NGVKFI 1720
E I H L TCP + W W + SS IR+Q SN N + F+
Sbjct: 1245 RENESINHALFTCPFATMAW-------WLS-----DSSLIRNQLMSNDFEENISNILNFV 1292
Query: 1721 VG--------------LWGVWKWRNNMI---FEDSPWSL-------EEAWRRVCHEHDEI 1756
+W +WK RNN++ F +SP W H +
Sbjct: 1293 QDTTMSDFHKLLPVWLIWRIWKARNNVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKT 1352
Query: 1757 VAVLGEEAGSMDCWLGSRWQPPMAGSIKLNVDGSYRDVDDSSGVGG-LARDPSG---NWL 1812
+ + A + W+ P A +K N D + DV GG + R+ G +W
Sbjct: 1353 PSPTRQIAENK-----IEWRNPPATYVKCNFDAGF-DVQKLEATGGWIIRNHYGTPISWG 1406
Query: 1813 FGFLAHRRGGNAFLAEAQALLLGLELVWARGYRDIVVEVDCADLLQSLD 1861
LAH N AE +ALL L+ W RGY + +E DC L+ ++
Sbjct: 1407 SMKLAHT--SNPLEAETKALLAALQQTWIRGYTQVFMEGDCQTLINLIN 1453
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 527 bits (1357), Expect = e-149
Identities = 385/1348 (28%), Positives = 622/1348 (45%), Gaps = 75/1348 (5%)
Query: 592 WNVRGALYANGKLAIRELVRSKKPDVFILMETRCQFRRVSAFWNSLGFFPIHIEEARGFS 651
WN +G + E+ R D+ L+ET+ Q +GF + I RG S
Sbjct: 368 WNCQGLGQPLTVRRLEEVQRVYFLDMLFLIETKQQDNYTRDLGVKMGFEDMCIISPRGLS 427
Query: 652 GGIWVLANTSVPYNFRVIDTHQQVVTFEVWKDNLSWVCSAVYASPIPANREILWRHMTLL 711
GG+ V + + +VI ++V V N ++ S +Y PIP+ R LW + +
Sbjct: 428 GGLVVYWKKHL--SIQVISHDVRLVDLYVEYKNFNFYLSCIYGHPIPSERHHLWEKLQRV 485
Query: 712 RRRFLLPWLLLGDFNEILFPSEVRGGDFLPNRAAM-FASVLDTCQLVDLGAVGRRFTWFQ 770
PW++ GDFNEIL +E +GG + F ++++ C + DL + G ++W
Sbjct: 486 SAHRSGPWMMCGDFNEILNLNEKKGGRRRSIGSLQNFTNMINCCNMKDLKSKGNPYSWVG 545
Query: 771 KANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCPILLQCGSPQTLAHNRPFR 830
K + I S LDR + +W+ +FP + L SDH P+++ + FR
Sbjct: 546 KRQNETIESC-LDRVFINSDWQASFPAFETEFLPIAGSDHAPVIIDIAE-EVCTKRGQFR 603
Query: 831 FLAAWADHPDFAAVVDNAWRSGEEC----ITSKLERVREASTVFNKEVFGNIFRRKRHVE 886
+ DF V W G KL R+ + + N + ++
Sbjct: 604 YDRRHFQFEDFVDSVQRGWNRGRSDSHGGYYEKLHCCRQELAKWKRRTKTNTAEKIETLK 663
Query: 887 ARLRGVQRELDRRVTSDMVLFEAELQREYRNILRQEELL*YQKARENRVHLGDRNTAYFH 946
R+ +R D + +L L+++ R EEL + K+R + LGDRNT +F+
Sbjct: 664 YRVDAAER--DHTLPHQTIL---RLRQDLNQAYRDEELYWHLKSRNRWMLLGDRNTMFFY 718
Query: 947 TQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSL 1006
T +R+ +NR+ + G ++ + +V +F +LFT + + P +
Sbjct: 719 ASTKLRKSRNRIKAITDAQGIENFRDDTIGKVAENYFADLFTTTQTSDWEEIISGIAPKV 778
Query: 1007 SVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQ 1066
+ + LLQ V EVR A+ ++ + APG DGF FY W+ +G+ + MV+ F+
Sbjct: 779 TEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWDLIGNDVCLMVRHFFE 838
Query: 1067 EGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPM 1126
++ + + LIPK+ P + D RPISLC ++K+I+K+L+ RL+ L +I
Sbjct: 839 SDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDS 898
Query: 1127 QSSFLPGRGTMDNAFLAQEVIHHM-SKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELY 1185
Q++F+PG+ DN +A E++H + S+ + G VA K D+ KAYD V W+FL++ +
Sbjct: 899 QAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQL 958
Query: 1186 GFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQ 1245
GF + IM+ VTS +L NG P RG+RQGDP+SPYLF+ C E LS +++
Sbjct: 959 GFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQGDPLSPYLFLFCAEVLSNMLR 1018
Query: 1246 SLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINL 1305
++++K+ ISHL FADD L FC AS + LA + + ++SG KIN
Sbjct: 1019 KAEVNKQIHGMKITKDCLAISHLLFADDSLFFCRASNQNIEQLALIFKKYEEASGQKINY 1078
Query: 1306 SKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKL 1365
+KS I + + + + + + I V GKYLG P + GR F +++ ++ +
Sbjct: 1079 AKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGGKYLGLPEQLGRRKVELFEYIVTKVKERT 1138
Query: 1366 GSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGW 1425
W N LS AG+ + K++ A+P Y+M F +P + +IN LI +F W GK
Sbjct: 1139 EGWAYNYLSPAGKEIVIKAIAMALPVYSMNCFLLPTLICNEINSLITAF-WWGK------ 1191
Query: 1426 HLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVL 1485
++ G LG +D N AL+ K W ++ P L R+ Y N++ L
Sbjct: 1192 -----------ENEGDLGFKDLHQFNRALLAKQAWRILTNPQSLLARLYKGLYYPNTTYL 1240
Query: 1486 QVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGETS-LWHDDWSGMGNIAPAVPYVDIHD 1544
+ S W + + + L G RLG+G+T+ +W D W + + P I D
Sbjct: 1241 RANKGGHASYGWNSIQEGKLLLQQGLRVRLGDGQTTKIWEDPW--LPTLPPRPARGPILD 1298
Query: 1545 IDRRLCDL--VDNGTWNMQVLYTSLPVDVLERLQKIKPTIVPN--RQDVWTWETNSMGIY 1600
D ++ DL + W+ + L E Q K + N +D + W Y
Sbjct: 1299 EDMKVADLWRENKREWDPVIFEGVLNP---EDQQLAKSLYLSNYAARDSYKWAYTRNTQY 1355
Query: 1601 TVRGAYRW------LQDQQSHLPVVDD---WNWIWKLKVPEKIRTFVWLTLQNSLQVNLH 1651
TVR Y W L +++ P+ D IW+LK+ KI+ F+W L +L
Sbjct: 1356 TVRSGY-WVATHVNLTEEEIINPLEGDVPLKQEIWRLKITPKIKHFIWRCLSGALSTTTQ 1414
Query: 1652 RFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELWLKFGAFAWPN---FTAG-DHSSWI 1707
+ A P+C RC +E I H + TC ++Q +W + F+ N FT + + +
Sbjct: 1415 LRNRNIPADPTCQRCCNADETINHIIFTCSYAQVVW-RSANFSGSNRLCFTDNLEENIRL 1473
Query: 1708 RSQARSNNGVKFIVGL------WGVWKWRNNMIFED---SPWSLEEAWRRVCHEHDEIV- 1757
Q + N + + GL W +WK RN +F+ PW + + + E E +
Sbjct: 1474 ILQGKKNQNLPILNGLMPFWIMWRLWKSRNEYLFQQLDRFPWKVAQKAEQEATEWVETMV 1533
Query: 1758 ---AVLGEEAGSMDCWL--GSRWQPPMAGSIKLNVDGSYRDVDDSSGVGGLARDPSGNWL 1812
A+ A S D L +W P G +K N D Y D + G + RD +G L
Sbjct: 1534 NDTAISHNTAQSNDRPLSRSKQWSSPPEGFLKCNFDSGYVQGRDYTSTGWILRDCNGRVL 1593
Query: 1813 F-GFLAHRRGGNAFLAEAQALLLGLELVWARGYRDIVVEVDCADLLQSLD-DEDRRRFLP 1870
G ++ +A AEA L L++VW RGY + E D +L ++ ED
Sbjct: 1594 HSGCAKLQQSYSALQAEALGFLHALQMVWIRGYCYVWFEGDNLELTNLINKTEDHHLLET 1653
Query: 1871 ILGDIRNMKDRGWRISLERVRRDCNAPA 1898
+L DIR + S+ V R+ N A
Sbjct: 1654 LLYDIRFWMTKLPFSSIGYVNRERNLAA 1681
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 526 bits (1356), Expect = e-149
Identities = 379/1302 (29%), Positives = 608/1302 (46%), Gaps = 62/1302 (4%)
Query: 615 PDVFILMETRCQFRRVSAFWNSLGFFPIHIEEARGFSGGIWVLANTSVPYNFRVIDTHQQ 674
PD+ LMET+ V + LG+ IH E G SGG+ + + + F D +
Sbjct: 7 PDILFLMETKNSQDFVYKVFCWLGYDFIHTVEPEGRSGGLAIFWKSHLEIEFLYAD--KN 64
Query: 675 VVTFEVWKDNLSWVCSAVYASPIPANREILWRHMTLLRRRFLLPWLLLGDFNEILFPSEV 734
++ +V N W S VY P+ R LW H+ + + W L+GDFN+I E
Sbjct: 65 LMDLQVSSRNKVWFISCVYGLPVTHMRPKLWEHLNSIGLKRAEAWCLIGDFNDIRSNDEK 124
Query: 735 RGGDFL-PNRAAMFASVLDTCQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKV 793
GG P+ F +L C + +LG+ G FTW ND+ + K LDR G+ W
Sbjct: 125 LGGPRRSPSSFQCFEHMLLNCSMHELGSTGNSFTWGGNRNDQWVQCK-LDRCFGNPAWFS 183
Query: 794 AFPDAVVDVLNRVHSDHCPILLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRS-- 851
FP+A L + SDH P+L++ + L + FR+ D P V+ +W S
Sbjct: 184 IFPNAHQWFLEKFGSDHRPVLVKFTNDNELFRGQ-FRYDKRLDDDPYCIEVIHRSWNSAM 242
Query: 852 --GEECITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEA 909
G L R A +V+ N ++R++ ++++LD + + +
Sbjct: 243 SQGTHSSFFSLIECRRAISVWKHSSDTN-------AQSRIKRLRKDLDAEKSIQIPCWPR 295
Query: 910 --ELQREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGT 967
++ + EEL QK+R+ + GD+NT +FH R KN + L E+
Sbjct: 296 IEYIKDQLSLAYGDEELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQ 355
Query: 968 WCGDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHAL 1027
++ FF NLFT L L ++ E L+Q V + EV +A+
Sbjct: 356 EFTRNSDKGKIASSFFENLFTSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELEVYNAV 415
Query: 1028 MSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQ 1087
S+N +APG DGF F++++W+ V + + F+ G + ++ + LIPK+
Sbjct: 416 FSINKESAPGPDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITS 475
Query: 1088 PSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVI 1147
P + DLRPISLC+V +K+I+K+L RL+ L ++ QS+F+P R DN +A E+I
Sbjct: 476 PQRMSDLRPISLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMI 535
Query: 1148 HHM-SKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSI 1206
H + + + +AFK D+ KAYD V W FL+ + GF I+ IM+ VTS S+
Sbjct: 536 HSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSV 595
Query: 1207 LWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPIS 1266
L NG P RG+RQGDP+SP LFVLC E L ++ G I+ ++
Sbjct: 596 LINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQDKKVSVN 655
Query: 1267 HLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISV 1326
HL FADD LL C+A+ + L + + SG INL+KS K V +KD I
Sbjct: 656 HLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQIKDWIKS 715
Query: 1327 VAPIPFVHDLGKYLGFP--LKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKS 1384
+ I GKYLG P L G + R+ F F+ E +Q +L W A LS G+ L KS
Sbjct: 716 RSGISLEGGTGKYLGLPECLSGSK--RDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKS 773
Query: 1385 VIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGV 1444
+ A+P Y M F +P+ + K+ ++ F W+ H ++W+++T PKD+GG G
Sbjct: 774 IALALPVYVMSCFKLPKNLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGF 833
Query: 1445 RDTELANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKAR 1504
+D + N AL+ K W ++ + L+ RV +Y SNS L + S W+ +L R
Sbjct: 834 KDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILFGR 893
Query: 1505 DRLHTGFVFRLGNGE-TSLWHDDWSGMGNIAPAVPYVDIHDIDRRLCDLVD--NGTWNMQ 1561
+ L G +GNG+ T +W D W G+ + ++D ++ L+D + WN+
Sbjct: 894 ELLMQGLRTVIGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSRNWNLN 953
Query: 1562 VLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWLQDQQSH------ 1615
+L P +E + K +P ++D + W + G+Y+V+ Y +L Q H
Sbjct: 954 MLRDLFPWKDVEIILKQRPLFF--KEDSFCWLHSHNGLYSVKTGYEFLSKQVHHRLYQEA 1011
Query: 1616 --LPVVDD-WNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCK-MAASPSCSRCSAPEE 1671
P V+ ++ IW L KIR F+W L ++ V R R + + + C C E
Sbjct: 1012 KVKPSVNSLFDKIWNLHTAPKIRIFLWKALHGAIPVE-DRLRTRGIRSDDGCLMCDTENE 1070
Query: 1672 DILHCLRTCPHSQELW-LKFGAFAWPNFTAGDHSSWIR-----SQARSNNGVKFIVG--L 1723
I H L CP ++++W + + A F+ +++ R Q + ++F+ L
Sbjct: 1071 TINHILFECPLARQVWAITHLSSAGSEFSNSVYTNMSRLIDLTQQNDLPHHLRFVSPWIL 1130
Query: 1724 WGVWKWRNNMIFEDS---PWSLEEAWRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPMA 1780
W +WK RN ++FE +L + HE + + + ++W PP+
Sbjct: 1131 WFLWKNRNALLFEGKGSITTTLVDKAYEAYHEWFSAQTHMQNDEKHLKI---TKWCPPLP 1187
Query: 1781 GSIKLNVDGSYRDVDDSSGVGGLARDPSGNWLFGFLAHRRGGN----AFLAEAQALLLGL 1836
G +K N+ ++ SG + RD G L L RR N + A+ ++ L
Sbjct: 1188 GELKCNIGFAWSKQHHFSGASWVVRDSQGKVL---LHSRRSFNEVHSPYSAKIRSWEWAL 1244
Query: 1837 ELVWARGYRDIVVEVDCADLLQSLDDEDRRRFLPILGDIRNM 1878
E + + ++ +++Q+L L LGDI +
Sbjct: 1245 ESMTHHHFDRVIFASSTHEIIQALHKPHEWPLL--LGDISEL 1284
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 509 bits (1310), Expect = e-144
Identities = 361/1222 (29%), Positives = 583/1222 (47%), Gaps = 83/1222 (6%)
Query: 681 WKDNLSWVCSAVYASP-IPANREILWRHMTLLRRRFLLPWLLLGDFNEILFPSEVRGGDF 739
WK+N+ + A+P NR + W ++ L + WLL GDFN+IL SE +GG
Sbjct: 38 WKENVE--VEILEAAPNFIDNRSVFWDKISSLGAQRSSAWLLTGDFNDILDNSEKQGGPL 95
Query: 740 LPNRAAM-FASVLDTCQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDA 798
+ F S + L D+ G +W I S RLDRALG+ W FP +
Sbjct: 96 RWEGFFLAFRSFVSQNGLWDINHTGNSLSWRGTRYSHFIKS-RLDRALGNCSWSELFPMS 154
Query: 799 VVDVLNRVHSDHCPILLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWR-SGEECIT 857
+ L SDH P++ G+P L ++PFRF + + A+V W + ++ +
Sbjct: 155 KCEYLRFEGSDHRPLVTYFGAPP-LKRSKPFRFDRRLREKEEIRALVKEVWELARQDSVL 213
Query: 858 SKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMV--LFEAELQREY 915
K+ R R++ + KE N + ++ Q+ L+ +++D+ + +E
Sbjct: 214 YKISRCRQSIIKWTKEQNSNSAKA-------IKKAQQALESALSADIPDPSLIGSITQEL 266
Query: 916 RNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVL 975
RQEEL Q +R ++ GDRN YFH T RR N + ++ G +EE +
Sbjct: 267 EAAYRQEELFWKQWSRVQWLNSGDRNKGYFHATTRTRRMLNNLSVIEDGSGQEFHEEEQI 326
Query: 976 KRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTA 1035
+ +F N+FT V L P +S L++ E++ AL S+++ A
Sbjct: 327 ASTISSYFQNIFTTSNNSDLQVVQEALSPIISSHCNEELIKISSLLEIKEALFSISADKA 386
Query: 1036 PGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLR 1095
PG DGF F+ YW+ + + ++ F + ++ L + V LIPK+ P V D R
Sbjct: 387 PGPDGFSASFFHAYWDIIEADVSRDIRSFFVDSCLSPRLNETHVTLIPKISAPRKVSDYR 446
Query: 1096 PISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKSTA 1155
PI+LCNV +K++ K+L RL+P+L +LI QS+F+PGR DN + E++H + S A
Sbjct: 447 PIALCNVQYKIVAKILTRRLQPWLSELISLHQSAFVPGRAIADNVLITHEILHFLRVSGA 506
Query: 1156 KRGM-VAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLP 1214
K+ +A K D+ KAYD + W+FLQE L GF + I +M V + S L NG
Sbjct: 507 KKYCSMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGSPQG 566
Query: 1215 AFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDV 1274
+ P RGLRQGDP+SPYLF+LC E LS L + +KG IR+++ P ++HL FADD
Sbjct: 567 SVVPSRGLRQGDPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVARGSPQVNHLLFADDT 626
Query: 1275 LLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVH 1334
+ FC+ + L++ ++ + +SG INL+KS S + +K + + I
Sbjct: 627 MFFCKTNPTCCGALSNILKKYELASGQSINLAKSAITFSSKTPQDIKRRVKLSLRIDNEG 686
Query: 1335 DLGKYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTM 1394
+GKYLG P GR R+ F+ +++ I+++ SW LS AG+ L K+V+++MP+Y M
Sbjct: 687 GIGKYLGLPEHFGRRKRDIFSSIVDRIRQRSHSWSIRFLSSAGKQILLKAVLSSMPSYAM 746
Query: 1395 QVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTAL 1454
F +P + +I ++ F W K V+W+K+T P + GGLG R+ E
Sbjct: 747 MCFKLPASLCKQIQSVLTRFWWDSKPDKRKMAWVSWDKLTLPINEGGLGFREIE------ 800
Query: 1455 MGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQ-DSQVWKGLLKARDRLHTGFVF 1513
K W ++ +P+ L RVL KY + SS + A P S W+G+L RD L G +
Sbjct: 801 -AKLSWRILKEPHSLLSRVLLGKYCNTSSFMDCSASPSFASHGWRGILAGRDLLRKGLGW 859
Query: 1514 RLGNGET-SLWHDDWSGMGNIAPAVPYVDI-----HDIDRRLCDLV--DNGTWNMQVLYT 1565
+G G++ ++W + W ++P+ P I + D + DL+ D +WN++ +
Sbjct: 860 SIGQGDSINVWTEAW-----LSPSSPQTPIGPPTETNKDLSVHDLICHDVKSWNVEAIRK 914
Query: 1566 SLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWLQDQQSHLPVVDDWNW- 1624
LP ++++KI +P QD W G YT + Y L S D+NW
Sbjct: 915 HLP-QYEDQIRKITINALP-LQDSLVWLPVKSGEYTTKTGYA-LAKLNSFPASQLDFNWQ 971
Query: 1625 --IWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPH 1682
IWK+ K++ F+W ++ +L V R + A +C RC E LH + CP+
Sbjct: 972 KNIWKIHTSPKVKHFLWKAMKGALPVGEALSRRNIEAEVTCKRCGQTESS-LHLMLLCPY 1030
Query: 1683 SQELWLKFGAFAWPNFTAGDHSSWIRSQARS---------NNGVKFIVGLWGVWKWRNNM 1733
++++W P+ + + A+ + + LW +WK RN +
Sbjct: 1031 AKKVWELAPVLFNPSEATHSSVALLLVDAKRMVALPPTGLGSAPLYPWLLWHLWKARNRL 1090
Query: 1734 IFEDSPWSLEEAWRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPMAG----SIKLNVDG 1789
IF++ C E ++ + + M+ L P++ + L V
Sbjct: 1091 IFDN----------HSCSEEGLVLKAILDARAWMEAQLLIHHPSPISDYPSPTPNLKVTS 1140
Query: 1790 SYRD----VDDSSGVGGLARDP------SGNWLFGFLAHRRGGNAFLAEAQALLLGLELV 1839
+ D G+G +DP F+ G+A +AE A+ L L
Sbjct: 1141 CFVDAAWTTSGYCGMGWFLQDPYKVKIKENQSSSSFV-----GSALMAETLAVHLALVDA 1195
Query: 1840 WARGYRDIVVEVDCADLLQSLD 1861
+ G R + V DC +L+ L+
Sbjct: 1196 LSTGVRQLNVFSDCKELISLLN 1217
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 482 bits (1240), Expect = e-135
Identities = 338/1130 (29%), Positives = 527/1130 (45%), Gaps = 56/1130 (4%)
Query: 808 SDHCPILLQCGSPQTLAHNRP-----FRFLAAWADHPDFAAVVDNAWR--SG--EECITS 858
SDH P++ T+A P FRF W + W SG E
Sbjct: 4 SDHSPVIA------TIADKIPRGKQNFRFDKRWIGKDGLLEAISQGWNLDSGFREGQFVE 57
Query: 859 KLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAELQREYRNI 918
KL R A + + K + + ++A L QR+ D R ++ L+ YR+
Sbjct: 58 KLTNCRRAISKWRKSLIPFGRQTIEDLKAELDVAQRD-DDRSREEITELTLRLKEAYRD- 115
Query: 919 LRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRV 978
EE YQK+R + LGD N+ +FH T RR +NR+ L E+G W +++ ++ +
Sbjct: 116 ---EEQYWYQKSRSLWMKLGDNNSKFFHALTKQRRARNRITGLHDENGIWSIEDDDIQNI 172
Query: 979 VHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGA 1038
+F NLFT P ++ ++ LL + EVR AL ++ APG
Sbjct: 173 AVSYFQNLFTTANPQVFDEALGEVQVLITDRINDLLTADATECEVRAALFMIHPEKAPGP 232
Query: 1039 DGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPIS 1098
DG F++K W + L +V QEG ++ L + LIPK ++P+ + +LRPIS
Sbjct: 233 DGMTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPIS 292
Query: 1099 LCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHM-SKSTAKR 1157
LCNV +KVI+K+L RL+ L LI QS+F+ GR DN +AQE+ H + + S+ K
Sbjct: 293 LCNVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKD 352
Query: 1158 GMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFK 1217
+A K D+ KAYD V W+F++ L GF E I+ IM +T+ Q +L NG
Sbjct: 353 KFMAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLII 412
Query: 1218 PGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLF 1277
P RGLRQGDP+SPYLF+LC E L I+ + I+++ P +SHL FADD L F
Sbjct: 413 PERGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVATPSPAVSHLLFADDSLFF 472
Query: 1278 CEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLG 1337
C+A+ Q ++ ++ + SG +IN SKS V + +K I ++ I + +G
Sbjct: 473 CKANKEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMG 532
Query: 1338 KYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVF 1397
YLG P G F+F+ + +Q ++ W A LS G+ + KSV A +P Y M F
Sbjct: 533 SYLGLPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCF 592
Query: 1398 NMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGK 1457
+P+ +T K+ + F WS G G H + W+K+ K GGLG R+ + N+AL+ K
Sbjct: 593 RLPKAITSKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAK 652
Query: 1458 AIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGN 1517
+W L+ P+ L+ +V +Y S+ L S W+ ++ AR ++ G + R+G+
Sbjct: 653 QLWRLITAPDSLFAKVFKGRYFRKSNPLDSIKSYSPSYGWRSMISARSLVYKGLIKRVGS 712
Query: 1518 GET-SLWHDDWSGMGNIAPAVPYVDIHDIDRRLCDLVDNGT--WNMQVLYTSL-PVDVLE 1573
G + S+W+D W PA I D ++ L+D+ + WN+ +L P DV
Sbjct: 713 GASISVWNDPWIPAQFPRPAKYGGSIVDPSLKVKSLIDSRSNFWNIDLLKELFDPEDV-- 770
Query: 1574 RLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRW--LQDQQSHLPVVDDWN----WIWK 1627
L P PN +D W G YTV+ Y L + + D +IWK
Sbjct: 771 PLISALPIGNPNMEDTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWK 830
Query: 1628 LKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELW 1687
++ P K+R F+W L + V+ + + + C C A EE I H L C ++++W
Sbjct: 831 VQCPPKLRHFLWQILSGCVPVSENLRKRGILCDKGCVSCGASEESINHTLFQCHPARQIW 890
Query: 1688 LKFGAFAWPN-------FTAGDHSSWIRSQARSNNGVKFIVGLWGVWKWRNNMIFE---- 1736
P FT DH W + +I+ W +WK RN +FE
Sbjct: 891 ALSQIPTAPGIFPSNSIFTNLDHLFWRIPSGVDSAPYPWII--WYIWKARNEKVFENVDK 948
Query: 1737 DSPWSLEEAWRRVCHEHDEIVAVLGEEAGSMDCWLGSR------WQPPMAGSIKLNVDGS 1790
D L A + + V + E GS+ + SR Q + +DGS
Sbjct: 949 DPMEILLLAVKEAQSWQEAQVELHSERHGSLS--IDSRIRVRDVSQDTTFSGFRCFIDGS 1006
Query: 1791 YRDVDDSSGVGGLARDPSG-NWLFGFLAHRRGGNAFLAEAQALLLGLELVWARGYRDIVV 1849
++ D SG G G + G RR + E +ALL ++ + +++
Sbjct: 1007 WKASDQFSGTGWFCLSSLGESPTMGAANVRRSLSPLHTEMEALLWAMKCMIGADNQNVAF 1066
Query: 1850 EVDCADLLQSLDD-EDRRRFLPILGDIRNMKDRGWRISLERVRRDCNAPA 1898
DC+DL++ + + F L ++++ ++ SL + R N A
Sbjct: 1067 FTDCSDLVKMVSSPTEWPAFSVYLEELQSDREEFTNFSLSLISRSANVKA 1116
>At1g24640 hypothetical protein
Length = 1270
Score = 480 bits (1235), Expect = e-135
Identities = 364/1240 (29%), Positives = 579/1240 (46%), Gaps = 95/1240 (7%)
Query: 637 LGFFPIHIEEARGFSGGIWVLANTSVPYNFRVIDTHQQVVTFEVWKDNLSWVCSAVYASP 696
L + ++ E G GG+ +L +SV + + +D + ++ +V +++ S VY P
Sbjct: 14 LEYDQVYTVEPVGKCGGLALLWKSSVQVDLKFVD--KNLMDAQVQFGAVNFCVSCVYGDP 71
Query: 697 IPANREILWRHMTLLRRRFLLPWLLLGDFNEILFPSEVRGGDFLPNRAAM----FASVLD 752
+ R W ++ + W + GDFN+IL E GG P R+ + F ++
Sbjct: 72 DRSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGG---PRRSDLDCKAFNEMIK 128
Query: 753 TCQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCP 812
C LV++ A G FTW + D I RLDRA G+ EW FP + L+ SDH P
Sbjct: 129 GCDLVEMPAHGNGFTWAGRRGDHWIQC-RLDRAFGNKEWFCFFPVSNQTFLDFRGSDHRP 187
Query: 813 ILLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRSGEE----CITSKLERVREAST 868
+L++ S Q ++ FRF + D + W G+ + +L R++ +
Sbjct: 188 VLIKLMSSQD-SYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNISVADRLRACRKSLS 246
Query: 869 VFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAELQREYRNILRQEELL*YQ 928
+ K+ N + +EA L E ++ + + + L+++ R+EE Q
Sbjct: 247 SWKKQNNLNSLDKINQLEAAL-----EKEQSLVWPIFQRVSVLKKDLAKAYREEEAYWKQ 301
Query: 929 KARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFT 988
K+R+ + G+RN+ YFH R++ R+ +LK +G E V +F NLF
Sbjct: 302 KSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLFK 361
Query: 989 GITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKK 1048
P L P +S L+ V E++ A+ S+ +APG DG F++
Sbjct: 362 SSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQH 421
Query: 1049 YWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVIT 1108
YW+ VG+ + VK+ F +G + + LIPK P+ + DLRPISLC+V +K+I+
Sbjct: 422 YWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIIS 481
Query: 1109 KVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMS-KSTAKRGMVAFKIDLE 1167
K++ RL+P+L +++ QS+F+ R DN +A E++H + +A K D+
Sbjct: 482 KIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMS 541
Query: 1168 KAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDP 1227
KAYD V WS+L+ L GF +N IM V+S S+L N C RGLRQGDP
Sbjct: 542 KAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDP 601
Query: 1228 MSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNL 1287
+SP+LFVLC E L+ L+ +G + I+ S+NGP + HL FADD L C+AS Q +
Sbjct: 602 LSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASREQSLV 661
Query: 1288 LASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFP--LK 1345
L ++++ +++G INL+KS + V E +K +I I G YLG P
Sbjct: 662 LQKILKVYGNATGQTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFS 721
Query: 1346 GGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTC 1405
G ++ + ++L + ++ KL W LS G+ L KSV AMP + M F +P
Sbjct: 722 GSKV--DMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCE 779
Query: 1406 KINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHK 1465
+ + SF W H +WE++ PKD GGLG RD + N AL+ K W L+H
Sbjct: 780 NLESAMASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHF 839
Query: 1466 PNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGET-SLWH 1524
P+ L R+L +Y + L + S W+ +L R+ L G R+G+G + +W
Sbjct: 840 PDCLLSRLLKSRYFDATDFLDAALSQRPSFGWRSILFGRELLSKGLQKRVGDGASLFVWI 899
Query: 1525 DDWSGMGNIAPAVPYVDIHDIDRRLCDLVD--NGTWNMQVLY-TSLPVDVLERLQKIKPT 1581
D W I+D+ ++ L++ G W+ +VL+ LP D+L R++ IKP
Sbjct: 900 DPWIDDNGFRAPWRKNLIYDVTLKVKALLNPRTGFWDEEVLHDLFLPEDIL-RIKAIKPV 958
Query: 1582 IVPNRQDVWTWETNSMGIYTVRGAYRWL------QDQQSHLPVVDDW----NWIWKLKVP 1631
I ++ D + W+ N G ++V+ AY WL Q+ +S + + +W L+
Sbjct: 959 I--SQADFFVWKLNKSGDFSVKSAY-WLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTD 1015
Query: 1632 EKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELW-LKF 1690
KI+ F+W C E H L CP S+++W L
Sbjct: 1016 PKIKIFLWKV------------------------CGELGESTNHTLFLCPLSRQIWALSD 1051
Query: 1691 GAFAWPNFTAGDHSSWIRSQARSNNGVKFIVG--------LWGVWKWRNNMIFEDSPWSL 1742
F F+ G S I + + ++ + LW +WK RN+ IFE +
Sbjct: 1052 YPFPPDGFSNGSIYSNINHLLENKDNKEWPINLRKIFPWILWRIWKNRNSFIFEGISYPA 1111
Query: 1743 EEAWRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPMAGSIKLNVDGSYRDVDDSSGVGG 1802
+ V D++V EA +D GS PP++ + + GS +
Sbjct: 1112 TDT---VIKIRDDVVEWF--EAQCLD-GEGSALNPPLSNGV--HFVGSVSE--------N 1155
Query: 1803 LARDPSGNWLFGFLAH-RRGGNAFLAEAQALLLGLELVWA 1841
L P NW L H RR ++A ALL L+WA
Sbjct: 1156 LWIKPPANWDGIVLLHSRRSFAPISSKADALLRC--LLWA 1193
>At4g10830 putative protein
Length = 1294
Score = 468 bits (1203), Expect = e-131
Identities = 306/946 (32%), Positives = 471/946 (49%), Gaps = 64/946 (6%)
Query: 606 IRELVRSKKPDVFILMETRCQFRRVSAFWNSLGFFPIHIEEARGFSGGIWVLANTSVPYN 665
+ L + K DV L+ET + +S + LGF + + +G SGG+ +L SV +
Sbjct: 381 LSNLCKVFKFDVLFLIETLNKCEVISNLASVLGFPNVITQPPQGHSGGLALLWKDSVRLS 440
Query: 666 F-----RVIDTHQQVVTFEVWKDNLSWVCSAVYASPIPANREILWRHMTLLRRRFLLPWL 720
R ID H + +N+++ S VY P + R LW H L + PW+
Sbjct: 441 NLYQDDRHIDVHISI-------NNINFYLSRVYGHPCQSERHSLWTHFENLSKTRNDPWI 493
Query: 721 LLGDFNEILFPSEVRGGDFLPNRAAM----FASVLDTCQLVDLGAVGRRFTWFQKANDRL 776
L+GDFNEIL +E GG P R F +++ TC L D+ ++G RF+W + +
Sbjct: 494 LIGDFNEILSNNEKIGG---PQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGERHSHT 550
Query: 777 ILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCPILLQCGSPQTLAHNRPFRFLAAWA 836
+ LDRA + E FP A ++ L SDH P+ L +T RPFRF
Sbjct: 551 VKCC-LDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFLSLEKTET-RKMRPFRFDKRLL 608
Query: 837 DHPDFAAVVDNAWRSGEECITSKL-ERVREASTVFNKEVFGNIFRRKRHVEARLR--GVQ 893
+ P F V W L ++VR K + K ++ +R+R +Q
Sbjct: 609 EVPHFKTYVKAGWNKAINGQRKHLPDQVRTCRQAMAK------LKHKSNLNSRIRINQLQ 662
Query: 894 RELDRRVTSDMVLFE---AELQREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTL 950
LD+ ++S + +QRE R EE QK+R + GDRNT +FH T
Sbjct: 663 AALDKAMSSVNRTERRTISHIQRELTVAYRDEERYWQQKSRNQWMKEGDRNTEFFHACTK 722
Query: 951 IRRRKNRVHRLKLEDGT-WCGDEEVLKRVVHG--FFVNL------------FTGITPLAS 995
R NR+ +K E+G + GD+E+ VH FF + F G P+ +
Sbjct: 723 TRFSVNRLVTIKDEEGMIYRGDKEI---GVHAQEFFTKVYESNGRPVSIIDFAGFKPIVT 779
Query: 996 PRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGD 1055
++ DL LS E+ +A+ + APG DG FYK W VG
Sbjct: 780 EQINDDLTKDLS------------DLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGP 827
Query: 1056 SLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRL 1115
+ VK F+ + +++ + +IPK+ P T+ D RPI+LCNV +K+I+K LV RL
Sbjct: 828 DVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSDYRPIALCNVLYKIISKCLVERL 887
Query: 1116 RPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHM-SKSTAKRGMVAFKIDLEKAYDSVS 1174
+ L ++ Q++F+PGR DN +A E++H + ++ + +A K D+ KAYD V
Sbjct: 888 KGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVE 947
Query: 1175 WSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFV 1234
W+FL+ T+ L+GF E I IM +V S S+L NG +P RG+RQGDP+SPYLF+
Sbjct: 948 WNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFI 1007
Query: 1235 LCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQL 1294
LC + L+ LI++ V +GD + IR+ P ++HL FADD L FC+++V L +
Sbjct: 1008 LCADILNHLIKNRVAEGDIRGIRIGNGVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDV 1067
Query: 1295 FCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRF 1354
+ SG KIN+SKS V ++ + + I GKYLG P + GR R+ F
Sbjct: 1068 YEYYSGQKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMF 1127
Query: 1355 NFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSF 1414
N+++E ++++ SW A LS AG+ + KSV +MP Y M F +P + +I L+ +F
Sbjct: 1128 NYIIERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNF 1187
Query: 1415 IWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVL 1474
W + W+++ K GGLG RD N AL+ K +W +++ PN L+ R++
Sbjct: 1188 WWEKNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIM 1247
Query: 1475 SHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGET 1520
+Y S+L + + S W +L D + G F +G+G+T
Sbjct: 1248 KARYFREDSILDAKRQRYQSYGWTSMLAGLDVIKKGSRFIVGDGKT 1293
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 462 bits (1190), Expect = e-130
Identities = 306/978 (31%), Positives = 472/978 (47%), Gaps = 34/978 (3%)
Query: 726 NEILFPSEVRGGDFLPNRAAM-FASVLDTCQLVDLGAVGRRFTWFQKANDRLILSKRLDR 784
NEIL SE RGG + + F S + L DL G F+W D + +RLDR
Sbjct: 36 NEILDNSEKRGGPPRDQGSFIDFRSFISKNGLWDLKYSGNPFSWRGMRYDWFV-RQRLDR 94
Query: 785 ALGDMEWKVAFPDAVVDVLNRVHSDHCPILLQCGSPQTLAHNRPFRFLAAWADHPDFAAV 844
A+ + W +FP + L SDH P+++ + + FRF D+ A+
Sbjct: 95 AMSNNSWLESFPSGRSEYLRFEGSDHRPLVVFVDEARVKRRGQ-FRFDNRLRDNDVVNAL 153
Query: 845 VDNAW-RSGEECITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSD 903
+ W +G+ + +K+ + R +E+ + + + Q+ L+ +T+D
Sbjct: 154 IQETWTNAGDASVLTKMNQCR-------REIINWTRLQNLNSAELIEKTQKALEEALTAD 206
Query: 904 ------MVLFEAELQREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNR 957
+ A L+ Y+ EE Q++R +H GDRNT YFH T RR +NR
Sbjct: 207 PPNPTTIGALTATLEHAYK----LEEQFWKQRSRVLWLHSGDRNTGYFHAVTRNRRTQNR 262
Query: 958 VHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQP 1017
+ ++ +G +E + +++ G+F +FT + V + P +S L +
Sbjct: 263 LTVMEDINGVAQHEEHQISQIISGYFQQIFTSESDGDFSVVDEAIEPMVSQGDNDFLTRI 322
Query: 1018 VRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDI 1077
+EV+ A+ S+N+ APG DGF FY YW+ + + ++ F + +
Sbjct: 323 PNDEEVKDAVFSINASKAPGPDGFTAGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNET 382
Query: 1078 LVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTM 1137
+ LIPK P V D RPI+LCN+ +K++ K++ R++ L KLI QS+F+PGR
Sbjct: 383 HIRLIPKDLGPRKVADYRPIALCNIFYKIVAKIMTKRMQLILPKLISENQSAFVPGRVIS 442
Query: 1138 DNAFLAQEVIHHMSKSTAKRGM-VAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIM 1196
DN + EV+H + S+AK+ +A K D+ KAYD V W FL++ L+ +GF + I+ ++
Sbjct: 443 DNVLITHEVLHFLRTSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVL 502
Query: 1197 SSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPI 1256
VTS S L NG P RGLRQGDP+SP LF+LC E LS L +
Sbjct: 503 ECVTSVSYSFLINGTPQGKVVPTRGLRQGDPLSPCLFILCTEVLSGLCTRAQRLRQLPGV 562
Query: 1257 RLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGV 1316
R+S NGP ++HL FADD + F ++ N L+ + + +SG IN KS S
Sbjct: 563 RVSINGPRVNHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQSINFHKSSVTFSSKT 622
Query: 1317 SEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLA 1376
VK + + I GKYLG P GR R+ F +++ I++K SW + LS A
Sbjct: 623 PRSVKGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIRQKSHSWASRFLSQA 682
Query: 1377 GRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQP 1436
G+ + K+V+A+MP Y+M F +P + KI L+ F W K V W K+T P
Sbjct: 683 GKQVMLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRFWWDTKPDVRKTSWVAWSKLTNP 742
Query: 1437 KDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQV 1496
K+ GGLG RD E N +L+ K W L++ P L R+L KY +SS ++ + Q S
Sbjct: 743 KNAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSSSFMECKLPSQPSHG 802
Query: 1497 WKGLLKARDRLHTGFVFRLGNGE-TSLWHDDWSGMGN-IAPAVPYVDIHDIDRRLCDLVD 1554
W+ ++ R+ L G + + NGE S+W+D W + + P P + H D R+ L++
Sbjct: 803 WRSIIAGREILKEGLGWLITNGEKVSIWNDPWLSISKPLVPIGPALREHQ-DLRVSALIN 861
Query: 1555 NGT--WNMQVLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWLQDQ 1612
T W+ + LP E L K P D W G YT R Y
Sbjct: 862 QNTLQWDWNKIAVILP--NYENLIKQLPAPSSRGVDKLAWLPVKSGQYTSRSGYGIASVA 919
Query: 1613 QSHLPVVDDWNW---IWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAP 1669
+P +NW +WKL+ KI+ +W +L V + R ++ S +C RC AP
Sbjct: 920 SIPIPQT-QFNWQSNLWKLQTLPKIKHLMWKAAMEALPVGIQLVRRHISPSAACHRCGAP 978
Query: 1670 EEDILHCLRTCPHSQELW 1687
E H C + ++W
Sbjct: 979 -ESTTHLFFHCEFAAQVW 995
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 443 bits (1140), Expect = e-124
Identities = 333/1207 (27%), Positives = 552/1207 (45%), Gaps = 59/1207 (4%)
Query: 637 LGFFPIHIEEARGFSGGIWVLANTSVPYNFRVIDTHQQVVTFEVWKDNLSWVCSAVYASP 696
LG+ H E G SGG+ + + +F D + ++ +V + SW S VY +P
Sbjct: 14 LGYDYFHTIEPVGKSGGLAIFWKNHLEIDFLFED--KNLLDLKVSQGKKSWFVSCVYGNP 71
Query: 697 IPANREILWRHMTLLRRRFLLPWLLLGDFNEILFPSEVRGGDF-LPNRAAMFASVLDTCQ 755
+ R +L ++ + + W ++GDFN+IL GG L + F ++L C
Sbjct: 72 VLHLRYLLLDKLSSIGVQRNSAWCMIGDFNDILSNDGKLGGPSRLISSFQPFKNMLLNCD 131
Query: 756 LVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCPILL 815
+ +G+ G FTW ND+ I K LDR G+ EW F ++ L ++ S H P+L+
Sbjct: 132 MHQMGSSGNSFTWGGTRNDQWIQCK-LDRCFGNSEWFTMFSNSHQWFLEKLGSHHRPVLV 190
Query: 816 QCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAW-RSGEECITSKLERVREASTVFNKEV 874
+ Q + + F + +A+ P AA ++W +G ++S + R+ + K +
Sbjct: 191 NFVNDQEVFRGQ-FCYDKRFAEDPQCAASTLSSWIGNGISDVSSSMLRMVKC----RKAI 245
Query: 875 FGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFE--AELQREYRNILRQEELL*YQKARE 932
G + + R+ ++ ELD + + + +Q + R+EE K+++
Sbjct: 246 SGWKKNSDFNAQNRILRLRSELDEEKSKQYPCWSRISVIQTQLGVAFREEESFWRLKSKD 305
Query: 933 NRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKR----VVHGFFVNLFT 988
+ GDRN+ +F R KN + L E+G +E L R + +F NLF
Sbjct: 306 KWLFGGDRNSKFFQAMVKANRTKNSLRFLVDENG----NEHTLNREKGNIASVYFENLFM 361
Query: 989 GITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKK 1048
P S +S E L Q V + E+ A+ S+N +AP + +
Sbjct: 362 SSYPANSQSALDGFKTRVSEEMNQELTQAVTELEIHSAVFSINVESAP--EKLECCQGSD 419
Query: 1049 YWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVIT 1108
Y +G F+ G + + + LIPK P + D+RPISLC+V +K+I+
Sbjct: 420 YIEILG---------FFETGVLPQEWNHTHLYLIPKFTNPQRMSDIRPISLCSVLYKIIS 470
Query: 1109 KVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHM-SKSTAKRGMVAFKIDLE 1167
K+L +L+ L ++ P QS+F R DN +A E++H + + + + FK D+
Sbjct: 471 KILSFKLKKHLPSIVSPSQSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMVFKTDMS 530
Query: 1168 KAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDP 1227
KAYD V WSFLQE L GF + I+ IM VTS S+L NG P RG+RQGDP
Sbjct: 531 KAYDRVEWSFLQEILVALGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIRQGDP 590
Query: 1228 MSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNL 1287
+SP+LFVLC E L ++Q + I+ + +GP ++HL F DD L C A+ +
Sbjct: 591 ISPFLFVLCTEALIHILQQAENSKKVSGIQFNGSGPSVNHLLFVDDTQLVCRATKSDCEQ 650
Query: 1288 LASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGG 1347
+ + + SG IN+ KS V E K I + I GKYLG P
Sbjct: 651 MMLCLSQYGHISGQLINVEKSSITFGVKVDEDTKRWIKNRSGIHLEGGTGKYLGLPENLS 710
Query: 1348 RIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKI 1407
++ F ++ E +Q L W LS G+ L KS+ A+P Y M F +P+G+ K+
Sbjct: 711 GSKQDLFGYIKEKLQSHLSGWYDKTLSQGGKEILLKSIALALPVYIMTCFRLPKGLCTKL 770
Query: 1408 NQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPN 1467
++ F W+ + H + +K+T PK GG G +D + N AL+ K W L
Sbjct: 771 TSVMMDFWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAKQAWRLFSDSK 830
Query: 1468 KLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGE-TSLWHDD 1526
+ ++ +Y N+ L + + S W+ +L R+ L+ G +GNGE T++W D
Sbjct: 831 SIVSQIFKSRYFMNTDFLNARQGTRPSYTWRSILYGRELLNGGLKRLIGNGEQTNVWIDK 890
Query: 1527 WSGMGNIAPAVPYVDIHDIDRRLCDLVDNGT--WNMQVLYTSLPVDVLERLQKIKPTIVP 1584
W G+ + + +I ++ L+D T WN++ L ++ + +P I
Sbjct: 891 WLFDGHSRRPMNLHSLMNIHMKVSHLIDPLTRNWNLKKLTELFHEKDVQLIMHQRPLI-- 948
Query: 1585 NRQDVWTWETNSMGIYTVRGAY--------RWLQDQQSHLPVVDD-WNWIWKLKVPEKIR 1635
+ +D + W + G+YTV+ Y + L + P V+ ++ +W L+ KI+
Sbjct: 949 SSEDSYCWAGTNNGLYTVKSGYERSSRETFKNLFKEADVYPSVNPLFDKVWSLETVPKIK 1008
Query: 1636 TFVWLTLQNSLQVNLHRFRCK-MAASPSCSRCSAPEEDILHCLRTCPHSQELW------- 1687
F+W L+ +L V R R + + + C C E I H L CP ++++W
Sbjct: 1009 VFMWKALKGALAVE-DRLRSRGIRTADGCLFCKEEIETINHLLFQCPFARQVWALSLIQA 1067
Query: 1688 --LKFGAFAWPNFTAGDHSSWIRSQARSNNGVKFIVGLWGVWKWRNNMIFEDSPWSLEEA 1745
FG + N +S R V + LW +WK RN +F+ + + E
Sbjct: 1068 PATGFGTSIFSNINHVIQNSQNFGIPRHMRTVSPWL-LWEIWKNRNKTLFQGTGLTSSEI 1126
Query: 1746 WRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPMAGSIKLNVDGSYRDVDDSSGVGGLAR 1805
+ E + + + +G + +W PP AG +K N+ ++ +GV + R
Sbjct: 1127 VAKAYEECNLWINAQEKSSGGVSP-SEHKWNPPPAGELKCNIGVAWSRQKQLAGVSWVLR 1185
Query: 1806 DPSGNWL 1812
D G L
Sbjct: 1186 DSMGQVL 1192
>At3g45550 putative protein
Length = 851
Score = 409 bits (1050), Expect = e-113
Identities = 259/797 (32%), Positives = 381/797 (47%), Gaps = 62/797 (7%)
Query: 982 FFVNLFTGITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGF 1041
FF ++FT SP D S++ S L Q R E+ A+ + APG DG
Sbjct: 13 FFTDIFTTNGIQVSPIDFADFPSSVTNIINSELTQDFRDSEIFEAICQIGDDKAPGPDGL 72
Query: 1042 QPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCN 1101
FYK+ W+ VG+ + VK F+ + ++ + +IPK+ P T+ D RPI+LCN
Sbjct: 73 TARFYKQCWDIVGNDVIKEVKLFFESSHMKTSVNHTNICMIPKIQNPQTLSDYRPIALCN 132
Query: 1102 VAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMS-KSTAKRGMV 1160
V +KVI+K +VNRL+ L ++ Q++F+PGR DN +A E++H + + + +
Sbjct: 133 VLYKVISKCMVNRLKAHLNSIVSDSQAAFIPGRIINDNVMIAHEIMHSLKVRKRVSKTYM 192
Query: 1161 AFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGR 1220
A K D+ KAYD V W FL+ T+ L+GF + I IM++V S S+L NG P R
Sbjct: 193 AVKTDVSKAYDRVEWDFLETTMRLFGFCDKWIGWIMAAVKSVHYSVLINGSPHGYISPTR 252
Query: 1221 GLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEA 1280
G+RQGDP+SPYLF+LC + LS LI+ GD + +R+ P I+HL FADD L FC+A
Sbjct: 253 GIRQGDPLSPYLFILCGDILSHLIKVKASSGDIRGVRIGNGAPAITHLQFADDSLFFCQA 312
Query: 1281 SVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYL 1340
+V L ++ SG KIN+ KS V + + + IP GKYL
Sbjct: 313 NVRNCQALKDVFDVYEYYSGQKINVQKSLITFGSRVYGSTQTRLKTLLNIPNQGGGGKYL 372
Query: 1341 GFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMP 1400
G P + GR + FN++++ ++ + SW A LS AG+ L KSV AMP Y M F +P
Sbjct: 373 GLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSPAGKEILLKSVALAMPVYAMSCFKLP 432
Query: 1401 RGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIW 1460
+G+ +I L+ +F W G V W+++ K GGLG RD N AL+ K W
Sbjct: 433 QGIVSEIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAW 492
Query: 1461 CLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGET 1520
++ PN L+ RV+ +Y ++S++ + + Q S W LL L G + +G+G+T
Sbjct: 493 RIIQYPNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLLSGIALLRKGTRYVIGDGKT 552
Query: 1521 SLWHDDWSGMGNIAPAVPYVDIHDIDRRLCDLVDNG--------------TWNMQVLYTS 1566
G+ N+ VD H L D NG W+ L T
Sbjct: 553 IR-----LGIDNV------VDSHPPRPLLTDEQHNGLSLDNLFQHRGHSRCWDNAKLQTF 601
Query: 1567 LPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWL--QDQQSHLPV------ 1618
+ + +++I + ++ D W NS G YTVR Y WL D + +P
Sbjct: 602 VDQSDHDYIKRIYLS-TRSKTDRLIWSYNSTGDYTVRSGY-WLSTHDPSNTIPTMAKPHG 659
Query: 1619 -VDDWNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCL 1677
VD IW L + K++ F+W L +L M P C RC E I H L
Sbjct: 660 SVDLKTKIWNLPIMPKLKHFLWRILSKALPTTDRLTTRGMRIDPGCPRCRRENESINHAL 719
Query: 1678 RTCPHSQELWLKFGAFAWPNFTAGDHSSWIRSQARSNNGVKFIV---------------- 1721
TCP F AW + S I S +N ++
Sbjct: 720 FTCP--------FATMAWRLSDTPLYRSSILSNNIEDNISNILLLLQNTTITDSQKLIPF 771
Query: 1722 -GLWGVWKWRNNMIFED 1737
LW +WK RNN++F +
Sbjct: 772 WLLWRIWKARNNVVFNN 788
>At1g25430 hypothetical protein
Length = 1213
Score = 399 bits (1024), Expect = e-110
Identities = 338/1193 (28%), Positives = 540/1193 (44%), Gaps = 72/1193 (6%)
Query: 589 LISWNVRGALYANGKLAIRELVRSKKPDVFILMETRCQFRRVSAFWNSL--GFFPIHIEE 646
L WN+RG + + ++ V++ KP ++ET + + F N+L G+ + E
Sbjct: 5 LFCWNIRGFNNVSHRSGFKKWVKANKPIFGGVIETHVKQPKDRKFINALLPGWSFV---E 61
Query: 647 ARGFS--GGIWVLANTSVPYNFRVIDTHQQVVTFEVW-KDNLSWVC-SAVYASPIPANRE 702
FS G IWV+ + SV V+ Q++T EV + SW+ S VYA+ A+R+
Sbjct: 62 NYAFSDLGKIWVMWDPSV--QVVVVAKSLQMITCEVLLPGSPSWIIVSVVYAANEVASRK 119
Query: 703 ILWRHMTLLRRRFLL---PWLLLGDFNEILFPSEVRGGDFLPNRAAM--FASVLDTCQLV 757
LW + + ++ PWL+LGDFN++L P E L M F L +L
Sbjct: 120 ELWIEIVNMVVSGIIGDRPWLVLGDFNQVLNPQEHSNPVSLNVDINMRDFRDCLLAAELS 179
Query: 758 DLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDH--CPILL 815
DL G FTW+ K++ + +K++DR L + W FP ++ + SDH C ++L
Sbjct: 180 DLRYKGNTFTWWNKSHTTPV-AKKIDRILVNDSWNALFPSSLGIFGSLDFSDHVSCGVVL 238
Query: 816 QCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAW------RSGEECITSKLERVREASTV 869
+ ++ RPF+F + DF +V + W S ++ KL+ +++
Sbjct: 239 E---ETSIKAKRPFKFFNYLLKNLDFLNLVRDNWFTLNVVGSSMFRVSKKLKALKKPIKD 295
Query: 870 FNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAELQREYRNILRQEELL*YQK 929
F++ + + +R + L G Q T FE E +R++ + EE QK
Sbjct: 296 FSRLNYSELEKRTKEAHDFLIGCQDRTLADPTPINASFELEAERKWHILTAAEESFFRQK 355
Query: 930 ARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTG 989
+R + GD NT YFH R N + L +G +E + + +F +L
Sbjct: 356 SRISWFAEGDGNTKYFHRMADARNSSNSISALYDGNGKLVDSQEGILDLCASYFGSLLGD 415
Query: 990 -ITPLASPRVTHDLFPSLSVEAKSL--LLQPVRKDEVRHALMSMNSYTAPGADGFQPFFY 1046
+ P + +L S + L +++R AL S+ + G DGF F+
Sbjct: 416 EVDPYLMEQNDMNLLLSYRCSPAQVCELESTFSNEDIRAALFSLPRNKSCGPDGFTAEFF 475
Query: 1047 KKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKV 1106
W+ VG + +KE F G + + +VLIPK+ P+ D RPIS N +KV
Sbjct: 476 IDSWSIVGAEVTDAIKEFFSSGCLLKQWNATTIVLIPKIVNPTCTSDFRPISCLNTLYKV 535
Query: 1107 ITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKST-AKRGMVAFKID 1165
I ++L +RL+ L +I QS+FLPGR +N LA +++H + S + RGM+ K+D
Sbjct: 536 IARLLTDRLQRLLSGVISSAQSAFLPGRSLAENVLLATDLVHGYNWSNISPRGML--KVD 593
Query: 1166 LEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQG 1225
L+KA+DSV W F+ L PE IN I +++ ++ NG FK +GLRQG
Sbjct: 594 LKKAFDSVRWEFVIAALRALAIPEKFINWISQCISTPTFTVSINGGNGGFFKSTKGLRQG 653
Query: 1226 DPMSPYLFVLCMERLSVLIQSLVDKG--DWKPIRLSKNGPPISHLFFADDVLLFCEASVA 1283
DP+SPYLFVL ME S L+ S + G + P + ISHL FADDV++F +
Sbjct: 654 DPLSPYLFVLAMEAFSNLLHSRYESGLIHYHP---KASNLSISHLMFADDVMIFFDGGSF 710
Query: 1284 QVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISV----VAPIPFVHDLGKY 1339
++ + T+ F SGLK+N KS ++ G++++ ++ + + +P +Y
Sbjct: 711 SLHGICETLDDFASWSGLKVNKDKSHLYLA-GLNQLESNANAAYGFPIGTLPI-----RY 764
Query: 1340 LGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNM 1399
LG PL ++ + LLE I + SW LS AGR+ L SVI + M F +
Sbjct: 765 LGLPLMNRKLRIAEYEPLLEKITARFRSWVNKCLSFAGRIQLISSVIFGSINFWMSTFLL 824
Query: 1400 PRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAI 1459
P+G +I L F+WSG V+W + PK GGLG+R N L + I
Sbjct: 825 PKGCIKRIESLCSRFLWSGNIEQAKGIKVSWAALCLPKSEGGLGLRRLLEWNKTLSMRLI 884
Query: 1460 WCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNG- 1518
W L + LW +LS S V+ DS WK LL R H V ++GNG
Sbjct: 885 WRLFVAKDSLWADWQHLHHLSRGSFWAVEGGQSDSWTWKRLLSLRPLAHQFLVCKVGNGL 944
Query: 1519 ETSLWHDDWSGM-------GNIAPAVPYVDIHDIDRRLCDLVDNGTWNMQVLYTSLPVDV 1571
+ W+D+W+ + G+I P+ V + ++ W + V ++ +
Sbjct: 945 KADYWYDNWTSLGPLFRIIGDIGPSSLRVPLL---AKVASAFSEDGWRLPVSRSAPAKGI 1001
Query: 1572 LERLQKIK-PTIVPNRQDVWTWETNSMGIYTVRGAYRWLQDQQSHLPVVDDW-NWIWKLK 1629
+ L + P+ D + W N A W + V W + IW
Sbjct: 1002 HDHLCTVPVPSTAQEDVDRYEWSVNGFLCQGFSAAKTW--EAIRPKATVKSWASSIWFKG 1059
Query: 1630 VPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDILHCLRTCPHSQELWLK 1689
K +W++ N L S +C CS E H L C S ++W
Sbjct: 1060 AVPKYAFNMWVSHLNRLLTRQRLASWGHIQSDACVLCSFASESRDHLLLICEFSAQVWRL 1119
Query: 1690 FGAFAWPN---FTA-GDHSSWIR-SQARSNNGVKFIVG---LWGVWKWRNNMI 1734
P F++ + SW+R S + ++ IV ++ +W+ RNN++
Sbjct: 1120 VFRRICPRQRLFSSWSELLSWVRQSSPEAPPLLRKIVSQVVVYNLWRQRNNLL 1172
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 385 bits (988), Expect = e-106
Identities = 275/966 (28%), Positives = 453/966 (46%), Gaps = 42/966 (4%)
Query: 884 HVEARLRGVQRELDRRVTSDMVLFE--AELQREYRNILRQEELL*YQKARENRVHLGDRN 941
+ ++R+ ++RELD+ ++ + + LQ + R+EE K+R+ + GD+N
Sbjct: 21 NAKSRITKLRRELDKEKSATFPSWTQISLLQDVLGDAYREEEDFWRLKSRDKWMVGGDKN 80
Query: 942 TAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTGITPLASPRVTHD 1001
+ +F R N + L E+G ++ FF +LF+ P + V
Sbjct: 81 SKFFQATVKANRVSNSLRFLVDENGNEQTVNREKGKIAVTFFEDLFSSSYPSSMDSVLEG 140
Query: 1002 LFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMV 1061
++ + L + V + E+ A+ S+N+ +APG DGF F+++ W V + + +
Sbjct: 141 FNKRVTEDMNQDLTKKVNEQEIYKAVFSINAESAPGPDGFTALFFQRQWPLVKNQIISDI 200
Query: 1062 KEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRK 1121
+ FQ G + E+ + LIPK+ +P+ + D+RPISLC+V +K+I+K+L RL+ +L
Sbjct: 201 ELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPISLCSVMYKIISKILSARLKKYLPV 260
Query: 1122 LIGPMQSSFLPGRGTMDNAFLAQEVIHHM-SKSTAKRGMVAFKIDLEKAYDSVSWSFLQE 1180
++ P QS+F+ R DN LA E++H++ + + + FK D+ KAYD V W FL+
Sbjct: 261 IVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDMSKAYDRVEWPFLKG 320
Query: 1181 TLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERL 1240
L GF IN +M+ V+S S+L NG P RGLRQGDP+SP+LFVLC E L
Sbjct: 321 ILLALGFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLRQGDPLSPFLFVLCTEAL 380
Query: 1241 SVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSG 1300
++ G I+ + GP ++HL FADD LL C+AS + + + + SG
Sbjct: 381 IHILNQAEKIGKISGIQFNGTGPSVNHLLFADDTLLICKASQLECAEIMHCLSQYGHISG 440
Query: 1301 LKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLES 1360
IN KS V+E K I + I GKYLG P + F F+ E
Sbjct: 441 QMINSEKSAITFGAKVNEETKQWIMNRSGIQTEGGTGKYLGLPECFQGSKQVLFGFIKEK 500
Query: 1361 IQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKG 1420
+Q +L W A LS G+ L KS+ A P Y M F + + + K+ ++ F W+
Sbjct: 501 LQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLSKTLCTKLTSVMMDFWWNSVQ 560
Query: 1421 GGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSHKYLS 1480
H + +K+ PK GG G +D + N AL+ K L + L ++L +Y
Sbjct: 561 DKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLHTDSDSLLSQILKSRYYM 620
Query: 1481 NSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNGE-TSLWHDDWSGMGNIAPAVPY 1539
NS L + S W+ +L R+ L +G +GNGE T +W D+W
Sbjct: 621 NSDFLSATKGTRPSYAWQSILYGRELLVSGLKKIIGNGENTYVWMDNWIFDDKPRRPESL 680
Query: 1540 VDIHDIDRRLCDLVD--NGTWNMQVLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSM 1597
+ DI ++ L+D + WN+ +L P ++ + + +P + +RQD + W +
Sbjct: 681 QIMVDIQLKVSQLIDPFSRNWNLNMLRDLFPWKEIQIICQQRP--MASRQDSFCWFGTNH 738
Query: 1598 GIYTVRGAYRWLQDQQSHLPVVDD----------WNWIWKLKVPEKIRTFVWLTLQNSLQ 1647
G+YTV+ Y L +Q H + + + IW L KI+ F+W L+ ++
Sbjct: 739 GLYTVKSEYD-LCSRQVHKQMFKEAEEQPSLNPLFGKIWNLNSAPKIKVFLWKVLKGAVA 797
Query: 1648 VNLHRFRCK-MAASPSCSRCSAPEEDILHCLRTCPHSQELWLKFGAFAWPN-------FT 1699
V R R + + CS C E + H L CP ++++W PN FT
Sbjct: 798 VE-DRLRTRGVLIEDGCSMCPEKNETLNHILFQCPLARQVW-ALTPMQSPNHGFGDSIFT 855
Query: 1700 AGDHSSWIRSQARSNNGVKFIVG--LWGVWKWRNNMIFED-SPWSLEEAWRRV--CHEHD 1754
+H + ++++ +W +WK RN +FE SL + + C E
Sbjct: 856 NVNHVIGNCHNTELSPHLRYVSPWIIWILWKNRNKRLFEGIGSVSLSIVGKALEDCKEWL 915
Query: 1755 EIVAVLGEEAGSMDCWLGSRWQPPMAGSIKLNVDGSYRDVDDSSGVGGLARDPSGNWLFG 1814
+ ++ + + D W PP+ +K N+ ++ +GV + R NW
Sbjct: 916 KAHELICSKEPTKDL----TWIPPLMNELKCNIGIAWSKKHQMAGVSWVVR----NWKGR 967
Query: 1815 FLAHRR 1820
L H R
Sbjct: 968 VLLHSR 973
>At2g11240 pseudogene
Length = 1044
Score = 372 bits (955), Expect = e-102
Identities = 240/828 (28%), Positives = 398/828 (47%), Gaps = 32/828 (3%)
Query: 754 CQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVVDVLNRVHSDHCPI 813
C L DL G +W K +D ++ RLDRAL + W +P + L SDH P+
Sbjct: 4 CDLYDLRHSGNFLSWRGKRHDHVVHC-RLDRALSNGAWAEDYPASRCIYLCFEGSDHRPL 62
Query: 814 LLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRSGE-ECITSKLERVREASTVFNK 872
L + FR+ ++ + A+V AW + + + K+ R R ++
Sbjct: 63 LTHFDLSKKKKKG-VFRYDRRLKNNDEVTALVQEAWNLYDTDIVEEKISRCRLEIVKWS- 120
Query: 873 EVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAELQREYRNIL---RQEELL*YQK 929
R K+ +L R+ S + L N+L + EE Q+
Sbjct: 121 -------RAKQQSSQKLIEENRQKLEEAMSSQDHNQELLSTINTNLLLAYKAEEEYWKQR 173
Query: 930 ARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTG 989
+R+ + LGD+N+ YFH T R N+ ++ EDG +E + V+ +F LF+
Sbjct: 174 SRQLWLALGDKNSGYFHAITRGRTVINKFSVIEKEDGVPEYEEAGILNVISEYFQKLFSA 233
Query: 990 ITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKY 1049
+ + + P +S E +E++ A S+++ APG DGF F++
Sbjct: 234 NEGARAATIKEAIKPFISPEQNP--------EEIKSACFSIHADKAPGPDGFSASFFQSN 285
Query: 1050 WNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITK 1109
W VG ++ ++ F + + + LIPK+ + D RPI+LC V +K+I+K
Sbjct: 286 WMTVGPNIVLEIQSFFSSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVFYKIISK 345
Query: 1110 VLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHM-SKSTAKRGMVAFKIDLEK 1168
+L RL+P L+++I QS+F+P R + DN + E +H++ S KR +A K ++ K
Sbjct: 346 LLSRRLQPILQEIISENQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMSK 405
Query: 1169 AYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPM 1228
AYD + W F++ ++ GF + I+ I+ +T+ S L NG A P RGLRQGDP+
Sbjct: 406 AYDRIEWDFIKLVMQEMGFHQTWISWILQCITTVSYSFLLNGSAQGAVTPERGLRQGDPL 465
Query: 1229 SPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLL 1288
SP+LF++C E LS L + G +R+SK P ++HL FADD + FC + +
Sbjct: 466 SPFLFIICSEVLSGLCRKAQLDGSLLGLRVSKGNPRVNHLLFADDTIFFCRSDLKSCKTF 525
Query: 1289 ASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGR 1348
++ + ++SG IN SKS S+ + +K + I V LGKYLG P GR
Sbjct: 526 LCILKKYEEASGQMINKSKSAITFSRKTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFGR 585
Query: 1349 IHRNRFNFLLESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKIN 1408
R+ FN +++ I+++ SW + LS AG+ + KSV+A+MPTYTM F + + +I
Sbjct: 586 KKRDLFNQIVDRIRQRSLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQ 645
Query: 1409 QLIRSFIWSGKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNK 1468
+ F W + W K+ + K GGLG +D N AL+ K W ++ P+
Sbjct: 646 SALTHFWWDSSADKKKMCWIAWSKMAKNKKEGGLGFKDITNFNDALLAKLSWRIVQSPSC 705
Query: 1469 LWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDRLHTGFVFRLGNG-ETSLWHDDW 1527
+ VR+L KY SS L S W+G+ +D + + +G+G +T +W++ W
Sbjct: 706 VLVRILLGKYCRTSSFLDCSVTAASSHGWRGICTGKDLIKSQLGKVIGSGLDTLVWNEPW 765
Query: 1528 SGMGN----IAPAVPYVDIHDIDRRLCDLVDN----GTWNMQVLYTSL 1567
+ + PA+ + + +C + W++ T+L
Sbjct: 766 LSLSTSSTPMGPALEQFKSMTVAQLICQTTKSWDREKVWDLSPFKTTL 813
Score = 38.5 bits (88), Expect = 0.038
Identities = 44/196 (22%), Positives = 79/196 (39%), Gaps = 38/196 (19%)
Query: 1723 LWGVWKWRNNMIFEDSPWSLEEAWRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPMA-- 1780
LW +W RN +IFE +H + ++ + WLG++ Q +
Sbjct: 849 LWNLWNCRNKLIFE--------------QKHISSMDLISQSISQSTEWLGAQIQASKSKI 894
Query: 1781 ------------GSIKLNVDGSYRDVDDSSGVGGLARDPSGNWLFGFLAHRRGG-----N 1823
+I+ + D S+R+ +G G + D S + +H + +
Sbjct: 895 VIPGISPSEIDLDTIQCSTDASWREETLQAGFGWVFVDHSNH----LESHHKAAAMNIRS 950
Query: 1824 AFLAEAQALLLGLELVWARGYRDIVVEVDCADLLQSLDDEDRRRFL-PILGDIRNMKDRG 1882
LA+A AL L ++ G++ +VV D L++ L+ E L I+ DI +
Sbjct: 951 PLLAKASALSLAIQHAADLGFKKLVVASDSQQLVKVLNGEPHPMELHGIVFDISVLSLNF 1010
Query: 1883 WRISLERVRRDCNAPA 1898
S V+R+ N+ A
Sbjct: 1011 EENSFSFVKRENNSKA 1026
>At1g31100 hypothetical protein
Length = 1090
Score = 362 bits (929), Expect = 1e-99
Identities = 298/1089 (27%), Positives = 478/1089 (43%), Gaps = 93/1089 (8%)
Query: 686 SWVCSAVYASPIPANREILWRHMTLLRRRFL---LPWLLLGDFNEILFPSEVRGGDFLP- 741
S V S VYA+ R+ LW + LL PW++LGDFN++L P+E L
Sbjct: 14 SVVVSIVYAANEAITRKELWEELLLLSVSLSGNGKPWIMLGDFNQVLCPAEHSQATSLNV 73
Query: 742 -NRAAMFASVLDTCQLVDLGAVGRRFTWFQKANDRLILSKRLDRALGDMEWKVAFPDAVV 800
R +F L +L DL G FTW+ K+ R + +K+LDR L + W FP A
Sbjct: 74 NRRMKVFRDCLFEAELCDLVFKGNTFTWWNKSATRPV-AKKLDRILVNESWCSRFPSAYA 132
Query: 801 DVLNRVHSDH--CPILLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRS----GEE 854
SDH C +++ +P RPFRF +PDF ++V W S G
Sbjct: 133 VFGEPDFSDHASCGVII---NPLMHREKRPFRFYNFLLQNPDFISLVGELWYSINVVGSS 189
Query: 855 C--ITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAELQ 912
++ KL+ ++ F+ E F N+ +R + + Q + T E E Q
Sbjct: 190 MFKMSKKLKALKNPIRTFSMENFSNLEKRVKEAHNLVLYRQNKTLSDPTIPNAALEMEAQ 249
Query: 913 REYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDE 972
R++ +++ EE Q++R + GD NT+YFH R+ N +H + ++G +
Sbjct: 250 RKWLILVKAEESFFCQRSRVTWMGEGDSNTSYFHRMADSRKAVNTIHIIIDDNGVKIDTQ 309
Query: 973 EVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSL-----SVEAKSLLLQPVRKDEVRHAL 1027
+K +F NL G + P + + F L S + K L + +++ A
Sbjct: 310 LGIKEHCIEYFSNLLGG--EVGPPMLIQEDFDLLLPFRCSHDQKKELAMSFSRQDIKSAF 367
Query: 1028 MSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQ 1087
S S G DGF F+K+ W+ +G + V E F + + +VLIPK+
Sbjct: 368 FSFPSNKTSGPDGFPVEFFKETWSVIGTEVTDAVSEFFTSSVLLKQWNATTLVLIPKITN 427
Query: 1088 PSTVRDLRPISLCN-----VAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFL 1142
S + D RPIS CN +KVI ++L NRL+ L ++I P QS+FLPGR +N L
Sbjct: 428 ASKMNDFRPIS-CNDFGPITLYKVIARLLTNRLQCLLSQVISPFQSAFLPGRFLAENVLL 486
Query: 1143 AQEVIHHMSKSTAK-RGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTS 1201
A E++ ++ RGM+ K+DL KA+DS+ W F+ L+ G P+ + I +++
Sbjct: 487 ATELVQGYNRQNIDPRGML--KVDLRKAFDSIRWDFIISALKAIGIPDRFVYWITQCIST 544
Query: 1202 SQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKG--DWKPIRLS 1259
S+ NG FK RGLRQG+P+SP+LFVL ME S L+ S G + P
Sbjct: 545 PTFSVCVNGNTGGFFKSTRGLRQGNPLSPFLFVLAMEVFSSLLNSRFQAGYIHYHP---- 600
Query: 1260 KNGP-PISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSE 1318
K P ISHL FADD+++F + + ++ ++ ++ F SGL +N K+ ++ G+
Sbjct: 601 KTSPLSISHLMFADDIMVFFDGGSSSLHGISEALEDFAFWSGLVLNREKTHLYLA-GLDR 659
Query: 1319 VVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAGR 1378
+ +I+ ++ + LLE + ++ SW LS AGR
Sbjct: 660 IEASTIA---------------------RKLRIAEYGPLLEKLAKRFRSWSVKCLSFAGR 698
Query: 1379 VCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKD 1438
V L SVI+ + + + F +P+G +I L F+WSG V W ++ PK+
Sbjct: 699 VQLIASVISGIINFWISTFILPKGCVKRIEALCARFLWSGNIDVKKGAKVAWSEVCLPKE 758
Query: 1439 RGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWK 1498
GG G +E +++W HK H + N S ++ DS W+
Sbjct: 759 EGGSGYYFSE-------KESLWAAWHK---------FHNF-RNKSFWEIDESATDSWTWR 801
Query: 1499 GLLKARDRLHTGFVFRLGNG-ETSLWHDDWSGMGNI----APAVPYVDIHDIDRRLCDLV 1553
LL+ R LG+G + S W D+WS +G + + P I ++ D
Sbjct: 802 SLLRLRTVAGRFLFSVLGDGKKISFWFDNWSPLGPLFKLFGSSGPRALCIPIQAKVADAC 861
Query: 1554 DNGTWNMQVLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWLQDQQ 1613
+ W + T + +L L I + D + W + + A W + +
Sbjct: 862 SDVGWLISPPRTDQALALLIHLTTIALPCFDSSPDTFVWIVDDFTCHGFSAARTW-EAMR 920
Query: 1614 SHLPVVDDWNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRCSAPEEDI 1673
PV D +W K +W++ N L + + C CS+ E
Sbjct: 921 PKKPVKDWTKSVWFKGSVPKHAFNMWVSHLNRLPTRQRLAAWGVTTTTDCCLCSSRPESR 980
Query: 1674 LHCLRTCPHSQELWLKFGAFAWPNFTAGDHSSWIRSQARSNNG--------VKFIVGLWG 1725
H L C S +W P+ + + + S R N+ + ++
Sbjct: 981 DHLLLYCVFSAVIWKLVFFRLTPSQAIFNSWAELLSWTRINSSKAPSLLRKIAAQASVFH 1040
Query: 1726 VWKWRNNMI 1734
+WK RNN++
Sbjct: 1041 LWKQRNNVL 1049
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 353 bits (905), Expect = 7e-97
Identities = 224/745 (30%), Positives = 357/745 (47%), Gaps = 41/745 (5%)
Query: 1138 DNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMS 1197
DN +A E+IH + + VA K+D+ KA+D + W F++ ++ GF E N IM+
Sbjct: 4 DNILIAHELIHSLHTKKLVQPFVATKLDITKAFDKIEWGFIEAIMKQMGFSEKWCNWIMT 63
Query: 1198 SVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIR 1257
+T++ SIL NG + P RG+RQGDP+SPYL++LC E LS LIQ+ + +
Sbjct: 64 CITTTTYSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLSALIQASIKAKQLHGFK 123
Query: 1258 LSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVS 1317
S+NGP ISHL FA D L+FC+A++ + L + ++L+ +SG +N KS + KG+
Sbjct: 124 ASRNGPAISHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASGQAVNFQKSAILFGKGLD 183
Query: 1318 EVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKLGSWRANMLSLAG 1377
+ +S + I G+YLG P GR N F+F+ +++ +K+ +W +LS AG
Sbjct: 184 FRTSEQLSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNKLLSPAG 243
Query: 1378 RVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPK 1437
+ L KS++ A+PTY+M F +P + +I +R F WS H V W K+ PK
Sbjct: 244 KEVLIKSIVTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWSKLNDPK 303
Query: 1438 DRGGLGVRDTELANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVW 1497
GGL +RD + N AL+ K W ++ +P L RV KY +L +A Q S W
Sbjct: 304 KMGGLAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKATSQSSYAW 363
Query: 1498 KGLLKARDRLHTGFVFRLGNGET-SLWHDDWSGMGNIAPAVPYVDIHDIDRRLCDLVDNG 1556
K +L + G + GNG LW D+W + P V D ++ DL+ G
Sbjct: 364 KSILHGTKLISRGLKYIAGNGNNIQLWKDNWLPLNPPRPPVGTCDSIYSQLKVSDLLIEG 423
Query: 1557 TWNMQVLYTSLPVDVLERLQKIKPTIVPNRQDVWTWETNSMGIYTVRGAYRWL----QDQ 1612
WN +L + + + ++ I+P+I D TW G Y+V+ Y L Q Q
Sbjct: 424 RWNEDLLCKLIHQNDIPHIRAIRPSIT-GANDAITWIYTHDGNYSVKSGYHLLRKLSQQQ 482
Query: 1613 QSHLPVVDD------WNWIWKLKVPEKIRTFVWLTLQNSLQVNLHRFRCKMAASPSCSRC 1666
+ LP ++ + IWK P KI+ F W + N+L + R ++ +C RC
Sbjct: 483 HASLPSPNEVSAQTVFTNIWKQNAPPKIKHFWWRSAHNALPTAGNLKRRRLITDDTCQRC 542
Query: 1667 SAPEEDILHCLRTCPHSQELWLKFGAFAWP--NFTAGDHSSWIRSQARSNNGVKFIVGL- 1723
ED+ H L C S+E+W + P + + + + S + N + V L
Sbjct: 543 GEASEDVNHLLFQCRVSKEIWEQAHIKLCPGDSLMSNSFNQNLESIQKLNQSARKDVSLF 602
Query: 1724 ----WGVWKWRNNMIFEDSPWSLEEAWRRVCHEHDEIVAVLGEEAGSMDCWLGSRWQPPM 1779
W +WK RN++IF + WS+ ++ ++ + + S++C + +P +
Sbjct: 603 PFIGWRIWKMRNDLIFNNKRWSIPDSIQKALIDQQ-------QWKESLNCNEQQQRKPQL 655
Query: 1780 AGSIKLNVDGSY----RDVDDSSGVGGLARDPSGNWLFGFLAHRRGGNAFLAEAQALLLG 1835
S G + R + + V L G + AEA+AL L
Sbjct: 656 HDSHNNKCYGGHLKTLRTLSKAQFV-----------LRGMSSIPPTSTPLEAEAEALKLA 704
Query: 1836 LELVWARGYRDIVVEVDCADLLQSL 1860
+ + GY DI+ D A L Q +
Sbjct: 705 MIHLQRLGYEDIIFHGDVAALFQPI 729
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.332 0.143 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,899,054
Number of Sequences: 26719
Number of extensions: 1838605
Number of successful extensions: 4838
Number of sequences better than 10.0: 126
Number of HSP's better than 10.0 without gapping: 102
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 4183
Number of HSP's gapped (non-prelim): 232
length of query: 1898
length of database: 11,318,596
effective HSP length: 114
effective length of query: 1784
effective length of database: 8,272,630
effective search space: 14758371920
effective search space used: 14758371920
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0114.7


