

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0114.12
(1414 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
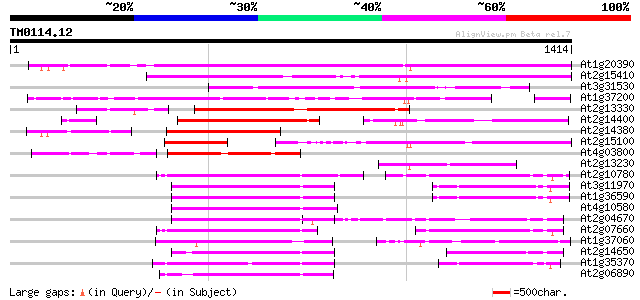
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 1020 0.0
At2g15410 putative retroelement pol polyprotein 832 0.0
At3g31530 hypothetical protein 568 e-162
At1g37200 hypothetical protein 490 e-138
At2g13330 F14O4.9 476 e-134
At2g14400 putative retroelement pol polyprotein 383 e-106
At2g14380 putative retroelement pol polyprotein 338 2e-92
At2g15100 putative retroelement pol polyprotein 322 1e-87
At4g03800 318 1e-86
At2g13230 pseudogene 254 2e-67
At2g10780 pseudogene 232 1e-60
At3g11970 hypothetical protein 219 7e-57
At1g36590 hypothetical protein 219 7e-57
At4g10580 putative reverse-transcriptase -like protein 219 1e-56
At2g04670 putative retroelement pol polyprotein 216 1e-55
At2g07660 putative retroelement pol polyprotein 210 5e-54
At1g37060 Athila retroelment ORF 1, putative 209 9e-54
At2g14650 putative retroelement pol polyprotein 207 3e-53
At1g35370 hypothetical protein 193 5e-49
At2g06890 putative retroelement integrase 181 3e-45
>At1g20390 hypothetical protein
Length = 1791
Score = 1020 bits (2638), Expect = 0.0
Identities = 581/1424 (40%), Positives = 818/1424 (56%), Gaps = 99/1424 (6%)
Query: 47 WCEYHKALGHTTDNCWNLRREIDRLIKAGHLA---------------------NFVKDTA 85
+CEYHK+ H+T+NC L+ + K+G + ++ D
Sbjct: 410 YCEYHKSRAHSTENCRFLQGLLMAKYKSGGITIECDRPPINNKNQRRNETTARQYLNDQT 469
Query: 86 TP----EVAKITQED------KGKGKEIVEELGDPVGECSSIAGGFGGGAISSRARKRY- 134
P E IT D + GK I E V + I GG + S R+ K+Y
Sbjct: 470 KPPTPAEQGIITSADDPAAKRQRNGKAIAAE-PVVVRQVHVIMGGLQNCSDSVRSIKQYR 528
Query: 135 ------VATVHSVQESNEGECWVNHSPIIFTPQDFAHVIPYDNDPIVVTIRVNNYVTKKV 188
VA S + +PI FT D + NDP+VV + +++ +V
Sbjct: 529 KKAEMVVAWPSSTSTTRNPN---QSAPISFTDVDLEGLDTPHNDPLVVELIISDSRVTRV 585
Query: 189 FLDQGSSADIIYGDAFDRLGLKESNLKPYKGTLVGFTGDRVSVRGYVEIPTAFGEGEFVK 248
+D GSS D+I+ D + + + +KP L GF GD V G +++P G G
Sbjct: 586 LIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTIKLPIFVG-GLIA- 643
Query: 249 KFQVKYLVLACRANYNVLLGRDTLNKVCAVISTAHLTVKYPACNGKVGILRVDQNAAREC 308
VK++V+ A YNV+LG ++++ A+ ST H VK+P NG + LR + A
Sbjct: 644 --WVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPTHNG-IFTLRAPKEA---- 696
Query: 309 YLRSVALYGRKAAKESHRITEIFPQEGFSLDPRDEADDFRPQPLEETKQVQVKDKFLKIG 368
K S+ +E+ E ++D D + + +G
Sbjct: 697 ----------KTPSRSYEESELCRTEMVNIDESDPT------------------RCVGVG 728
Query: 369 SGLTTEQEDRLITLLGDNLDLFAWTINDVPGIDPKVITHKLAIRPGATPVIQPMRRMSEE 428
+ ++ LI LL N FAW+I D+ GIDP + H+L + P PV Q R++ E
Sbjct: 729 AEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLGPE 788
Query: 429 KHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPL 488
+ +AV E EKL+KA I EV+YP WLAN V+VKK NGKWR+C DYT LNK CPKDSYPL
Sbjct: 789 RARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPL 848
Query: 489 PNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAG 548
P++D+LV+ SGN LLS MDA+SGYNQI+MH D+E T+F+T++ YCYK M FGLKNAG
Sbjct: 849 PHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAG 908
Query: 549 ATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCS 608
ATYQR ++K+ + Q+GR +EVY+DDM+VKS + DH L + FD L TY MKLNP KC+
Sbjct: 909 ATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCT 968
Query: 609 FGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGRMAALSRFLPMAGDK 668
FG+ G+FLG+++T RGIE NP + RAILE+ SP + +EVQRLTGR+AAL+RF+ + DK
Sbjct: 969 FGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDK 1028
Query: 669 AAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTV 728
PF+ LK+ ++F W ++ E+AF KLK+ L+T P+L KP G L LY+AV+D AVS+V
Sbjct: 1029 CLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHAVSSV 1088
Query: 729 LLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYFQSFQVKVKTDVPL 788
L++E+ +Q+ I++ S +L AE RY IEKAALA++ +AR+LRPYFQS + V TD PL
Sbjct: 1089 LVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLTDQPL 1148
Query: 789 RQVLQKPDLSGRLVSWSVELSEYDIQYEPRGQVTVQSLIDFVAELTPTEGEKTQG----- 843
R L P SGR+ W+VELSEYDI + PR + Q L DF+ EL E+
Sbjct: 1149 RVALHSPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLADFLIELPLQSAERAVSGNRGE 1208
Query: 844 EWVLSVDGSSNNTGSGAGITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGLRLAIELG 903
EW L VDGSS+ GSG GI + SP ++EQS + F A+NN +EYE LIAGLRLA +
Sbjct: 1209 EWSLYVDGSSSARGSGIGIRLVSPTAEVLEQSFRLRFVATNNVAEYEVLIAGLRLAAGMQ 1268
Query: 904 VQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPRGQNERADV 963
+ + DSQL+ Q+ GEY+ K+ ++ YL++V+ + + + K+ +PRG N AD
Sbjct: 1269 ITTIHAFTDSQLIAGQLSGEYEAKNEKMDAYLKIVQLMTKDFENFKLSKIPRGDNAPADA 1328
Query: 964 LAKLASTGRLGNYQTVIQETLPRPSIDLVEIKLKVVKSVNEGELP-------WMESIKTF 1016
LA LA T + + E++ +PSID + +++V ++ P W I+ +
Sbjct: 1329 LAALALTSDSDLRRIIPVESIDKPSIDSTD-AVEIVNTIRSSNAPDPADPTDWRVEIRDY 1387
Query: 1017 L-ENPPKEDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDALGIMAEVHEGVC 1075
L + D R R +A+ YTL+ L + ML C+ + IM E HEG
Sbjct: 1388 LSDGTLPSDKWTARRLRIKAAKYTLMKEHLLKVSAFGAMLNCLHGTEINEIMKETHEGAA 1447
Query: 1076 SSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPF 1135
+H GGR+LA+K+ + GFYWPTM DC + KCE+CQ + P E L +AP+PF
Sbjct: 1448 GNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTELLRAGVAPYPF 1507
Query: 1136 AMWGTDILGPFPVAKAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRFGVP 1195
W DI+GP P A Q ++I+V DYFTKW+EAE+ ATI A V+NF+W+ I+CR G+P
Sbjct: 1508 MRWAMDIVGPMP-ASRQKRFILVMTDYFTKWVEAESYATIRANDVQNFVWKFIICRHGLP 1566
Query: 1196 MALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAK 1255
++ DNG+QF S FCA I + ++ +PQ NGQAE+ NK IL GLKK+LDE K
Sbjct: 1567 YEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNKTILSGLKKRLDEKK 1626
Query: 1256 GLWAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEIENQSWRVARFNEND--NGE 1313
G WA+EL GVLW+Y TT +S+T +TP+ YG +AM E+ S R + +N N
Sbjct: 1627 GAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYSSLRRSMMVKNPELNDR 1686
Query: 1314 NLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDLVL*K---NTIPDKHN 1370
++ L L E + A R + ++ A+ ++ KV R VGDLVL K NT
Sbjct: 1687 MMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGDLVLRKVFENTAEINAG 1746
Query: 1371 KLSPNWGGPYRIIGDVGGEAYKLEQLSGQKVPRTWNASHLKQYF 1414
KL NW G Y++ V Y+L +SG VPRTWN+ HLK+Y+
Sbjct: 1747 KLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTWNSMHLKRYY 1790
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 832 bits (2149), Expect = 0.0
Identities = 469/1138 (41%), Positives = 655/1138 (57%), Gaps = 126/1138 (11%)
Query: 346 DFRPQPLEETK-QVQVKD----KFLKIGSGLTTEQEDRLITLLGDNLDLFAWTINDVPGI 400
D RP P + T QV + + + + I L +E ++ L+ L N FAW++ D+PGI
Sbjct: 706 DERPNPQKGTVVQVNIDESDPSRCVGIRIDLPSELQNELVNFLRQNAATFAWSVEDMPGI 765
Query: 401 DPKVITHKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVM 460
D V H+L + P P+ Q R++ ++ K V E +KL+ A I EV+YP WL N V+
Sbjct: 766 DSAVTCHELNVDPTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVV 825
Query: 461 VKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHP 520
VKK NGKWR+C D+T LNK CPKDS+PLP++D+LV+ +GNELLS MDA+SGYNQI+MH
Sbjct: 826 VKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQ 885
Query: 521 SDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSAR 580
+D E T F+T+Q YCYK MPFGLKNAGATY RL++++F+ Q+ +MEVY+DDM+VKS R
Sbjct: 886 NDREKTVFITDQGTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLR 945
Query: 581 ASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMK 640
A +H L++ F L Y MKLNP KC+FG+ G+FLG+++T RGIE NP + AI+++
Sbjct: 946 AEEHITHLRQCFQVLNRYNMKLNPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLP 1005
Query: 641 SPTSVKEVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLA 700
SP + +EVQRL GR+AAL+RF+ + DK PF+ L+ N +F+W E+CE+
Sbjct: 1006 SPRNTREVQRLIGRIAALNRFISRSTDKCLPFYQLLRANKRFEWDEKCEE---------- 1055
Query: 701 TLPVLSKPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKA 760
G L LY+AV+ AVS VL++E+ +Q I++VS TL GAELRY +EK
Sbjct: 1056 ----------GETLYLYIAVSTSAVSGVLVREDRGEQHPIFYVSKTLDGAELRYPTLEKL 1105
Query: 761 ALAILKTARRLRPYFQSFQVKVKTDVPLRQVLQKPDLSGRLVSWSVELSEYDIQYEPRGQ 820
A A++ +AR+LRPYF+S+ V+V T+ PLR +L P SGRL W+VELSEYDI Y+ R
Sbjct: 1106 AFAVVISARKLRPYFKSYTVEVLTNQPLRTILHSPSQSGRLAKWAVELSEYDIAYKNRTC 1165
Query: 821 VTVQSLIDFVAELTPTEGEKTQGEWVLSVDGSSNNTGSGAGITIESPDKMIIEQSLKFEF 880
+ PT + I D F
Sbjct: 1166 AK--------SHRAPTRPYR-------------------RSIPCRKVDLA--------RF 1190
Query: 881 KASNNQSEYEALIAGLRLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRR 940
ASNN++EYEALIAGLRLA + V+K+ DSQLV Q G Y+ K+ ++ YL+VVR
Sbjct: 1191 PASNNEAEYEALIAGLRLAHGIEVKKIQAYCDSQLVASQFSGNYEAKNERMDAYLKVVRE 1250
Query: 941 LMMEVKEIKIEHVPRGQNERADVLAKLASTGRLGNYQTV-----------IQETLPRPS- 988
L + ++ +PR N AD LA LAST + + IQ T+ +PS
Sbjct: 1251 LSYNFEVFELTKIPRSDNAPADALAVLASTSDPDLRRVIPVECIDVPSIKIQGTITKPSE 1310
Query: 989 ---IDLVEIKLKVV------------------------------------------KSVN 1003
IDL +L +V S +
Sbjct: 1311 YTVIDLTAEQLSIVMVILPPATDRSEDAGPSGNLVCSSDPGQSSGCDRSDDSNRSADSDS 1370
Query: 1004 EGELPWMESIKTFLEN--PPKEDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTK 1061
W+E I++++ + PK+ R + R A YTL L R +L C+D +
Sbjct: 1371 PASTNWIEEIRSYIADRIVPKDKWAARRLRARSAQ-YTLPHEHLLRWSATGVLLSCLDDE 1429
Query: 1062 DALGIMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKA 1121
+A +M E+HEG +H GGR+LA+K+ + G YWPTM DC ++V +CEKCQ +
Sbjct: 1430 EAQQVMREIHEGAGGNHSGGRALALKIRKHGQYWPTMNADCEKFVARCEKCQGHAPFIHI 1489
Query: 1122 PPEELTTMMAPWPFAMWGTDILGPFPVAKAQMKYIIVAVDYFTKWIEAEAVATITAAKVR 1181
P E L T M P+PF W DI+ PFP A Q KYI+V DYFTKW EAE+ A I + +V+
Sbjct: 1490 PSEVLQTAMPPYPFMRWAMDIIRPFP-ASRQKKYILVMTDYFTKWFEAESYARIQSKEVQ 1548
Query: 1182 NFLWQRIVCRFGVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANK 1241
NF+W+ I+C G+P +V DNGTQFTS FCA+ I + ++ +PQ NGQAE+ NK
Sbjct: 1549 NFVWKNIICHHGLPYEIVTDNGTQFTSLQFEGFCAKWKIRLSKSTPRYPQCNGQAEATNK 1608
Query: 1242 VILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEIENQSW 1301
IL GLKK+LDE KG WA+EL GVLW+Y TT + ST TP+ LTYG +A+ E +
Sbjct: 1609 TILDGLKKRLDEKKGAWADELDGVLWSYRTTPRRSTDRTPFSLTYGMEALAPCEAGLPTV 1668
Query: 1302 RVARFNEND--NGENLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDLV 1359
R + + N + L+ NL L E++ +A +R + + A+ ++ KV R GDLV
Sbjct: 1669 RRSMLINDPTLNDQMLLDNLDTLEEQRDQALLRIQNYQQAAAKFYNKKVKNRLFAEGDLV 1728
Query: 1360 L*K---NTIPDKHNKLSPNWGGPYRIIGDVGGEAYKLEQLSGQKVPRTWNASHLKQYF 1414
L K NT+ KL +W GPY I V Y+L + G +PR+WN+++LK+Y+
Sbjct: 1729 LRKVFENTVEQDAGKLGAHWEGPYLISKVVKPGVYELLTMDGTPIPRSWNSANLKRYY 1786
>At3g31530 hypothetical protein
Length = 831
Score = 568 bits (1465), Expect = e-162
Identities = 328/822 (39%), Positives = 475/822 (56%), Gaps = 95/822 (11%)
Query: 501 NELL--SLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKI 558
N+LL S +++ + +MM+P D+E TAF T Q +CY+ MPFGLKNAGATY+R ++KI
Sbjct: 77 NQLLETSYCHSWTLFVVMMMNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKI 136
Query: 559 FSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLG 618
F+ Q+G+ MEVY+DDM+VKS DH L+E F QL Y +KLNP KC FG++ G
Sbjct: 137 FALQIGKTMEVYIDDMLVKSMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRSG---- 192
Query: 619 FMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGRMAALSRFLPMAGDKAAPFFTCLKK 678
IE NP + A+L M SP + +EVQ LTGR+AAL+RF+ + +K F+ L++
Sbjct: 193 -------IEANPKQIEALLGMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRR 245
Query: 679 NSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQK 738
N KF+WT CE+AF +LK+ LAT P+L+KP G P LY+AV+D AVS L++E+ +QK
Sbjct: 246 NKKFEWTTRCEEAFQELKKYLATPPILAKPVIGEPQYLYVAVSDTAVSGELVREDRGEQK 305
Query: 739 VIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYFQSFQVKVKTDVPLRQVLQKPDLS 798
+I++VS T AE RY ++EK ALA++ +A++LRPYFQS + V +PLR +L P S
Sbjct: 306 LIFYVSQTFTSAESRYPQMEKLALAVVMSAQKLRPYFQSHSIIVMGSMPLRVILHSPSQS 365
Query: 799 GRLVSWSVELSEYDIQYEPRGQVTVQSLIDFVAELTPTEGEKTQGE--WVLSVDGSSNNT 856
GRL W++ELSEYDI+Y+ + Q L DF+ EL E + + W+L VDGSS+
Sbjct: 366 GRLAKWTIELSEYDIEYQNKTCAKSQVLADFIVELPTKEARENPLDTTWLLHVDGSSSKQ 425
Query: 857 GSGAGITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGLRLAIELGVQKLFIKGDSQLV 916
GSG GI + SP +A+NN +EYEAL+AGL LA L + K DSQL+
Sbjct: 426 GSGVGIRLTSPTG-----------EATNNVAEYEALVAGLNLAWGLKIGKTRAFCDSQLI 474
Query: 917 VKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPRGQNERADVLAKLASTGRLGNY 976
Q GEY +D ++ YL V+ L E ++ +PRG+N AD LA LAST
Sbjct: 475 ANQFNGEYTTQDKKMEAYLIHVQNLAKNFDEFELTRIPRGENTSADALAALASTSDTSLK 534
Query: 977 QTVIQETLPRPSIDL--VEIKLKVVKSVNEGE-----LPWMESIKTFL-ENPPKEDDLNT 1028
+ + E + +PSI+L E L + S ++ + WME I +++ E D
Sbjct: 535 RVIPVEFIEKPSIELGKEEHVLPIQISADQDDPDDCNSEWMEPIISYISEGKLPSDKWKA 594
Query: 1029 RTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDALGIMAEVHEGVCSSHIGGRSLAVKV 1088
R + +A+ + LVD +LY+ + P++ CV+ + IM E+H G SH
Sbjct: 595 RKLKAQAARFVLVDTKLYKWRLSGPLMTCVEAEAICKIMKEIHSG---SHA--------- 642
Query: 1089 IRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMWGTDILGPFPV 1148
PT+ + P E L+++ +P+PF W DI+GP
Sbjct: 643 -------PTIHQ---------------------PAELLSSIASPYPFMRWSMDIIGPMHP 674
Query: 1149 AKAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTS 1208
+K Q K ++V DYF+KWIEAE+ A+I A+V NF+ + I+CR G+P +V DNG+QF S
Sbjct: 675 SK-QKKLVLVLTDYFSKWIEAESYASIKDAQVENFVLKHILCRHGIPYEIVTDNGSQFIS 733
Query: 1209 SVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWA 1268
+ + FC + GI + ++ +PQ NGQAE+AN EL GVLW
Sbjct: 734 TRFQGFCDKWGIRLSKSTPRYPQGNGQAEAAN--------------------ELEGVLWL 773
Query: 1269 YNTTEQSSTKETPYRLTYGTDAMLSVEIENQSWRVARFNEND 1310
+ TT + +T+ETP+ YGT+ ++ E+ S R + END
Sbjct: 774 HRTTPRRATRETPFASIYGTECVIPAEMIVPSLRRSLSPEND 815
Score = 30.4 bits (67), Expect = 7.6
Identities = 13/27 (48%), Positives = 18/27 (66%)
Query: 263 YNVLLGRDTLNKVCAVISTAHLTVKYP 289
YN ++G LN+ AV ST HL +K+P
Sbjct: 30 YNAIIGTPWLNQFRAVASTYHLCLKFP 56
>At1g37200 hypothetical protein
Length = 1564
Score = 490 bits (1262), Expect = e-138
Identities = 368/1246 (29%), Positives = 577/1246 (45%), Gaps = 198/1246 (15%)
Query: 46 RWCEYHKALGHTTDNCWNLRREIDRLIKAGHL----ANFVKDTATPEVAKITQEDKGKGK 101
++C+YH GH+T+ C R + +LI AG +N +T P+ + Q K K +
Sbjct: 322 KYCKYHNKRGHSTEEC----RAVKKLIAAGGKTKKGSNPKVETPPPDEQEEEQTPKQKKR 377
Query: 102 EIVEELGD--PVGECSSIAGGFGGGAISSRARKRYVATVHSVQESNEGECWVNHSPIIFT 159
E E GD P I F + + R V T S + S F
Sbjct: 378 ERTLEGGDSPPPARRERIDLVFTELDLGGKTG-RSVTTPPSAPHKKNMRFPLRLSK--FC 434
Query: 160 PQDFAHVIPYDNDPIVVTIRVNNYVTKKVFLDQGSSADIIYGDAF-----DRLGLKESNL 214
H+ P D I+ ++ N + Q + G + ++ L ES
Sbjct: 435 RSATEHLSPKRIDFIMGGSQLCNDSINSIKTHQRKADSYTKGKSLMMGPDHKITLWESET 494
Query: 215 ----KPYKGTLVGFTGDRVSVRGYVEIPTAFGEGEFVKKFQVKYLVLACRANYNVLLGRD 270
KP+ +V R+ V Y G V K A +N +LGR
Sbjct: 495 TDLDKPHDDAIV----IRIDVGNYKLSRIMIDTGSSVDK-----------APFNAILGRP 539
Query: 271 TLNKVCAVISTAHLTVKYPACNGKVGILRVDQNAARECYLRSVALYGRKAAKESHRITEI 330
L+ + AV ST H +K+P+ G + ++ Q ++R CY+ S L K++ + +
Sbjct: 540 WLHAMKAVPSTYHQCIKFPSEKG-IAVIYGSQRSSRRCYMGSYELI-----KKAELVVLM 593
Query: 331 FPQEGFSLDPRDEADDFRPQPLEET-KQVQVKDKFLK----IGSGLTTEQEDRLITLLGD 385
+ + +D + P + + QV + + K +G L + IT L +
Sbjct: 594 IEDKLAEMKTVRSSDPSQCGPQKSSITQVYIDESDPKQCVGVGQDLDPAIREDFITFLKE 653
Query: 386 NLDLFAWTINDVPGIDPKVITHKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARF 445
N D FAW+ ++ GI +V +H+L + P P+ Q R++ E+ KAVQ E ++L+K
Sbjct: 654 NKDSFAWSSANLQGISLEVTSHELNVDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGS 713
Query: 446 IREVQYPTWLANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLS 505
IREV+YP WLAN V+VK NGKWR+C D+ LNK CPKDS+PLP++D+LV +G+ELLS
Sbjct: 714 IREVKYPDWLANPVVVKNKNGKWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLS 773
Query: 506 LMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGR 565
MDA+SGYNQI+M P D+E TAF+T+ CYK MPFGLKN GATYQRL++++F+ Q+G+
Sbjct: 774 FMDAFSGYNQILMRPDDQEKTAFITD----CYKVMPFGLKNTGATYQRLVNRMFADQLGK 829
Query: 566 NMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRG 625
ME+Y+DDM+VKSA DH L+E F L ++MKLNPEKCSFG+ G+FLG+++T RG
Sbjct: 830 TMELYIDDMLVKSAHEKDHLPQLRECFKILNKFEMKLNPEKCSFGVPSGEFLGYLVTQRG 889
Query: 626 IEVNPDKGRAILEMKSPTSVKEVQRLTGRMAALSRFLPMAGDKAAPFF----TCLKKNSK 681
IE NP + A ++M SP ++ R R + P + P + T + +
Sbjct: 890 IEANPKQIAAFIDMPSPKMARD--RPYRRPQQVIYRAPPDHEGQKPIYYVSKTLIDAETH 947
Query: 682 FQWTEECEQAFT----KLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQ 737
++ E+ A KL++ L + P+ + +T + + T+L
Sbjct: 948 YRAMEKLALAVVMSARKLRQYLQSHPI-------------VVMTSQPIRTIL-------- 986
Query: 738 KVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYFQSFQVKVKTDVPLRQVLQKPDL 797
T Q + L IE + I R SF+ +V D + L D
Sbjct: 987 ------HSTTQSSRLAKWAIELSEYDIEYRTR------TSFKAQVLADFVIELPLADLDG 1034
Query: 798 SGRLVSWSVELSEYDIQYEPRGQVTVQSLIDFVAELTPTEGEKTQGEWVLSVDGSSNNTG 857
+ W + + G Q + + +LT GE + + L + S+N +
Sbjct: 1035 TNSNKKWLLHVD---------GSSNRQGSGEGI-QLTSPTGEVIEQSFRLGFNASNNES- 1083
Query: 858 SGAGITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGLRLAIELGVQKLFIKGDSQLVV 917
EYEALI G++LA + ++ + DSQLV
Sbjct: 1084 ------------------------------EYEALIDGIKLAQGMRIRDIHAHSDSQLVT 1113
Query: 918 KQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPRGQNERADVLAKLASTGRLGNYQ 977
Q GEY+ KD ++ YLE+V+ L + + ++ +PRG+N D LA LAST +
Sbjct: 1114 SQFHGEYEAKDERMEAYLELVKTLTQQFESFELTRIPRGENTSTDALAALASTLDPFVKR 1173
Query: 978 TVIQETLPRPSIDLV---------------------------------------EIKLKV 998
+ E + PSIDL E KL+
Sbjct: 1174 IIPVEGIEHPSIDLTVKHAGMETSATCNFTRVTRSTTAAARALAAAAEAGAAEEEAKLRT 1233
Query: 999 VKSV--------NEGELPWMESIKTFLENPPKEDDLNTRTKRREASFYTLVDGELYRRGI 1050
+ + N+ +P ++ I+ + P D TR + +++ Y +++G L +R +
Sbjct: 1234 PEQLASYEPEPYNDWRIPIIDYIERGITPP---DKWETRKLKAQSARYCIMEGRLMKRSV 1290
Query: 1051 MSPMLKCVDTKDALGIMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCE 1110
P + C + +M +HEG C SH GR+L M DC+ + +C+
Sbjct: 1291 AGPYMVCTYGQQTKDLMKSMHEGQCGSHYSGRTLRC----------IMLADCIAHSLRCD 1340
Query: 1111 KCQVFSDLHKAPPEELTTMMAPWPFAMWGTDILGPFPVA--KAQMKYIIVAVDYFTKWIE 1168
KCQ + PPEE++++ +P+PF W D++ P + K ++K ++V DY TKWIE
Sbjct: 1341 KCQRHAPTLHQPPEEMSSISSPYPFMKWSMDVVRPMEASGGKKRLKNLLVLTDYSTKWIE 1400
Query: 1169 AEAVATITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTSSVTREF 1214
+A +T +V FL + IV R G+P +V DNG ++ F
Sbjct: 1401 GKAFQQVTEKQVEVFLLENIVYRHGIPYEIVTDNGLTSPREKSKHF 1446
Score = 52.8 bits (125), Expect = 1e-06
Identities = 35/96 (36%), Positives = 50/96 (51%), Gaps = 4/96 (4%)
Query: 1322 LPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDLVL*K---NTIPDKHNKLSPNWGG 1378
+ E + A I+ + + R +++ + R+ VG+LVL K NT KL NW G
Sbjct: 1467 IDERRDHAMIQVQNYQQPAVRYYNSNIKIRRFEVGELVLRKVFSNTRELNAGKLGTNWEG 1526
Query: 1379 PYRIIGDVGGEAYKLEQL-SGQKVPRTWNASHLKQY 1413
PYRI V YKL ++ +G R WNA HLK+Y
Sbjct: 1527 PYRITEVVRDGIYKLVKVFNGVPELRPWNAMHLKKY 1562
>At2g13330 F14O4.9
Length = 889
Score = 476 bits (1224), Expect = e-134
Identities = 246/546 (45%), Positives = 336/546 (61%), Gaps = 37/546 (6%)
Query: 465 NGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEE 524
NGKWR+C D+ LNK CPKDS+PLP++D+LV+ +E LS MDA+ GYNQI+M +E
Sbjct: 291 NGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQE 350
Query: 525 STAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDH 584
TAF+T++ Y YK MPFGLKNAG TYQRL++++F Q+G+ +EVY+DDM+VKSA DH
Sbjct: 351 KTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDH 410
Query: 585 GGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTS 644
L+E F L ++MKLNPEKCSF +Q +FL +++T RGIE NP + A +EM SP
Sbjct: 411 VPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSPKM 470
Query: 645 VKEVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPV 704
+EVQRLTGR+ AL+ F+ + +K PF+ L+K +F W ++CEQAF +LK L P+
Sbjct: 471 AREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTEPPI 530
Query: 705 LSKPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAI 764
L+KP G PL LY + R +EK AL +
Sbjct: 531 LAKPEKGEPLYLY------------------------------TNRDKRCPAMEKLALTV 560
Query: 765 LKTARRLRPYFQSFQVKVKTDVPLRQVLQKPDLSGRLVSWSVELSEYDIQYEPRGQVTVQ 824
+ + R+LR YFQS + V T P+R +L SGRL W++EL EYDI+Y R + Q
Sbjct: 561 VMSVRKLRLYFQSHPIVVMTSQPIRTILHSLTQSGRLAKWAIELREYDIEYRTRTSLKAQ 620
Query: 825 SLIDFVAE--LTPTEGEKTQGEWVLSVDGSSNNTGSGAGITIESPDKMIIEQSLKFEFKA 882
L DFV + L +G + +W+L VDGSSN GSG GI + SP +IEQSL+ F A
Sbjct: 621 VLADFVIKLPLADLDGTNSNKKWLLHVDGSSNRQGSGVGIQLTSPTGEVIEQSLQLGFNA 680
Query: 883 SNNQSEYEALIAGLRLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRRLM 942
SNN+SEYEALIAG++LA E G++++ DSQLV Q GEY+ KD ++ YLE+V+ L
Sbjct: 681 SNNESEYEALIAGIKLAQEKGIREIHAYSDSQLVTSQFHGEYEAKDERMEAYLELVKTLA 740
Query: 943 MEVKEIKIEHVPRGQNERADVLAKLASTGRLGNYQTVIQETLPRPSIDLVEIKLKVVKSV 1002
+ + K+ +PRG+N AD LA LAST ++ +P I+ I L V +
Sbjct: 741 QQFESFKLTRIPRGENTSADTLAALASTS-----DPFVKRIIPVEGIEHTSIDLTVKHAG 795
Query: 1003 NEGELP 1008
E E P
Sbjct: 796 MEPEAP 801
Score = 92.8 bits (229), Expect = 1e-18
Identities = 76/246 (30%), Positives = 119/246 (47%), Gaps = 32/246 (13%)
Query: 168 PYDNDPIVVTIRVNNYVTKKVFLDQGSSADIIYGDAFDRLGLKESNLKPYKGTLVGFTGD 227
P+D D +V+ I V NY + +D GSS D+++ DAF R G +S L+ K L GF GD
Sbjct: 60 PHD-DALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTGFAGD 118
Query: 228 RVSVRGYVEIPT-AFGEGEFVKKFQVKYLVLACRANYNVLLGRDTLNKVCAVISTAHLTV 286
G +++PT A G + +LV+ +A +N +LGR L+ + AV ST H +
Sbjct: 119 TTFSIGTIQLPTIARGVRQL-----TNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQCI 173
Query: 287 KYPACNGKVGILRVDQNAARECYLRS---------VALYGRKAAKESHRITEIFP-QEGF 336
K+P+ G + ++ Q ++R+CY+ S V L E + + P Q G
Sbjct: 174 KFPSYKG-IAVVYGSQRSSRKCYMGSYEDIKKADPVVLMIEDELAEMKTVRSLDPSQRGT 232
Query: 337 --SLDPRDEADDFRPQPLEETKQVQVKDKFLKIGSGLTTEQEDRLITLLGDNLDLFAWTI 394
SL + D+ P+ + + IG L + LIT L +N D FAW+
Sbjct: 233 RKSLITQVCIDESDPK------------RCVGIGHDLDLTVREDLITFLKENKDSFAWSS 280
Query: 395 NDVPGI 400
++ GI
Sbjct: 281 ANLQGI 286
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 383 bits (984), Expect = e-106
Identities = 188/358 (52%), Positives = 256/358 (70%), Gaps = 2/358 (0%)
Query: 423 RRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKWRMCTDYTSLNKVCP 482
R++ E+ KAV E +KL+K IREVQYP WLAN V+VKK NGK R+C D+T LNK CP
Sbjct: 470 RKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACP 529
Query: 483 KDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPF 542
KDS+PLP++D+LV+ +GNELLS MDA+SGYNQIMM+P D+E T F+T++ YCYK MPF
Sbjct: 530 KDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPF 589
Query: 543 GLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKL 602
GL+NAGATY RL++K+FS+ VG+ MEVY+DDM++KS + DH L+E F L YQMKL
Sbjct: 590 GLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKL 649
Query: 603 NPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGRMAALSRFL 662
NP KC+FG+ G+FLG+++T RGIE NP++ A L M SP + KEVQRLTGR+AAL+RF+
Sbjct: 650 NPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFI 709
Query: 663 PMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTD 722
+ DK+ PF+ LK N +F W E+CE+AF +LK L + PVL KP L LY++V++
Sbjct: 710 SRSTDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSN 769
Query: 723 KAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYFQSFQV 780
AVS VL++E+ +QK IY++ + Y+ + A L + L F+ F++
Sbjct: 770 HAVSGVLIREDRGEQKPIYYIIVNQFNGD--YEAKDSRMEAYLHVVKDLAKNFRKFEL 825
Score = 261 bits (668), Expect = 2e-69
Identities = 174/571 (30%), Positives = 271/571 (46%), Gaps = 118/571 (20%)
Query: 893 IAGLRLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEH 952
++G+ + + G QK ++V Q G+Y+ KD ++ YL VV+ L ++ ++
Sbjct: 772 VSGVLIREDRGEQKPIY----YIIVNQFNGDYEAKDSRMEAYLHVVKDLAKNFRKFELIR 827
Query: 953 VPRGQNERADVLAKLASTG----------RLGNYQTVIQE-------------------- 982
+PRGQN AD LA LAST + +++ +E
Sbjct: 828 IPRGQNTTADALAALASTSDPEVNSIIPVECISERSIKEEKEAFVVTRSRAAARDRGDTA 887
Query: 983 --------------TLPRPS------IDLVEIKLKVVKSVNEGELPWMESIKTFLENPPK 1022
T+P+P+ I+L+E + K E E PW++ P+
Sbjct: 888 VELPPVKRRKSKEATVPKPAVQENIGIELIEEVISDAKIEAETE-PWVDEDILAPNEHPE 946
Query: 1023 EDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDALGIMAEVHEGVCSSHIGGR 1082
+ LN+ LYRRG+ P L + +M+EVHEG+C SH GR
Sbjct: 947 PEALNS----------------LYRRGVSDPYLLSIFGPVVEIVMSEVHEGLCGSHSSGR 990
Query: 1083 SLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMWGTDI 1142
++A K+ + +YWPTM DC++Y ++C++CQ+ + L P E +++ AP+PF W DI
Sbjct: 991 AMAFKIKKMCYYWPTMITDCVKYAQRCKRCQLHAPLIHQPSELFSSISAPYPFMRWSMDI 1050
Query: 1143 LGPFPVAKAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRFGVPMALVMDN 1202
+GP + ++Y++V DYF+KWIEAEA+
Sbjct: 1051 IGPLHRSTRGVQYLLVLTDYFSKWIEAEAI------------------------------ 1080
Query: 1203 GTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEEL 1262
GI++ ++++ +PQ NGQAE+ANK IL LKK+L+ KG W +EL
Sbjct: 1081 ------------LLNWGIKVSYSTLRYPQGNGQAEAANKTILSNLKKRLNHLKGGWYDEL 1128
Query: 1263 PGVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEIENQSWR--VARFNENDNGENLIANLI 1320
VLWAY TT + ST ETP+ L YG A++ E+ R A NE +N L +L
Sbjct: 1129 QPVLWAYRTTPRRSTGETPFSLVYGMKAVVPAELNVPGLRRTEAPLNEKENSAMLDDSLD 1188
Query: 1321 MLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDLVL*K---NTIPDKHNKLSPNWG 1377
+ E + +A I+ + + AR +++KV R VGD VL + N + KL NW
Sbjct: 1189 TINERRDQALIQIQNYQHAAARYYNSKVKSRPFFVGDYVLKRVFDNKKEEGAGKLGINWE 1248
Query: 1378 GPYRIIGDVGGEAYKLEQLSGQKVPRTWNAS 1408
GPY + V Y+L+ L + V R WN +
Sbjct: 1249 GPYIVTEVVRNGVYRLKDLEDRPVQRPWNVN 1279
Score = 43.5 bits (101), Expect = 9e-04
Identities = 23/87 (26%), Positives = 46/87 (52%), Gaps = 1/87 (1%)
Query: 132 KRYVATVHSVQESNEGECWVNHSPIIFTPQDFAHVIPYDNDPIVVTIRVNNYVTKKVFLD 191
K++ VH +E E + I+F ++ H+ +D +VVT+ V N+ ++ +D
Sbjct: 386 KKHSRNVHLKLSLSE-EVDFQSTSILFDEKETQHLERSHDDALVVTLDVANFEVSRILID 444
Query: 192 QGSSADIIYGDAFDRLGLKESNLKPYK 218
GSS D+I+ +R+G+ +++ K
Sbjct: 445 TGSSVDLIFLSTLERMGISRADVNRRK 471
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 338 bits (866), Expect = 2e-92
Identities = 157/287 (54%), Positives = 214/287 (73%)
Query: 395 NDVPGIDPKVITHKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTW 454
+D+ GIDP+V H+L + P V Q R++ E+ KAV E +KL+ A I EV+YP W
Sbjct: 475 HDMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEW 534
Query: 455 LANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYN 514
LAN V+VKK N KWR+C D+T LNK CPKDS+PLP++D++V+ +GNELLS MDA+SGYN
Sbjct: 535 LANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYN 594
Query: 515 QIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDM 574
QI MH D+E T+F+ ++ YCYK MPFGLKN GA YQRL++++F+ Q+G+ MEVY+DDM
Sbjct: 595 QIPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDM 654
Query: 575 IVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGR 634
+VKS R++DH LK F+ L Y MKLNP KC FG+ G+FLG+++T RGIE NP + R
Sbjct: 655 LVKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIR 714
Query: 635 AILEMKSPTSVKEVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSK 681
AIL+++SP + KEVQRLTGR+A L+RF+ + DK+ PF+ L+ +K
Sbjct: 715 AILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLRSANK 761
Score = 80.5 bits (197), Expect = 6e-15
Identities = 70/306 (22%), Positives = 129/306 (41%), Gaps = 52/306 (16%)
Query: 43 RIWRWCEYHKALGHTTDNCWNLRREIDRLIKAGHL---------------------ANFV 81
R+ ++C++HK GH+T C +L+ + K G + N V
Sbjct: 182 RLDQYCDFHKRSGHSTAACRHLQSILLNKYKKGDIEVQHRQYKSHNNTYAARGGRDGNNV 241
Query: 82 KDTATPEVAKITQ-------------EDKGKGKEIVEELGD---PVGECSSIAGGFGGGA 125
P + + E K +E ++ D P + I GG
Sbjct: 242 FHRLGPHTGRQQEAPPANEEERHPDMEPPKKNRENDQQHNDAPVPRRRVNMIMGGLTACR 301
Query: 126 ISSRARKRYV----ATVHSVQESNEGECWVNHSPIIFTPQDFAHVIPYDNDPIVVTIRVN 181
S R+ K Y+ AT+ S + E +P+ FT +D V NDP+V+ + +
Sbjct: 302 DSFRSIKEYIKSGAATLWSSPATKE------MTPLTFTSEDLFGVDLPHNDPLVIELHIG 355
Query: 182 NYVTKKVFLDQGSSADIIYGDAFDRLGLKESNLKPYKGTLVGFTGDRVSVRGYVEIPTAF 241
++ +D GSS ++++ D ++ + + ++KP L GF G+ + G +++P
Sbjct: 356 ESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTNGTIKLPIYL 415
Query: 242 GEGEFVKKFQVKYLVLACRANYNVLLGRDTLNKVCAVISTAHLTVKYPACNGKVGILRVD 301
G KF +V+ YN++LG ++ + A+ S+ H +K P G + +R +
Sbjct: 416 GGAATWHKF----VVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG-IETIRGN 470
Query: 302 QNAARE 307
QN A +
Sbjct: 471 QNLAHD 476
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 322 bits (824), Expect = 1e-87
Identities = 244/805 (30%), Positives = 366/805 (45%), Gaps = 154/805 (19%)
Query: 671 PFFTCLKKNSK-FQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTVL 729
PF K SK F W E CE+AF +LK L+ VL+K G L LY+AV++ AV+
Sbjct: 617 PFLHFTAKISKGFIWNESCEEAFKQLKRYLSEPSVLAKREFGEQLFLYIAVSESAVT--- 673
Query: 730 LQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYFQSFQVKVKTDVPLR 789
G ++R ++ +K RP F VKT P+
Sbjct: 674 -------------------GVQVRVERSDK------------RPIFY-----VKTRYPMM 697
Query: 790 QVLQKPDLSGRLVSWSVELSEYDIQYEPRGQVTVQSLIDFVAELTPTEGEKTQGEWVLSV 849
+ L+ +V+ + +L Y ++ V + SL +PT+ + G+W + +
Sbjct: 698 E-----KLALAVVTAARKLRPY---FQSHPIVVLTSLPLRTILHSPTQSGRL-GKWAIEL 748
Query: 850 DGSSNNTGSGAGITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGLRLAIELGVQKLFI 909
E L+F + S N + + + + G K ++
Sbjct: 749 S----------------------EFDLEFRARTSLNST--------MDTSRQWGFIKTWV 778
Query: 910 KG---DSQLVVKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPRGQNERADVLAK 966
+ + GEY+ KD + YL +VR + ++ ++ +PR +N A+ LA
Sbjct: 779 RSRNLSDAPTANRYSGEYEAKDACMEAYLNLVREVSGRFEQFELTRIPRAENSAANALAA 838
Query: 967 LASTGRLGNYQTVIQETLPRPSIDLVEIKLKVVKS------------------------- 1001
LAST + + + ET+ +PSI L EI ++
Sbjct: 839 LASTFEVTLPRVIPVETISQPSIRLDEISFVTTRAMRRRLDAQSAENGLHQLGDDEEISD 898
Query: 1002 -------VNEGELP------------------WMESIKTFLENP--PKEDDLNTRTKRRE 1034
V LP W E I+ ++ N P E + K
Sbjct: 899 AVHPTEIVENQSLPDNHNAPLPDQPPHDWGADWREPIRDYILNGTLPAEKWAARKLKATC 958
Query: 1035 ASFYTLVDGELYRRGIMSPMLKCVDTKDALGIMAEVHEGVCSSHIGGRSLAVKVIRAGFY 1094
A F + + LYRR +P C+ + +M EVH+G C +H GGRSLA KV + G+Y
Sbjct: 959 ARF-CIANDILYRRIFSAPDAVCIFGEQTRTVMKEVHDGTCGNHTGGRSLAFKVRKYGYY 1017
Query: 1095 WPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMWGTDILGPFPVAKAQMK 1154
WPT+ DC Y +KCE+CQ + L P E LTT+ AP+PF W DI+GP V+
Sbjct: 1018 WPTLVADCEAYARKCEQCQKHAPLILQPAELLTTVSAPYPFMKWLMDIVGPLHVS----- 1072
Query: 1155 YIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTSSVTREF 1214
T+ +EA A + IT +V NF+W+ I+CR G+P +V DNG+QF S F
Sbjct: 1073 ---------TRGVEAAAYSNITHVQVWNFIWKDIICRHGLPYEIVTDNGSQFISEQFEVF 1123
Query: 1215 CAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQ 1274
C E I + ++ +PQ NGQAE+ NK I+ LKKKL+ KG W EL VLWA TT +
Sbjct: 1124 CEEWQIRLSHSTPRYPQGNGQAEAMNKTIISNLKKKLNAYKGAWFGELQNVLWAVRTTPR 1183
Query: 1275 SSTKETPYRLTYGTDAMLSVEIENQSWRVAR--FNENDNGENLIANLIMLPEEQREAHIR 1332
+T ETP+ L YG +A++ EI+ S R R NE +N E +I + + E + A R
Sbjct: 1184 RATDETPFSLIYGMEAVIPAEIKVPSARRIRNPQNETENNEMIIDVIDTIDERRNRALAR 1243
Query: 1333 NEAGKVKVARKFSTKVVPRKMRVGDLVL*K---NTIPDKHNKLSPNWGGPYRIIGDVGGE 1389
+ AR +++ V R VG LVL + N KL +W GPY+I V
Sbjct: 1244 MQNYHNAGARYYNSNVRNRSFEVGTLVLRRVQQNKAEKGAGKLGISWEGPYKITHVVRNG 1303
Query: 1390 AYKLEQLSGQKVPRTWNASHLKQYF 1414
Y+L + G+ V R WN+ HLK+++
Sbjct: 1304 VYRLINMEGKTVRRAWNSMHLKRFY 1328
Score = 180 bits (457), Expect = 5e-45
Identities = 81/158 (51%), Positives = 114/158 (71%)
Query: 390 FAWTINDVPGIDPKVITHKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREV 449
F + + GI +VI+H+L + P PV Q R++ ++ +AV +E +L++ IREV
Sbjct: 405 FPTPVGKMTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREV 464
Query: 450 QYPTWLANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDA 509
+YP WLAN V+VKK NGKWR+C D+T LNK C KD +PLP++D+LV+ +G+E+LS MDA
Sbjct: 465 KYPEWLANPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDA 524
Query: 510 YSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNA 547
+SGYNQI+M+P D+E T+F+T Y YK MPFGLKNA
Sbjct: 525 FSGYNQILMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
Score = 32.7 bits (73), Expect = 1.5
Identities = 30/82 (36%), Positives = 39/82 (46%), Gaps = 6/82 (7%)
Query: 215 KPYKGTLVGFTGDRVSVRGYVEIP-TAFGEGEFVKKFQVKYLVLACRANYNVLLGRDTLN 273
KP GT + G V +P TA G VK V++ V A YNV+LG L
Sbjct: 336 KPAAGTCTTMLCETSMSLGTVVLPVTAQG---VVK--MVEFTVFDRPAAYNVILGTPWLY 390
Query: 274 KVCAVISTAHLTVKYPACNGKV 295
++ V ST H VK+P GK+
Sbjct: 391 EMKVVPSTYHQCVKFPTPVGKM 412
>At4g03800
Length = 637
Score = 318 bits (815), Expect = 1e-86
Identities = 164/337 (48%), Positives = 228/337 (66%), Gaps = 21/337 (6%)
Query: 397 VPGIDPKVITHKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLA 456
+P IDP +I H+L + P P+ Q R++ E+ KAV + +KL+K IREVQYP W+A
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 457 NVVMVKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQI 516
V+VKK NGK R+C D+T LNK CPKDS+PLP++D+LV+ +GNELL+ MDA+ GYNQI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 517 MMHPSDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIV 576
MM+P D+E T+F+T++ GATYQ L++K+F++ + + MEV +DD +V
Sbjct: 441 MMNPEDQEKTSFITDR---------------GATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 577 KSARASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAI 636
KS + DH L E F+ L YQMKLN KC+FG+ G+FLG+++T RGIE NP++ A
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINAF 545
Query: 637 LEMKSPTSVKEVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLK 696
L+ S + KEVQRLTGR+A+ S DK+ PF+ LK N+ F W E+CE+AF +LK
Sbjct: 546 LKTPSLRNFKEVQRLTGRIASRST------DKSLPFYQILKGNNGFLWDEKCEEAFRQLK 599
Query: 697 ETLATLPVLSKPTPGVPLVLYLAVTDKAVSTVLLQEE 733
L T PVLSKP L LY+ V++ AVS VL++E+
Sbjct: 600 AYLTTPPVLSKPEADEKLYLYVFVSNHAVSGVLVRED 636
Score = 69.3 bits (168), Expect = 1e-11
Identities = 84/325 (25%), Positives = 134/325 (40%), Gaps = 47/325 (14%)
Query: 56 HTTDNCWNLRREIDRLIKAGHLA-----NFVKDTATPEVAKITQEDKGKGKEIVEELGDP 110
H T +C L+R + L +G L+ +FVK TQ K EE
Sbjct: 81 HLTKDCIVLKRYLAELWASGDLSKFNIDDFVKQYHEARDNSETQNSKRPRLTNEEEPRSS 140
Query: 111 VGECSSIAGGFGGGAISSRARKRYVATV---HSVQESNEGECWVNHSPIIFTPQDFAHVI 167
G+ + I GG S A K++ V S+ E +C S + F ++ H+
Sbjct: 141 KGKINVILGGSKLCHNSVSAIKKHRPNVLLKSSLSEEVNYQC----SSVSFDEEETRHIE 196
Query: 168 PYDNDPIVVTIRVNNYVTKKVFLDQGSSADIIYGDAFDRLGLKESNLKPYKGTLVGFTGD 227
+D +++T+ V N+ ++ +D GSS D+I+ L P LV FT +
Sbjct: 197 RPHDDALIITLDVANFKISRILVDTGSSVDLIF-------------LGP-PSPLVAFTSE 242
Query: 228 RVSVRGYVEIPTAFGEGEFVKKFQVKYLVLACRANYNVLLGRDTLNKVCAVISTAHLTVK 287
G +++P G+ + V ++V A YN++LG + ++ AV ST H +K
Sbjct: 243 SAMSLGTIKLPVLAGKMSKI----VDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWLK 298
Query: 288 YPACNGKVGILRVDQNAARECYLRSVALYGRKAAKESHRITEIFPQEGFSLDPRDE--AD 345
+P NG I + + Y+ + I E ++DPR +
Sbjct: 299 FPTSNGVETIWGDLEGSRTSAYMPDID-------------PSIICHE-LNVDPRFKPLKQ 344
Query: 346 DFRPQPLEETKQVQVK-DKFLKIGS 369
R +E K V K DK LKIGS
Sbjct: 345 KRRKLGVERAKAVNGKIDKLLKIGS 369
>At2g13230 pseudogene
Length = 353
Score = 254 bits (649), Expect = 2e-67
Identities = 136/361 (37%), Positives = 215/361 (58%), Gaps = 22/361 (6%)
Query: 931 LSKYLEVVRRLMMEVKEIKIEHVPRGQNERADVLAKLASTGRLGNYQTVIQETLPRPSID 990
+ YL +V+ + E K+ +PRG+N AD LA LAST L + + E + +PSI+
Sbjct: 1 MEAYLALVQDHAKQFHEFKLTRIPRGENTSADALAALASTSDLNMRRVIPVEFIEKPSIE 60
Query: 991 LVEIK----LKVVKSVNEG---------ELPWMESIKTFL-ENPPKEDDLNTRTKRREAS 1036
L +++ ++VV + + W+E I+ F+ + + D R + EA+
Sbjct: 61 LDKVEHAFPIQVVAGYEDASDNSPSLGCDSEWIEPIRNFISKRKVRLDKWEARKLKAEAA 120
Query: 1037 FYTLVDGELYRRGIMSPMLKCVDTKDALGIMAEVHEGVCSSHIGGRSLAVKVIRAGFYWP 1096
+ LV+ +L++ + P++ CV+ + A +M EVH G C ++ GGR+LA+K+ R G++WP
Sbjct: 121 RFVLVEEKLFKWRLSGPLMTCVEKEAARKVMKEVHGGSCGNNSGGRALAIKIKRLGYFWP 180
Query: 1097 TMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMWGTDILGPFPVAKAQMKYI 1156
TM KDC EKCQ + P E L+++ +P+P W DI+GP +K Q K +
Sbjct: 181 TMIKDC-------EKCQRNAPTIHLPAELLSSIASPYPLMRWSMDIIGPMHASK-QKKLV 232
Query: 1157 IVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTSSVTREFCA 1216
+V DYF+K EAE+ A+I A+V +F+W+ I+CR GVP +V DNG+QF S+ + FC
Sbjct: 233 LVLTDYFSKLREAESYASIKEAQVESFMWKHILCRHGVPYEIVTDNGSQFISTRFQGFCD 292
Query: 1217 EMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSS 1276
+ GI + ++ + Q NGQA++ANK IL GLKK+L KG W +EL GVLW++ TT + +
Sbjct: 293 KWGIRLSKSTPRYLQGNGQADAANKTILDGLKKRLTAKKGSWVDELEGVLWSHRTTPRRA 352
Query: 1277 T 1277
T
Sbjct: 353 T 353
>At2g10780 pseudogene
Length = 1611
Score = 232 bits (592), Expect = 1e-60
Identities = 159/535 (29%), Positives = 267/535 (49%), Gaps = 27/535 (5%)
Query: 370 GLTTEQEDRLITLLGDNLDLFAWTINDVPGIDP-KVITHKLAIRPGATPVIQPMRRMSEE 428
G + E +D I ++ + D+FA V G+ P + + + PG TP+ + RM+
Sbjct: 617 GASAELKD--IPIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPA 670
Query: 429 KHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPL 488
+ ++ + E+L+ FIR P W A V+ VKK +G +R+C DY LNKV K+ YPL
Sbjct: 671 EMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPL 729
Query: 489 PNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAG 548
P +D+L+D G + S +D SGY+QI + P+D TAF T ++ + MPFGL NA
Sbjct: 730 PRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAP 789
Query: 549 ATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCS 608
A + ++M+ +F + + ++++D++V S H L+ ++LR +++ KCS
Sbjct: 790 AAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCS 849
Query: 609 FGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGRMAALSRFLPMAGDK 668
F + FLG +++ +G+ V+P+K R+I E P + E++ G RF+
Sbjct: 850 FWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASM 909
Query: 669 AAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTV 728
A P K++ F W++ECE++F +LK L PVL P G P +Y + + V
Sbjct: 910 AQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCV 969
Query: 729 LLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYFQSFQVKVKTD-VP 787
L+Q K VI + S L+ E Y + A++ + R Y +V++ TD
Sbjct: 970 LMQ----KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKS 1025
Query: 788 LRQVLQKPDLSGRLVSWSVELSEY--DIQYEPRGQVTVQSLIDFVAELTPTEGEKTQGEW 845
L+ + +P+L+ R W +++Y DI Y P V + + E E++Q +
Sbjct: 1026 LKYIFTQPELNLRQRRWMELVADYNLDIAYHPGKANQVADALS--RRRSEVEAERSQVDL 1083
Query: 846 V-----LSVDGSSNNT---GSGAGITIESPDKMIIEQSLKFEFK--ASNNQSEYE 890
V L V+ S G GA + ++ + Q E K A NN++EY+
Sbjct: 1084 VNMMGTLHVNALSKEVEPLGLGAADQADLLSRIRLAQERDEEIKGWAQNNKTEYQ 1138
Score = 106 bits (264), Expect = 1e-22
Identities = 113/477 (23%), Positives = 200/477 (41%), Gaps = 43/477 (9%)
Query: 948 IKIEHVPRGQNERADVLAKLASTGRLGNYQTVIQETLPRPSIDLVEIKLKVVKSVNEGEL 1007
+ I + P N+ AD L++ S Q + + ++ + K V+ + G
Sbjct: 1051 LDIAYHPGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALS---KEVEPLGLGAA 1107
Query: 1008 PWMESIKTFLENPPKEDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDALG-- 1065
+ + +++++ + + + T +G + G + CV AL
Sbjct: 1108 DQADLLSRIRLAQERDEEIKGWAQNNKTEYQTSNNGTIVVNGRV-----CVPNDRALKEE 1162
Query: 1066 IMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEE 1125
I+ E H+ S H G + + ++ ++W MKKD +V KC CQ+ H+ P
Sbjct: 1163 ILREAHQSKFSIHPGSNKM-YRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGL 1221
Query: 1126 LTTMMAP-WPFAMWGTDILGPFPVA-KAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNF 1183
L + P W + D + P K++ + V VD TK A++ A++
Sbjct: 1222 LQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAE 1281
Query: 1184 LW-QRIVCRFGVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKV 1242
+ IV G+P+++V D T+FTS + F +G + ++ HPQT+ Q+E +
Sbjct: 1282 KYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQT 1341
Query: 1243 ILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYG----TDAMLSVEIEN 1298
+ L+ + + G W + L V +AYN + Q+S +PY YG T + E
Sbjct: 1342 LEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGER 1401
Query: 1299 QSWRVARFNENDNGENLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDL 1358
+ + +E + L + +EA R ++ K ++ + +VGDL
Sbjct: 1402 RLFGPTIVDETTE------RMKFLKIKLKEAQDRQKSYANKRRKEL-------EFQVGDL 1448
Query: 1359 VL*KNTIP------DKHNKLSPNWGGPYRIIGDVGGEAYKLEQLSGQKVPRTWNASH 1409
V K KLSP + GPY++I VG AYKL+ +P NA H
Sbjct: 1449 VYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLD------LPPKLNAFH 1499
>At3g11970 hypothetical protein
Length = 1499
Score = 219 bits (559), Expect = 7e-57
Identities = 132/413 (31%), Positives = 216/413 (51%), Gaps = 7/413 (1%)
Query: 407 HKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANG 466
HK+ + G+ PV Q R S + + E L+ ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 467 KWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEEST 526
WR+C DY LN + KDS+P+P ++ L+D G + S +D +GY+Q+ M P D + T
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 527 AFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGG 586
AF T+ ++ Y MPFGL NA AT+Q LM+ IF + + + V+ DD++V S+ +H
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 587 DLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVK 646
LK+ F+ +R ++ KC+F + ++LG ++++GIE +P K +A+ E PT++K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 647 EVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLS 706
+++ G RF+ G A P L K F+WT +QAF LK L PVLS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGPLH-ALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 707 KPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILK 766
P V+ + + VL+QE + ++S L+G +L EK LA++
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQE----GHPLAYISRQLKGKQLHLSIYEKELLAVIF 944
Query: 767 TARRLRPYFQSFQVKVKTDV-PLRQVLQKPDLSGRLVSWSVELSEYDIQYEPR 818
R+ R Y +KTD L+ +L++ + W +L E+D + + R
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQYR 997
Score = 107 bits (268), Expect = 4e-23
Identities = 96/360 (26%), Positives = 156/360 (42%), Gaps = 30/360 (8%)
Query: 1066 IMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEE 1125
I+ +H H GR + + ++ FYW M KD Y++ C CQ A P
Sbjct: 1084 ILLWLHGSGVGGH-SGRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGL 1142
Query: 1126 LTTMMAPWPFAMWGT---DILGPFPVAKAQMKYIIVAVDYFTKWIEAEAVA-TITAAKVR 1181
L + P P +W D + PV+ + I+V VD +K A++ +A V
Sbjct: 1143 LQPL--PIPDTIWSEVSMDFIEGLPVSGGKT-VIMVVVDRLSKAAHFIALSHPYSALTVA 1199
Query: 1182 NFLWQRIVCRFGVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANK 1241
+ + G P ++V D FTS REF G+ ++ S HPQ++GQ E N+
Sbjct: 1200 HAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNR 1259
Query: 1242 VILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEI---EN 1298
+ L+ + LW++ L + YNT SS++ TP+ + YG + + E+
Sbjct: 1260 CLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGES 1319
Query: 1299 QSWRVARFNENDNGENLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDL 1358
+ VAR + L ++ + R ++ A + + R+F +GD
Sbjct: 1320 KVAVVARSLQEREDMLLFLKFHLMRAQHR---MKQFADQHRTEREF---------EIGDY 1367
Query: 1359 VL*K-------NTIPDKHNKLSPNWGGPYRIIGDVGGEAYKLEQLSGQKVPRTWNASHLK 1411
V K + + + KLSP + GPY+II G AYKL S +V ++ S LK
Sbjct: 1368 VYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSYSQVHPVFHVSQLK 1427
>At1g36590 hypothetical protein
Length = 1499
Score = 219 bits (559), Expect = 7e-57
Identities = 132/413 (31%), Positives = 216/413 (51%), Gaps = 7/413 (1%)
Query: 407 HKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANG 466
HK+ + G+ PV Q R S + + E L+ ++ P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 467 KWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEEST 526
WR+C DY LN + KDS+P+P ++ L+D G + S +D +GY+Q+ M P D + T
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 527 AFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGG 586
AF T+ ++ Y MPFGL NA AT+Q LM+ IF + + + V+ DD++V S+ +H
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 587 DLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVK 646
LK+ F+ +R ++ KC+F + ++LG ++++GIE +P K +A+ E PT++K
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLK 829
Query: 647 EVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLS 706
+++ G RF+ G A P L K F+WT +QAF LK L PVLS
Sbjct: 830 QLRGFLGLAGYYRRFVRSFGVIAGPLH-ALTKTDAFEWTAVAQQAFEDLKAALCQAPVLS 888
Query: 707 KPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILK 766
P V+ + + VL+QE + ++S L+G +L EK LA++
Sbjct: 889 LPLFDKQFVVETDACGQGIGAVLMQE----GHPLAYISRQLKGKQLHLSIYEKELLAVIF 944
Query: 767 TARRLRPYFQSFQVKVKTDV-PLRQVLQKPDLSGRLVSWSVELSEYDIQYEPR 818
R+ R Y +KTD L+ +L++ + W +L E+D + + R
Sbjct: 945 AVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDYEIQYR 997
Score = 102 bits (253), Expect = 2e-21
Identities = 95/360 (26%), Positives = 154/360 (42%), Gaps = 30/360 (8%)
Query: 1066 IMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEE 1125
I+ +H H GR + + ++ FY M KD Y++ C CQ A P
Sbjct: 1084 ILLWLHGSGVGGH-SGRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGL 1142
Query: 1126 LTTMMAPWPFAMWGT---DILGPFPVAKAQMKYIIVAVDYFTKWIEAEAVA-TITAAKVR 1181
L + P P +W D + PV+ + I+V VD +K A++ +A V
Sbjct: 1143 LQPL--PIPDTIWSEVSMDFIEGLPVSGGKT-VIMVVVDRLSKAAHFIALSHPYSALTVA 1199
Query: 1182 NFLWQRIVCRFGVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANK 1241
+ G P ++V D FTS REF G+ ++ S HPQ++GQ E N+
Sbjct: 1200 QAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNR 1259
Query: 1242 VILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEI---EN 1298
+ L+ + LW++ L + YNT SS++ TP+ + YG + + E+
Sbjct: 1260 CLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGES 1319
Query: 1299 QSWRVARFNENDNGENLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDL 1358
+ VAR + L ++ + R ++ A + + R+F +GD
Sbjct: 1320 KVAVVARSLQEREDMLLFLKFHLMRAQHR---MKQFADQHRTEREF---------EIGDY 1367
Query: 1359 VL*K-------NTIPDKHNKLSPNWGGPYRIIGDVGGEAYKLEQLSGQKVPRTWNASHLK 1411
V K + + + KLSP + GPY+II G AYKL S +V ++ S LK
Sbjct: 1368 VYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSYSQVHPVFHVSQLK 1427
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 219 bits (557), Expect = 1e-56
Identities = 132/422 (31%), Positives = 219/422 (51%), Gaps = 9/422 (2%)
Query: 409 LAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKW 468
+ + PG P+ + RM+ + ++ + + L+ FIR P W A V+ VKK +G +
Sbjct: 483 IELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 541
Query: 469 RMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAF 528
R+C DY LN+V K+ YPLP +D+L+D G S +D SGY+QI + +D TAF
Sbjct: 542 RLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 601
Query: 529 MTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDL 588
T ++ + MPFGL NA A + RLM+ +F + + + +++DD++V S + L
Sbjct: 602 RTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHL 661
Query: 589 KEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEV 648
+ ++LR ++ KCSF + FLG ++++ G+ V+P+K AI + PT+ E+
Sbjct: 662 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 721
Query: 649 QRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKP 708
+ G RF+ A P K+ F W++ECE+ F LKE L + PVL+ P
Sbjct: 722 RSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALP 781
Query: 709 TPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTA 768
G P ++Y + + VL+Q KVI + S L E Y + A++
Sbjct: 782 EHGQPYMVYTDASRVGLGCVLMQH----GKVIAYASRQLMKHEGNYPTHDLEMAAVIFAL 837
Query: 769 RRLRPYFQSFQVKVKTD-VPLRQVLQKPDLSGRLVSWSVELSEYDIQ--YEP-RGQVTVQ 824
+ R Y +V+V TD L+ + +P+L+ R W +++YD++ Y P + V V
Sbjct: 838 KIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAYHPGKANVVVD 897
Query: 825 SL 826
+L
Sbjct: 898 AL 899
Score = 33.9 bits (76), Expect = 0.68
Identities = 36/144 (25%), Positives = 60/144 (41%), Gaps = 7/144 (4%)
Query: 1093 FYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAP-WPFAMWGTDILGPFPVAKA 1151
+ W MK D +V +C+ CQ+ H+ L ++ P W + D++ V++
Sbjct: 969 YQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGLRVSRT 1028
Query: 1152 QMKYIIVAVDYFTKWIEAEAV-ATITAAKVRNFLWQRIVCRFGVPMAL--VMDNGTQFTS 1208
+ I V VD TK A+ T AA + IV GVP+ + D +
Sbjct: 1029 K-DAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMKEAQDRQRSYAD 1087
Query: 1209 SVTREFCAEMG--IEMRFASVEHP 1230
RE E+G + ++ A + P
Sbjct: 1088 KRRRELEFEVGDRVYLKMAMLRGP 1111
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 216 bits (549), Expect = 1e-55
Identities = 128/412 (31%), Positives = 213/412 (51%), Gaps = 8/412 (1%)
Query: 409 LAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKW 468
+ + PG P+ + RM+ + ++ + E L+ FIR P W A V+ VKK +G +
Sbjct: 509 IELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 567
Query: 469 RMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAF 528
R+C DY LN V K+ YPLP +D+L+D G S +D SGY+QI + +D TAF
Sbjct: 568 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 627
Query: 529 MTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDL 588
T ++ + MPF L NA A + RLM+ +F + + + +++DD++V S +H L
Sbjct: 628 RTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHL 687
Query: 589 KEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEV 648
+ ++LR ++ KCSF + FLG ++++ G+ V+P+K AI + PT+ E+
Sbjct: 688 RRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 747
Query: 649 QRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKP 708
+ RF+ A P K+ F W+ ECE+ F LKE L + PVL+ P
Sbjct: 748 RSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 807
Query: 709 TPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTA 768
G P ++Y + + VL+Q + KVI + S L+ E Y + ++
Sbjct: 808 EHGQPYMVYTDASRVGLGCVLMQ----RGKVIAYASRQLRKHEGNYPTHDLEMAVVIFAL 863
Query: 769 RRLRPYFQSFQVKVKTD-VPLRQVLQKPDLSGRLVSWSVELSEYDIQ--YEP 817
+ R Y +V+V TD L+ + +P+L+ R + W +++YD++ Y P
Sbjct: 864 KIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADYDLEIAYHP 915
Score = 105 bits (261), Expect = 2e-22
Identities = 156/686 (22%), Positives = 286/686 (40%), Gaps = 110/686 (16%)
Query: 737 QKVIYFVSHTL--QGAELRYQKIEKAA-------LAILKTARRLRPYFQSFQVKVKTDVP 787
Q+ I F+ H + +G + +KIE +++ RL Y++ F VK
Sbjct: 710 QREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRF---VKGFAS 766
Query: 788 LRQVLQKPDLSGRLVS--WSVELSEYDIQYEPRGQVTVQSLIDFVAELT-PTEGEKTQGE 844
+ Q + K L+G+ V WS E E G V+++ ++ L P G+
Sbjct: 767 MAQPMTK--LTGKDVPFVWSPECEE--------GFVSLKEMLTSTPVLALPEHGQP---- 812
Query: 845 WVLSVDGSSNNTGSGAGITIESPDKMIIEQSLKFEFKASNNQSEYEALIAGLRLAIELGV 904
+++ D S G G + K+I S + K N ++ +A + A+++
Sbjct: 813 YMVYTDASR----VGLGCVLMQRGKVIAYASRQLR-KHEGNYPTHDLEMAVVIFALKIWR 867
Query: 905 QKLFIKGDSQLVVKQVKGEYQVKDPQLS----KYLEVVRRLMMEVKEIKIEHVPRGQNER 960
L+ G Q+ +Y P+L+ +++E+V +++I + P N
Sbjct: 868 SYLY-GGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADY-----DLEIAYHPGKANVV 921
Query: 961 ADVLAKLASTGRLGNYQTVIQETLPRPSIDLVEIKLKVVKSVNEGELPWMESIKTFLENP 1020
AD L++ G + + + +V + +++V+ +L +
Sbjct: 922 ADALSRKRVGAAPGQSVEALVSEIGALRLCVVAREPLGLEAVDRADLLTRARLAQ----- 976
Query: 1021 PKEDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDALG--IMAEVHEGVCSSH 1078
K++ L +K + + +G ++ G + CV + L I++E H + S H
Sbjct: 977 EKDEGLIAASKAEGSEYQFAANGTIFVYGRV-----CVPKDEELRREILSEAHASMFSIH 1031
Query: 1079 IGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMW 1138
G + + ++ + W MK+D +V +C+ CQ+ H+ P A+W
Sbjct: 1032 PGATKM-YRDLKRYYQWVGMKRDVANWVAECDVCQLVKAEHQVPD------------AIW 1078
Query: 1139 GTDILGPFPVAKAQMKYIIVAVDYFTKWIEAEAVA-TITAAKVRNFLWQRIVCRFGVPMA 1197
V +D TK A+ T AA + IV GVP++
Sbjct: 1079 -------------------VIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVS 1119
Query: 1198 LVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGL 1257
+V D ++FT + R F A+MG +++ ++ HPQT+GQ+E + + L+ + + G
Sbjct: 1120 IVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGH 1179
Query: 1258 WAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAMLSV---EIENQSWRVARFNENDNGEN 1314
WA+ L V +AYN + Q+S P+ YG + ++E +S A D +
Sbjct: 1180 WADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPLRWTQVEERSIYGA-----DYVQE 1234
Query: 1315 LIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDLVL*KNTI---PDK--- 1368
+ +L +EA R + K R+ + VGD V K + P++
Sbjct: 1235 TTERIRVLKLNMKEAQARQRSYADKRRREL-------EFEVGDRVYLKMAMLRGPNRSIL 1287
Query: 1369 HNKLSPNWGGPYRIIGDVGGEAYKLE 1394
KLSP + GP+RI+ VG AY+LE
Sbjct: 1288 ETKLSPRYMGPFRIVERVGPVAYRLE 1313
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 210 bits (534), Expect = 5e-54
Identities = 128/406 (31%), Positives = 210/406 (51%), Gaps = 12/406 (2%)
Query: 370 GLTTEQEDRLITLLGDNLDLFAWTINDVPGIDP-KVITHKLAIRPGATPVIQPMRRMSEE 428
G + E +D LI + + D+FA V G+ P + + + PG TP+ + RM+
Sbjct: 112 GASAELKDILI--VNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPA 165
Query: 429 KHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPL 488
+ ++ + E+L+ FIR P W A V+ VKK +G +R+C DY LNKV K+ YPL
Sbjct: 166 EMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPL 224
Query: 489 PNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAG 548
P +D+L+D G + S +D SGY+QI + P+D TAF T ++ + MPFGL NA
Sbjct: 225 PRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAP 284
Query: 549 ATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCS 608
A + ++M+ +F + + +++DD++V S H L+ ++LR +++ K S
Sbjct: 285 AAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHELFAKLSKFS 344
Query: 609 FGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGRMAALSRFLPMAGDK 668
F + FLG +++ +G+ V+P+K R+I E P + E++ G RF+
Sbjct: 345 FWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASM 404
Query: 669 AAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTV 728
A P K++ F W++ECE++F +LK L PVL P G P +Y + + V
Sbjct: 405 AQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGEPYTVYTDASIVGLGCV 464
Query: 729 LLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPY 774
L+Q K VI + S L+ E Y + A++ + R Y
Sbjct: 465 LMQ----KGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSY 506
Score = 103 bits (258), Expect = 5e-22
Identities = 96/388 (24%), Positives = 171/388 (43%), Gaps = 34/388 (8%)
Query: 1022 KEDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDALG--IMAEVHEGVCSSHI 1079
+++++ T + + T +G + G + CV AL I+ E H+ S H
Sbjct: 514 RDEEIKGWTLNNKTEYQTSNNGTIVVNGRV-----CVPNDRALKEEILREAHQSKFSIHP 568
Query: 1080 GGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTM-MAPWPFAMW 1138
G + + ++ ++W M+KD +V KC CQ+ H+ P L + ++ W +
Sbjct: 569 GSNKM-YRDLKRYYHWVGMRKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHI 627
Query: 1139 GTDILGPFPVA-KAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRF-GVPM 1196
D + P K++ + V VD TK A++ A++ + + R G+P+
Sbjct: 628 TMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPV 687
Query: 1197 ALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKG 1256
++V D T+FTS F +G + ++ HPQT+GQ+E + + L+ + + G
Sbjct: 688 SIVSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGG 747
Query: 1257 LWAEELPGVLWAYNTTEQSSTKETPYRLTYG----TDAMLSVEIENQSWRVARFNENDNG 1312
W + L + +AYN + Q+S +PY YG T + E + + +E
Sbjct: 748 NWEKYLRLIEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTE- 806
Query: 1313 ENLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKMRVGDLVL*KNTIP------ 1366
+ L + +EA R ++ K ++ + +VGDLV K
Sbjct: 807 -----RMKFLKIKLKEAQDRQKSYANKRRKEL-------EFQVGDLVYLKAMTYKGAGRF 854
Query: 1367 DKHNKLSPNWGGPYRIIGDVGGEAYKLE 1394
KLSP + GPY++I VG AYKL+
Sbjct: 855 TSRKKLSPRYVGPYKVIERVGAVAYKLD 882
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 209 bits (532), Expect = 9e-54
Identities = 134/467 (28%), Positives = 217/467 (45%), Gaps = 47/467 (10%)
Query: 367 IGSGLTTEQEDRLITLLGDNLDLFAWTINDVPGIDPKVITHKLAIRPGATPVIQPMRRMS 426
+ LT +Q + LIT L ++++D+ GI P + TH++ + + I+P RR++
Sbjct: 849 VNDELTADQVNLLITELMKYRKAIGYSLDDIKGISPTLCTHRIHLENESYSSIEPQRRLN 908
Query: 427 EEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKW------------------ 468
+ V+ E KL+ A I + TW++ V V K G
Sbjct: 909 PNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGH 968
Query: 469 RMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAF 528
RMC +Y LN K+ +PLP +D +++ + + +D+YSG+ QI +HP+D+ T F
Sbjct: 969 RMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTF 1028
Query: 529 MTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDL 588
+ YK MPFGL NA AT+QR M IFS + +EV++DD V + S +L
Sbjct: 1029 TCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNL 1088
Query: 589 KEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEV 648
+ + LN EKC F ++ G LG ++ GIEV+ K +++++ P +VK++
Sbjct: 1089 CRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDI 1148
Query: 649 QRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKP 708
+ G F+ A P L K ++F + +EC AF +KE L T P++ P
Sbjct: 1149 RSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEALITAPIVQAP 1208
Query: 709 TPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTA 768
P F T+ A++RY EK LA++
Sbjct: 1209 NWDFP----------------------------FEIITMDDAQVRYATTEKELLAVVFAF 1240
Query: 769 RRLRPYFQSFQVKVKTD-VPLRQVLQKPDLSGRLVSWSVELSEYDIQ 814
+ R Y +V + TD LR + K D RL+ W + L E+D++
Sbjct: 1241 EKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFDME 1287
Score = 167 bits (424), Expect = 3e-41
Identities = 135/507 (26%), Positives = 226/507 (43%), Gaps = 74/507 (14%)
Query: 924 YQVKD--PQLSKYLEVVRRLMMEVKEIKIEHVPRGQNERADVLAKLASTGRLGNYQTVIQ 981
Y KD P+L +++ +++ ME+ + K +N AD L+++ + +I
Sbjct: 1265 YAKKDTKPRLLRWILLLQEFDMEIVDKK-----GIENGVADHLSRMRIED-----EVLID 1314
Query: 982 ETLPRPSIDLVEIKLKVVKSVNEGELPWMESIKTFLENPPKEDDLNTRTKRR-------- 1033
+++P E +L ++ +NE +LPW +L + + +L++ K++
Sbjct: 1315 DSMP-------EEQLMAIQQLNEKKLPWYADHVNYLVSGEEPPNLSSYEKKKFFKDINHF 1367
Query: 1034 ---EASFYTLVDGELYRRGIMSPMLKCVDTKDALGIMAEVHEGVCSSHIGGRSLAVKVIR 1090
E YTL ++YR CV + GI+ H H K+++
Sbjct: 1368 YWDEPYLYTLCKDKIYRT--------CVSEDEIEGILLHCHGFAYGGHFATFKTMSKILQ 1419
Query: 1091 AGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMWGTDILGPFPVAK 1150
AGF+WP+M KD E++ KC+ + F +WG D +GPFP +
Sbjct: 1420 AGFWWPSMFKDAQEFISKCDSFENFD--------------------VWGIDFMGPFPSSY 1459
Query: 1151 AQMKYIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTSSV 1210
KYI+VA+DY +KW+EA A T A V I RFGVP ++ D G F +
Sbjct: 1460 GN-KYILVAIDYVSKWVEAIASHTNDARVVLKLFKTIIFPRFGVPRIVISDGGKHFINKG 1518
Query: 1211 TREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYN 1270
+ G++ + T+GQ E +N+ I L+K + + W+ +L LWAY
Sbjct: 1519 FENLLKKHGVKHK--------TSGQVEISNREIKAILEKTVGSTRKDWSAKLNDTLWAYR 1570
Query: 1271 TTEQSSTKETPYRLTYGTDAMLSVEIENQS-W--RVARFNENDNGENLIANLIMLPEEQR 1327
T ++ TP+ L YG L VE+E ++ W ++ F+ E + L L + +
Sbjct: 1571 TAFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLNDLNKIRL 1630
Query: 1328 EAHIRNEAGKVKVARKFSTKVVPRKMRVGDLVL*KNT-IPDKHNKLSPNWGGPYRIIGDV 1386
EA+ ++ K + K+V R +VGD VL N+ + KL W GP+ +
Sbjct: 1631 EAYESSKIYKERTKSFHDKKIVSRDFKVGDQVLLFNSRLRLFPGKLKSRWSGPFSV---T 1687
Query: 1387 GGEAYKLEQLSGQKVPRTWNASHLKQY 1413
Y L+G+ T N LK+Y
Sbjct: 1688 AVRPYGAITLAGKNGDFTVNGQRLKKY 1714
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 207 bits (528), Expect = 3e-53
Identities = 125/412 (30%), Positives = 209/412 (50%), Gaps = 20/412 (4%)
Query: 409 LAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKW 468
+ + PG P+ + RM+ + ++ + E L+ A V+ VKK +G +
Sbjct: 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAP-------------VLFVKKKDGSF 516
Query: 469 RMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAF 528
R+C DY LN V K+ YPLP +D+L+D G S +D SGY+ I + +D TAF
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 529 MTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDL 588
T ++ + MPFGL NA A + RLM+ +F + + + +++DD++V S +H L
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 589 KEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEV 648
+ ++LR ++ KCSF + FLG ++++ G+ V+P+K AI + +PT+ E+
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEI 696
Query: 649 QRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKP 708
+ G RF+ A P K+ F W+ ECE+ F LKE L + PVL+ P
Sbjct: 697 RSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 756
Query: 709 TPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTA 768
G P ++Y + + VL+Q + KVI + S L+ E Y + A++
Sbjct: 757 EHGEPYMVYTDASGVGLGCVLMQ----RGKVIAYASRQLRKHEGNYPTHDLEMAAVIFAL 812
Query: 769 RRLRPYFQSFQVKVKTD-VPLRQVLQKPDLSGRLVSWSVELSEYDIQ--YEP 817
+ R Y V+V TD L+ + +P+L+ R W +++YD++ Y P
Sbjct: 813 KIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLEIAYHP 864
Score = 97.1 bits (240), Expect = 7e-20
Identities = 86/303 (28%), Positives = 139/303 (45%), Gaps = 18/303 (5%)
Query: 1100 KDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAP-WPFAMWGTDILGPFPVAKAQMKYIIV 1158
+D +V +C+ CQ+ H+ P L ++ P W + D + PV++ + I V
Sbjct: 941 EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTK-DAIWV 999
Query: 1159 AVDYFTKWIEAEAVA-TITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTSSVTREFCAE 1217
VD TK A+ T AA + IV GVP+++V D ++FTS+ R F AE
Sbjct: 1000 IVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAE 1059
Query: 1218 MGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSST 1277
MG +++ ++ HPQT GQ+E + + L+ + + G WA+ L V +AYN + +S
Sbjct: 1060 MGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASI 1119
Query: 1278 KETPYRLTYGTDAMLSVEIENQSWRVARFNENDNGENLIANLIMLPEEQREAHIRNEAGK 1337
P+ Y + + R D + + +L +EA R +
Sbjct: 1120 GMAPFEALYERPCRTPLCLTQVGER--SIYGADYVQETTERIRVLKLNMKEAQDRQRSYA 1177
Query: 1338 VKVARKFSTKVVPRKMRVGDLVL*KNTI---PDK---HNKLSPNWGGPYRIIGDVGGEAY 1391
K R+ + VGD V K + P++ KLSP + GP+RI+ VG AY
Sbjct: 1178 DKRRREL-------EFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPVAY 1230
Query: 1392 KLE 1394
+LE
Sbjct: 1231 RLE 1233
>At1g35370 hypothetical protein
Length = 1447
Score = 193 bits (491), Expect = 5e-49
Identities = 139/464 (29%), Positives = 234/464 (49%), Gaps = 21/464 (4%)
Query: 360 VKDKFLKIGS--GLTTE--QEDRLITLLGDNLDLFAWTINDVPGIDPKVITHKLAIRPGA 415
V D+ +IGS LT++ +E + ++ + D+FA D+P K HK+ + GA
Sbjct: 521 VSDEEQEIGSISALTSDVVEESVVQNIVEEFPDVFAEP-TDLPPFREKH-DHKIKLLEGA 578
Query: 416 TPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANGKWRMCTDYT 475
PV Q R + + + +IK+ I+ P + + VV+VKK +G WR+C DYT
Sbjct: 579 NPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSP-FASPVVLVKKKDGTWRLCVDYT 637
Query: 476 SLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQANY 535
LN + KD + +P ++ L+D G+ + S +D +GY+Q+ M P D + TAF T+ ++
Sbjct: 638 ELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHF 697
Query: 536 CYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQL 595
Y M FGL NA AT+Q LM+ +F + + + V+ DD+++ S+ +H L+ F+ +
Sbjct: 698 EYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVM 757
Query: 596 RTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGRM 655
R +++ F + LG +++R IE +P K +A+ E +PT+VK+V+ G
Sbjct: 758 RLHKL--------FAKGSKEHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFA 809
Query: 656 AALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLV 715
RF+ G A P L K F W+ E + AF LK L PVL+ P +
Sbjct: 810 GYYRRFVRNFGVIAGPLH-ALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFM 868
Query: 716 LYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYF 775
+ + + VL+Q K + ++S L+G +L EK LA + R+ R Y
Sbjct: 869 VETDACGQGIRAVLMQ----KGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYL 924
Query: 776 QSFQVKVKTDV-PLRQVLQKPDLSGRLVSWSVELSEYDIQYEPR 818
+KTD L+ +L++ + W +L E+D + + R
Sbjct: 925 LPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPKLLEFDYEIQYR 968
Score = 98.6 bits (244), Expect = 2e-20
Identities = 87/320 (27%), Positives = 140/320 (43%), Gaps = 27/320 (8%)
Query: 1081 GRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMW-- 1138
GR + + +++ FYW M KD +++ C CQ + A P L + P P +W
Sbjct: 1069 GRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPL--PIPDKIWCD 1126
Query: 1139 -GTDILGPFPVAKAQMKYIIVAVDYFTKWIEAEAVA-TITAAKVRNFLWQRIVCRFGVPM 1196
D + P + + I+V VD +K A+A +A V + G P
Sbjct: 1127 VSMDFIEGLPNSGGK-SVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHGCPT 1185
Query: 1197 ALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKG 1256
++V D FTS +EF G+E+R +S HPQ++GQ E N+ + L+
Sbjct: 1186 SIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHARPH 1245
Query: 1257 LWAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEIENQSWRVARFNEN-DNGENL 1315
LW + LP + YNT SS++ TP+ L YG + + +VA + EN+
Sbjct: 1246 LWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQERENM 1305
Query: 1316 IANLIMLPEEQREAHIRNEAGKVK-VARKFSTKVVPRKMRVGDLVL*K-------NTIPD 1367
+ L + H+ ++K A + T+ R +GD V K + +
Sbjct: 1306 LLFL--------KFHLMRAQHRMKQFADQHRTE---RTFDIGDFVYVKLQPYRQQSVVLR 1354
Query: 1368 KHNKLSPNWGGPYRIIGDVG 1387
+ KLSP + GPY+II G
Sbjct: 1355 VNQKLSPKYFGPYKIIEKCG 1374
>At2g06890 putative retroelement integrase
Length = 1215
Score = 181 bits (459), Expect = 3e-45
Identities = 131/443 (29%), Positives = 212/443 (47%), Gaps = 39/443 (8%)
Query: 379 LITLLGDNLDLFAWTINDVP-GIDP-KVITHKLAIRPGATPVIQPMRRMSEEKHKAVQLE 436
+ +LL D D+F D P G+ P + I H++ PGA+ +P R + + K +Q
Sbjct: 408 MTSLLQDYKDVFP---EDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELQ-- 462
Query: 437 TEKLIKARFIREVQYPTWLANVVMVKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVD 496
++ +G WRMC D ++N V K +P+P +D ++D
Sbjct: 463 -------------------------RQKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLD 497
Query: 497 GASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMD 556
G+ + S +D SGY+QI M+ DE TAF T Y + MPFGL +A +T+ RLM+
Sbjct: 498 ELHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMN 557
Query: 557 KIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKF 616
+ +G + VY DD++V S +H L + LR ++ N +KC+F F
Sbjct: 558 HVLRAFIGIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVF 617
Query: 617 LGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGRMAALSRFLPMAGDKAAPFFTCL 676
LGF++++ G++V+ +K +AI + SP +V EV+ G RF AP +
Sbjct: 618 LGFVVSADGVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVM 677
Query: 677 KKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGVPLVLYLAVTDKAVSTVLLQEEGKK 736
KK+ F+W + E+AF LK+ L PVL + + + VL+Q+
Sbjct: 678 KKDVGFKWEKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQD---- 733
Query: 737 QKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLRPYFQSFQVKVKTD-VPLRQVLQKP 795
QK+I F S L GA L Y +K A+++ +R + Y + TD L+ + +
Sbjct: 734 QKLIAFFSEKLGGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQ 793
Query: 796 DLSGRLVSW--SVELSEYDIQYE 816
L+ R W +E Y I+Y+
Sbjct: 794 KLNKRHARWVEFIETFAYVIKYK 816
Score = 30.0 bits (66), Expect = 9.9
Identities = 25/78 (32%), Positives = 39/78 (49%), Gaps = 5/78 (6%)
Query: 1322 LPEEQREAHIRNEAGKVKVARKFSTKVVPRKM-RVGDLV---L*KNTIP-DKHNKLSPNW 1376
L ++ E RN K K+ K + K ++ VGD+V L K P + +KL P
Sbjct: 1023 LVQQIHENARRNIEEKTKLYAKQANKGRREQIFEVGDMVWIHLRKERFPAQRKSKLMPRI 1082
Query: 1377 GGPYRIIGDVGGEAYKLE 1394
GP++II + AY+L+
Sbjct: 1083 DGPFKIIKRINDNAYQLD 1100
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,526,955
Number of Sequences: 26719
Number of extensions: 1381160
Number of successful extensions: 3715
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 3344
Number of HSP's gapped (non-prelim): 278
length of query: 1414
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1302
effective length of database: 8,326,068
effective search space: 10840540536
effective search space used: 10840540536
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0114.12


