

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0113.5
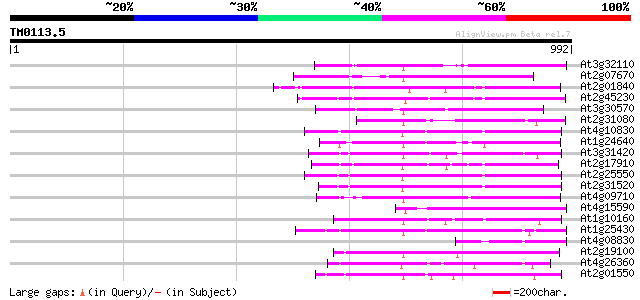
(992 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g32110 non-LTR reverse transcriptase, putative 263 4e-70
At2g07670 putative non-LTR retrolelement reverse transcriptase 236 4e-62
At2g01840 putative non-LTR retroelement reverse transcriptase 214 3e-55
At2g45230 putative non-LTR retroelement reverse transcriptase 203 3e-52
At3g30570 putative reverse transcriptase 201 2e-51
At2g31080 putative non-LTR retroelement reverse transcriptase 199 6e-51
At4g10830 putative protein 183 4e-46
At1g24640 hypothetical protein 182 6e-46
At3g31420 hypothetical protein 182 1e-45
At2g17910 putative non-LTR retroelement reverse transcriptase 182 1e-45
At2g25550 putative non-LTR retroelement reverse transcriptase 181 2e-45
At2g31520 putative non-LTR retroelement reverse transcriptase 177 2e-44
At4g09710 RNA-directed DNA polymerase -like protein 174 2e-43
At4g15590 reverse transcriptase like protein 171 1e-42
At1g10160 putative reverse transcriptase 154 2e-37
At1g25430 hypothetical protein 143 4e-34
At4g08830 putative protein 139 6e-33
At2g19100 putative non-LTR retroelement reverse transcriptase 139 6e-33
At4g26360 putative protein 131 2e-30
At2g01550 putative non-LTR retroelement reverse transcriptase 130 4e-30
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 263 bits (672), Expect = 4e-70
Identities = 163/456 (35%), Positives = 242/456 (52%), Gaps = 45/456 (9%)
Query: 539 LALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHL 598
LA++E + +I + LGFE+ F V+A G SGG+W+LW G+V ++ S QF+H
Sbjct: 597 LAIFETHAGGDQASRICQGLGFENSFRVDAVGHSGGLWLLWRTGIGEVSVVDSTDQFIHA 656
Query: 599 R-IHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKM 657
+ ++ K+NV N +V VY +P AS R+ LW L ++ + P V GDFNT ++ ++
Sbjct: 657 KDVNGKDNV-NLVV--VYAAPTASRRSGLWDRLGDVIRSMDGPVVIGGDFNTIVRLDERS 713
Query: 658 GGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRG------VKERIDWALGNARWIS 711
GG F + ++ SL DLGFKG FTWK RG V +R+D L A
Sbjct: 714 GGNGRLSSDSLAFGEWINDHSLIDLGFKGNKFTWK-RGREERFFVAKRLDRVLCCAHARL 772
Query: 712 SFPEATVLHLPKLKYDHKPILIQLGPEHMDS-SQRPFRFLAAWLTHEDFPRLVKSSWDSH 770
+ EA+VLHLP L DH P+ +QL PE + +RPFRF AAWL+H F L+ +SW
Sbjct: 773 KWQEASVLHLPFLASDHAPLYVQLTPEVSGNRGRRPFRFEAAWLSHPGFKELLLTSW--- 829
Query: 771 HDSWIPASEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLW 830
N+D+ T L + L + S L K ++ L
Sbjct: 830 ------------------NKDI-----STPEALK-----VQELLDLHQSDDLLKKEEELL 861
Query: 831 KEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQ 890
K++ VL +EE++W QKSR W HGDRNT FFHT+T++ R+RN+IE L ++ G +++
Sbjct: 862 KDFDVVLEQEEVVWMQKSREKWFVHGDRNTKFFHTSTIIRRRRNQIEMLQDNDGRWLSNA 921
Query: 891 DQLRSMAVTSFQNLYTSPGPGEPYH--VRNAFPSLPRETLDEIMGPLEAAEIRVAVFSMG 948
+L + A+ ++ LY+ + F +L + P E+ A+ SMG
Sbjct: 922 QELETHAIDYYKRLYSLDDLDAVVEQLPQEGFTALSEADFSSLTKPFSPLEVEGAIRSMG 981
Query: 949 AMKAPGPDDLNPLFFQSQWQVIGPSVVKLAMTVYSN 984
KAPGPD P+F+Q W+V+G SV K M +S+
Sbjct: 982 KYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFFSS 1017
>At2g07670 putative non-LTR retrolelement reverse transcriptase
Length = 913
Score = 236 bits (603), Expect = 4e-62
Identities = 147/430 (34%), Positives = 217/430 (50%), Gaps = 44/430 (10%)
Query: 503 LFMMLYNLFSWNIICAGAQGVPLLLKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFED 562
++ L + WN ++ ++ +HR+ LAL+E +I + LGFE+
Sbjct: 500 IYRYLMDCLKWNCRGVNKPNFRRSIRYMLKKHRIDVLALFETHAGGEKAVRIYQNLGFEN 559
Query: 563 FFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASI 622
F V+A G SGGIW+LW GDV I+ S QF+H ++ N + + VY +P+
Sbjct: 560 SFRVDARGQSGGIWLLWKSEVGDVSIVESAEQFIHAKVG--NGLAAIHLLAVYAAPSTRR 617
Query: 623 RNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDL 682
+N GG P+ L+ F + ++ SL D+
Sbjct: 618 KN------------------------------GGNGGLSPDSLA---FGEWINELSLIDM 644
Query: 683 GFKGPPFTWKGRG------VKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLG 736
GFKG FTWK RG V +R+D L + + EA+V HLP DH PI IQL
Sbjct: 645 GFKGNKFTWK-RGRVESTFVAKRLDRVLCRPQTRLKWQEASVTHLPFFASDHAPIYIQLE 703
Query: 737 PE-HMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAFCEEAASWNRDVFGE 795
PE + +RPFRF AAWLTH F L+++SW++ ++ + A A + WNR+VFG+
Sbjct: 704 PEVRSNPLRRPFRFEAAWLTHSGFKDLLQASWNTEGETPV-ALAALKSKLKKWNREVFGD 762
Query: 796 IGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNH 855
+ R K LM ++ + L ++ + L ++ L KE+ VL +EE+LW QKSR W+
Sbjct: 763 VNRRKESLMNEIKVVQELLEINQTDNLLSKEEELIKEFDVVLEQEEVLWFQKSREKWVEL 822
Query: 856 GDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYH 915
GDRNT +FHT T+V R+RN+IE L D G ++ Q +L MAV + LY+ E
Sbjct: 823 GDRNTKYFHTMTVVRRRRNRIEMLKADDGSWVSQQQELEKMAVDYYSRLYSMEDVDEVVE 882
Query: 916 VRNAFPSLPR 925
P PR
Sbjct: 883 RLPTNPPNPR 892
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 214 bits (544), Expect = 3e-55
Identities = 152/520 (29%), Positives = 246/520 (47%), Gaps = 28/520 (5%)
Query: 467 SNAPGDMGGRTFRERDRSLSPRKHRSS*RAFPSISTLFMMLYNLFSWNIICAGAQGVPLL 526
+NA GD+ G + L RKH R +T M + WN C G G PL
Sbjct: 326 ANATGDILGPV-ELLSQELRKRKHNFESRGGAGTTTSPM---RVGFWN--CQGL-GQPLT 378
Query: 527 LKDIVMRHRV---SCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHW 583
++ + RV L L E + Q N + +GFED ++ G SGG+ V W KH
Sbjct: 379 VRRLEEVQRVYFLDMLFLIETKQQDNYTRDLGVKMGFEDMCIISPRGLSGGLVVYWKKHL 438
Query: 584 GDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVT 643
++++S + V L + KN NF ++ +YG P S R+ LW++LQ +++ + PW+
Sbjct: 439 S-IQVISHDVRLVDLYVEYKNF--NFYLSCIYGHPIPSERHHLWEKLQRVSAHRSGPWMM 495
Query: 644 VGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRGVKERIDWA 703
GDFN L ++K GG ++ S+ F + ++ C++ DL KG P++W G+ E I+
Sbjct: 496 CGDFNEILNLNEKKGGRRRSIGSLQNFTNMINCCNMKDLKSKGNPYSWVGKRQNETIESC 555
Query: 704 LG----NARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDF 759
L N+ W +SFP LP DH P++I + E + + + FR+ EDF
Sbjct: 556 LDRVFINSDWQASFPAFETEFLPIAGSDHAPVIIDIA-EEVCTKRGQFRYDRRHFQFEDF 614
Query: 760 PRLVKSSWD----SHHDSWIPASEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLR 815
V+ W+ H + +E A W R L R++
Sbjct: 615 VDSVQRGWNRGRSDSHGGYYEKLHCCRQELAKWKRRTKTNTAEKIETLKYRVDAAERDHT 674
Query: 816 MSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNK 875
+ H L +L++ L + Y+ EEL W KSR W+ GDRNT FF+ +T + + RN+
Sbjct: 675 LPHQTIL-RLRQDLNQAYRD----EELYWHLKSRNRWMLLGDRNTMFFYASTKLRKSRNR 729
Query: 876 IEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAF-PSLPRETLDEIMGP 934
I+A+ + G D + +A F +L+T+ + + + P + + E++
Sbjct: 730 IKAITDAQGIENFRDDTIGKVAENYFADLFTTTQTSDWEEIISGIAPKVTEQMNHELLQS 789
Query: 935 LEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSV 974
+ E+R AVF++GA +APG D F+ W +IG V
Sbjct: 790 VTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWDLIGNDV 829
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 203 bits (517), Expect = 3e-52
Identities = 131/484 (27%), Positives = 228/484 (47%), Gaps = 22/484 (4%)
Query: 510 LFSWNIICAGAQGVPLL--LKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVE 567
+ SWN C G P + L++I + + L E + + N L ++ LGF D VE
Sbjct: 3 ILSWN--CQGVGNTPTVRHLREIRGLYFPEVIFLCETKKRRNYLENVVGHLGFFDLHTVE 60
Query: 568 AEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLW 627
G SGG+ ++W K +++L S ++ + + ++ F +T +YG P + R LW
Sbjct: 61 PIGKSGGLALMW-KDSVQIKVLQSDKRLIDALLIWQDK--EFYLTCIYGEPVQAERGELW 117
Query: 628 QELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGP 687
+ L + + PW+ GDFN + S+K+GG S +FR L++C L ++ G
Sbjct: 118 ERLTRLGLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCLEFRQMLNSCGLWEVNHSGY 177
Query: 688 PFTWKGRGVKE----RIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSS 743
F+W G E R+D + N W+ FP+A +L K+ DH P++ L ++
Sbjct: 178 QFSWYGNRNDELVQCRLDRTVANQAWMELFPQAKATYLQKICSDHSPLINNLVGDNW-RK 236
Query: 744 QRPFRFLAAWLTHEDFPRLVKSSWDSHH---DSWIPASEAFCE-EAASWNRDVFGEIGRT 799
F++ W+ E F L+ + W ++ + A C E + W R
Sbjct: 237 WAGFKYDKRWVQREGFKDLLCNFWSQQSTKTNALMMEKIASCRREISKWKRVSKPSSAVR 296
Query: 800 KRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRN 859
+ L +L+ + L +L+K L +EY EE W++KSR+ W+ +GDRN
Sbjct: 297 IQELQFKLDAATKQIPFDRRE-LARLKKELSQEYNN----EEQFWQEKSRIMWMRNGDRN 351
Query: 860 THFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPY-HVRN 918
T +FH T R +N+I+ L ++ G T + L +A F+ L+ S G + N
Sbjct: 352 TKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYFKKLFASEDVGYTVEELEN 411
Query: 919 AFPSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVVKLA 978
P + + + ++ P+ E++ A FS+ K PGPD +N +Q W+ +G + ++
Sbjct: 412 LTPLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQITEMV 471
Query: 979 MTVY 982
+
Sbjct: 472 QAFF 475
>At3g30570 putative reverse transcriptase
Length = 1099
Score = 201 bits (511), Expect = 2e-51
Identities = 136/424 (32%), Positives = 206/424 (48%), Gaps = 35/424 (8%)
Query: 541 LYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRI 600
++E + +I + L FE+ F V+AEG SGG+WVLW GDV I+ QF+H R+
Sbjct: 83 VFETHAGGDKAGRICQGLDFENSFWVDAEGQSGGLWVLWRTCIGDVVIVEFAYQFIHARV 142
Query: 601 HPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGG 660
+ + +V VY P S R+ LW L + V P + DFN ++ ++ GG
Sbjct: 143 VNGDAAVDLIV--VYAPPTPSRRSKLWDTLSAVIRGVKGPVIIGEDFNMIVRVDERSGGS 200
Query: 661 PPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRG--------------VKERIDWALGN 706
P F + ++ SL TW R V +R+D L
Sbjct: 201 GPLSPDSIAFGEWINDSSL---------LTWVSRVTNSVGSGSVRRDFFVAKRLDRVLCC 251
Query: 707 ARWISSFPEATVLHLPKLKYDHKPILIQLG-PEHMDSSQRPFRFLAAWLTHEDFPRLVKS 765
+ + EAT+ HLP L DH P+ +QL + +RPFRF AAWL+H F L+ +
Sbjct: 252 SHARLKWQEATLTHLPFLASDHAPLYLQLALGVGKNRFRRPFRFEAAWLSHPGFKELLLA 311
Query: 766 SWDSHHDSWIPASEAFC---EEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYL 822
SW + IP EA WN++VFG+I K +LM ++ + + L + + L
Sbjct: 312 SWKGN----IPTPEALKGIQTTLRKWNKEVFGDIQEKKEKLMVEIKTVQDSLYSNQTDDL 367
Query: 823 EKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNND 882
+ + L E+ VL +EE LW KSR W GDRNT FFHT+T++ R+ N+IE L N+
Sbjct: 368 LRRESELIIEFDVVLEQEETLWLHKSREKWFVQGDRNTSFFHTSTIIRRRHNRIEMLKNE 427
Query: 883 VGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNA--FPSLPRETLDEIMGPLEAAEI 940
G I++ ++L +A+ ++ LY+ E A F L +E + P A E+
Sbjct: 428 KGGWISNTNELEKLAIDYYKRLYSLDDVDEVVERLPAGRFLGLNQEEQIRLNKPFSAMEV 487
Query: 941 RVAV 944
A+
Sbjct: 488 ESAL 491
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 199 bits (506), Expect = 6e-51
Identities = 130/380 (34%), Positives = 191/380 (50%), Gaps = 57/380 (15%)
Query: 614 VYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDC 673
VY +P+ S R+ LW EL+++ + + P + GDFNT L ++MGG F D
Sbjct: 6 VYAAPSVSRRSGLWGELKDVVNGLEGPLLIGGDFNTILWVDERMGGNGRLSPDSLAFGDW 65
Query: 674 LDACSLSDLGFKGPPFTWKGRG------VKERIDWALGNARWISSFPEATVLHLPKLKYD 727
++ SL DLGFKG FTW+ RG V +R+D A + EA V HLP + D
Sbjct: 66 INELSLIDLGFKGNKFTWR-RGRQESTVVAKRLDRVFVCAHARLKWQEAVVSHLPFMASD 124
Query: 728 HKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAFCEEAAS 787
H P+ +QL P QR R
Sbjct: 125 HAPLYVQLEP----LQQRKLR--------------------------------------K 142
Query: 788 WNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQK 847
WNR+VFG+I K +L+ ++ + + L + S L ++ L KE VL +EE LW QK
Sbjct: 143 WNREVFGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEVLLKEMDLVLEQEETLWFQK 202
Query: 848 SRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTS 907
SR ++ GDRNT FFHT+T++ R+RN+IE+L D +TD+ +L +MA+T ++ LY+
Sbjct: 203 SREKYIELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMALTYYKRLYSL 262
Query: 908 PGPGEPYHVRNAFPSLPRETLDE-----IMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLF 962
E VRN P+ ++ E ++ AE+ AV SMG KAPGPD P+F
Sbjct: 263 EDVSE---VRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPDGYQPVF 319
Query: 963 FQSQWQVIGPSVVKLAMTVY 982
+Q W+ +GPSV + + +
Sbjct: 320 YQQCWETVGPSVTRFVLEFF 339
>At4g10830 putative protein
Length = 1294
Score = 183 bits (465), Expect = 4e-46
Identities = 130/467 (27%), Positives = 209/467 (43%), Gaps = 20/467 (4%)
Query: 522 GVPLL---LKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVL 578
GVPL L ++ + L L E + + + LGF + +G SGG+ +L
Sbjct: 373 GVPLTQSQLSNLCKVFKFDVLFLIETLNKCEVISNLASVLGFPNVITQPPQGHSGGLALL 432
Query: 579 WSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVT 638
W V + + ++ H+ +H N NF ++ VYG P S R+ LW +N++
Sbjct: 433 WKD---SVRLSNLYQDDRHIDVHISINNINFYLSRVYGHPCQSERHSLWTHFENLSKTRN 489
Query: 639 KPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG----R 694
PW+ +GDFN L ++K+GG + + FR+ + C L D+ G F+W G
Sbjct: 490 DPWILIGDFNEILSNNEKIGGPQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGERHSH 549
Query: 695 GVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWL 754
VK +D A N+ FP A + L DHKP+ + L + RPFRF L
Sbjct: 550 TVKCCLDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFLSL-EKTETRKMRPFRFDKRLL 608
Query: 755 THEDFPRLVKSSWD---SHHDSWIPASEAFCEEAASWNRDVFGEIGRTK-RRLMRRLEGI 810
F VK+ W+ + +P C +A + + R + +L L+
Sbjct: 609 EVPHFKTYVKAGWNKAINGQRKHLPDQVRTCRQAMAKLKHKSNLNSRIRINQLQAALDKA 668
Query: 811 NNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVH 870
+ + + + +Q+ L Y+ EE W+QKSR W+ GDRNT FFH T
Sbjct: 669 MSSVNRTERRTISHIQRELTVAYRD----EERYWQQKSRNQWMKEGDRNTEFFHACTKTR 724
Query: 871 RKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPG-PGEPYHVRNAFPSLPRETLD 929
N++ + ++ G + ++ A F +Y S G P P + + D
Sbjct: 725 FSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVYESNGRPVSIIDFAGFKPIVTEQIND 784
Query: 930 EIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVVK 976
++ L EI A+ +G KAPGPD L F++S W+++GP V+K
Sbjct: 785 DLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIK 831
>At1g24640 hypothetical protein
Length = 1270
Score = 182 bits (463), Expect = 6e-46
Identities = 132/451 (29%), Positives = 198/451 (43%), Gaps = 50/451 (11%)
Query: 549 NNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKH------WGDVEILSSHRQFVHLRIHP 602
++L I L ++ + VE G GG+ +LW + D ++ + QF
Sbjct: 5 DDLVDIQSWLEYDQVYTVEPVGKCGGLALLWKSSVQVDLKFVDKNLMDAQVQF------- 57
Query: 603 KNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPP 662
NF V+ VYG P+ S R+ W+ + I W GDFN L +K GG
Sbjct: 58 --GAVNFCVSCVYGDPDRSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGGPRR 115
Query: 663 NLLSMAKFRDCLDACSLSDLGFKGPPFTWKGR----GVKERIDWALGNARWISSFPEATV 718
+ L F + + C L ++ G FTW GR ++ R+D A GN W FP +
Sbjct: 116 SDLDCKAFNEMIKGCDLVEMPAHGNGFTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQ 175
Query: 719 LHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWD-SHHDSWIPA 777
L DH+P+LI+L DS + FRF +L ED + +W H + I
Sbjct: 176 TFLDFRGSDHRPVLIKL-MSSQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNISV 234
Query: 778 SE---AFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQ 834
++ A + +SW K+ + L+ IN LEK Q +W +Q
Sbjct: 235 ADRLRACRKSLSSWK----------KQNNLNSLDKIN-----QLEAALEKEQSLVWPIFQ 279
Query: 835 TVLI----------REELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVG 884
V + EE W+QKSR WL G+RN+ +FH +R+R +IE L + G
Sbjct: 280 RVSVLKKDLAKAYREEEAYWKQKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNG 339
Query: 885 ELITDQDQLRSMAVTSFQNLYTSPGP-GEPYHVRNAFPSLPRETLDEIMGPLEAAEIRVA 943
+ T + +A F NL+ S P G P + + ++G + A EI+ A
Sbjct: 340 NMQTSEAAKGEVAAAYFGNLFKSSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEA 399
Query: 944 VFSMGAMKAPGPDDLNPLFFQSQWQVIGPSV 974
VFS+ APGPD ++ LFFQ W +G V
Sbjct: 400 VFSIKPASAPGPDGMSALFFQHYWSTVGNQV 430
>At3g31420 hypothetical protein
Length = 1491
Score = 182 bits (461), Expect = 1e-45
Identities = 125/465 (26%), Positives = 222/465 (46%), Gaps = 31/465 (6%)
Query: 529 DIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEI 588
+I ++ + + L E + Q + + + LGF + V +G SGG+ V W+ D+ +
Sbjct: 202 EICRKNLLDIICLSETKQQDDRIRDVGAELGFCNHVTVPPDGLSGGLVVFWNSSV-DIFL 260
Query: 589 LSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNI-ASQVTKPWVTVGDF 647
S+ V L H K+N +F ++FVYG PN S R+ LW+ L+ + ++ W +GDF
Sbjct: 261 CFSNSNLVDL--HVKSNEGSFYLSFVYGHPNPSHRHHLWERLERLNTTRQGTAWFIMGDF 318
Query: 648 NTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRG----VKERIDWA 703
N L +K GG + FR+ + C+ +DL G F+W G+ V +D
Sbjct: 319 NEILSNREKRGGRLRPERTFQDFRNMVRGCNFTDLKSVGDRFSWAGKRGDHHVTCSLDRT 378
Query: 704 LGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTH-EDFPRL 762
+ N W + FPE+ + + + DH+P++ + + + +R F + LTH E F ++
Sbjct: 379 MANNEWHTLFPESETVFMEYGESDHRPLVTNISAQKEE--RRGFFSYDSRLTHKEGFKQV 436
Query: 763 VKSSWDSHH-----DSWIPASEAFCEEAAS-WNRDVFGEIGRTKRRLMRRLEGINNHL-- 814
V + W + DS + C +A S W ++ + R++ I + L
Sbjct: 437 VLNQWHRRNGSFEGDSSLNRKLVECRQAISRWKKN-------NRVNAEERIKIIRHKLDR 489
Query: 815 RMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRN 874
++ + ++++ ++ EE+ W+ KSR WLN GDRNT +FH+TT R RN
Sbjct: 490 AIATGTATPRERRQMRQDLNQAYADEEIYWQTKSRSRWLNAGDRNTRYFHSTTKTRRCRN 549
Query: 875 KIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAFPSLPRETLDEI--- 931
++ ++ + G++ + + +A+ F +LY S P + F ++ DEI
Sbjct: 550 RLLSVQDSDGDICRGDENIAKVAINYFDDLYKST-PNTSLRYADVFQGFQQKITDEINED 608
Query: 932 -MGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
+ P+ EI +VFS+ + P PD F+Q W I V+
Sbjct: 609 LIRPVTELEIEESVFSVAPSRTPDPDGFTADFYQQFWPDIKQKVI 653
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 182 bits (461), Expect = 1e-45
Identities = 130/448 (29%), Positives = 207/448 (46%), Gaps = 22/448 (4%)
Query: 534 HRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHR 593
H L L E + + + K+ LG++ VE EG SGG+ + W H ++E L + +
Sbjct: 5 HFPDILFLMETKNSQDFVYKVFCWLGYDFIHTVEPEGRSGGLAIFWKSHL-EIEFLYADK 63
Query: 594 QFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQA 653
+ L++ +N V + ++ VYG P +R LW+ L +I + + W +GDFN
Sbjct: 64 NLMDLQVSSRNKV--WFISCVYGLPVTHMRPKLWEHLNSIGLKRAEAWCLIGDFNDIRSN 121
Query: 654 SDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG----RGVKERIDWALGNARW 709
+K+GG + S F L CS+ +LG G FTW G + V+ ++D GN W
Sbjct: 122 DEKLGGPRRSPSSFQCFEHMLLNCSMHELGSTGNSFTWGGNRNDQWVQCKLDRCFGNPAW 181
Query: 710 ISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDS 769
S FP A L K DH+P+L++ ++ + + FR+ ++ SW+S
Sbjct: 182 FSIFPNAHQWFLEKFGSDHRPVLVKFTNDN-ELFRGQFRYDKRLDDDPYCIEVIHRSWNS 240
Query: 770 H-----HDSWIPASEAFCEEAAS-WNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLE 823
H S+ E C A S W +RL + L+ +++ P +E
Sbjct: 241 AMSQGTHSSFFSLIE--CRRAISVWKHSSDTNAQSRIKRLRKDLDA-EKSIQIPCWPRIE 297
Query: 824 KLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDV 883
++ +L Y EEL WRQKSR WL GD+NT FFH T R +N++ L ++
Sbjct: 298 YIKDQLSLAYGD----EELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDEN 353
Query: 884 GELITDQDQLRSMAVTSFQNLYTSPG-PGEPYHVRNAFPSLPRETLDEIMGPLEAAEIRV 942
+ T +A + F+NL+TS H+ + E ++ + E+
Sbjct: 354 DQEFTRNSDKGKIASSFFENLFTSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELEVYN 413
Query: 943 AVFSMGAMKAPGPDDLNPLFFQSQWQVI 970
AVFS+ APGPD LFFQ W ++
Sbjct: 414 AVFSINKESAPGPDGFTALFFQQHWDLV 441
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 181 bits (458), Expect = 2e-45
Identities = 127/466 (27%), Positives = 218/466 (46%), Gaps = 20/466 (4%)
Query: 522 GVPLLLKDIVMRHRV---SCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVL 578
G+PL + R+ L L E Q + + K+ LGF + G SGG+ ++
Sbjct: 393 GMPLTQSRLFRLFRMYNYDILFLVETLNQCDKVCKLAYDLGFPNVITQPPNGRSGGLALM 452
Query: 579 WSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVT 638
W K+ + ++S + + H N +F ++ VYG P S R+ LWQ L++I+
Sbjct: 453 W-KNNVSLSLISQDERLIDS--HVTFNNKSFYLSCVYGHPTQSERHQLWQTLEHISDNRN 509
Query: 639 KPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG----R 694
W+ VGDFN L ++K+GG + FR+ + C + D+ KG F+W G
Sbjct: 510 AEWLLVGDFNEILSNAEKIGGPMREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGERHTH 569
Query: 695 GVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWL 754
VK +D N+ W ++FP A + L DHKP+L+ E + FRF +
Sbjct: 570 TVKCCLDRVFINSAWTATFPYAEIEFLDFTGSDHKPVLVHFN-ESFPRRSKLFRFDNRLI 628
Query: 755 THEDFPRLVKSSWDSHHDS-WIPASEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNH 813
F R+V++SW ++ +S P +E + R +++R+ + +N
Sbjct: 629 DIPTFKRIVQTSWRTNRNSRSTPITERISSCRQAMARLKHASNLNSEQRIKKLQSSLNRA 688
Query: 814 L---RMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVH 870
+ R + +LQ+ L K + EE+ W+QKSR W+ GD+NT +FH T
Sbjct: 689 MESTRRVDRQLIPQLQESLAKAFSD----EEIYWKQKSRNQWMKEGDQNTGYFHACTKTR 744
Query: 871 RKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAFPSLPRETLD- 929
+N++ + +D G + T ++ + A F N++++ G F S T++
Sbjct: 745 YSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNL 804
Query: 930 EIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
++ EI A+ +G KAPGPD L F+++ W ++G V+
Sbjct: 805 DLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVI 850
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 177 bits (450), Expect = 2e-44
Identities = 120/438 (27%), Positives = 207/438 (46%), Gaps = 17/438 (3%)
Query: 547 QTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNV 606
Q + + K+ LGF + G SGG+ ++W K+ + ++S + + H N
Sbjct: 195 QCDKVCKLAYDLGFPNVITQPPNGRSGGLALMW-KNNVSLSLISQDERLIDS--HVTFNN 251
Query: 607 PNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLS 666
+F ++ VYG P S R+ LWQ L++I+ W+ VGDFN L ++K+GG +
Sbjct: 252 KSFYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKIGGPMREEWT 311
Query: 667 MAKFRDCLDACSLSDLGFKGPPFTWKG----RGVKERIDWALGNARWISSFPEATVLHLP 722
FR+ + C + D+ KG F+W G VK +D N+ W ++FP A L
Sbjct: 312 FRNFRNMVSHCDIEDMRSKGDRFSWVGERHTHTVKCCLDRVFINSAWTATFPYAETEFLD 371
Query: 723 KLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDS-WIPASEAF 781
DHKP+L+ E + FRF + F R+V++SW ++ +S P +E
Sbjct: 372 FTGSDHKPVLVHFN-ESFPRRSKLFRFDNRLIDIPTFKRIVQTSWRTNRNSRSTPITERI 430
Query: 782 CEEAASWNRDVFGEIGRTKRRLMRRLEGINNHL---RMSHSPYLEKLQKRLWKEYQTVLI 838
+ R +++R+ + +N + R + +LQ+ L K +
Sbjct: 431 SSCRQAMARLKHASNLNSEQRIKKLQSSLNRAMESTRRVDRQLIPQLQESLAKAFSD--- 487
Query: 839 REELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAV 898
EE+ W+QKSR W+ GD+NT +FH T +N++ + +D G + T ++ + A
Sbjct: 488 -EEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQ 546
Query: 899 TSFQNLYTSPGPGEPYHVRNAFPSLPRETLD-EIMGPLEAAEIRVAVFSMGAMKAPGPDD 957
F N++++ G F S T++ ++ EI A+ +G KAPGPD
Sbjct: 547 DFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDG 606
Query: 958 LNPLFFQSQWQVIGPSVV 975
L F+++ W ++G V+
Sbjct: 607 LTARFYKNCWDIVGYDVI 624
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 174 bits (442), Expect = 2e-43
Identities = 120/438 (27%), Positives = 196/438 (44%), Gaps = 37/438 (8%)
Query: 543 EPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHP 602
E + Q + K +G+ F + EG SGG+ + W ++ +VEIL +
Sbjct: 2 ETKNQDEFISKTFDWMGYAHRFTIPPEGLSGGLALYWKENV-EVEILEA----------- 49
Query: 603 KNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPP 662
PNF+ R+ W ++ ++ +Q + W+ GDFN L S+K GG
Sbjct: 50 ---APNFIDN----------RSVFWDKISSLGAQRSSAWLLTGDFNDILDNSEKQGGPLR 96
Query: 663 NLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRG----VKERIDWALGNARWISSFPEATV 718
FR + L D+ G +W+G +K R+D ALGN W FP +
Sbjct: 97 WEGFFLAFRSFVSQNGLWDINHTGNSLSWRGTRYSHFIKSRLDRALGNCSWSELFPMSKC 156
Query: 719 LHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWD-SHHDSWIPA 777
+L DH+P++ G + S +PFRF E+ LVK W+ + DS +
Sbjct: 157 EYLRFEGSDHRPLVTYFGAPPLKRS-KPFRFDRRLREKEEIRALVKEVWELARQDSVLYK 215
Query: 778 SEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVL 837
+ W ++ + ++ + LE +S L + +E +
Sbjct: 216 ISRCRQSIIKWTKEQNSNSAKAIKKAQQALESA-----LSADIPDPSLIGSITQELEAAY 270
Query: 838 IREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMA 897
+EEL W+Q SRV WLN GDRN +FH TT R N + + + G+ +++Q+ S
Sbjct: 271 RQEELFWKQWSRVQWLNSGDRNKGYFHATTRTRRMLNNLSVIEDGSGQEFHEEEQIASTI 330
Query: 898 VTSFQNLYTSPGPGEPYHVRNAF-PSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPD 956
+ FQN++T+ + V+ A P + +E++ EI+ A+FS+ A KAPGPD
Sbjct: 331 SSYFQNIFTTSNNSDLQVVQEALSPIISSHCNEELIKISSLLEIKEALFSISADKAPGPD 390
Query: 957 DLNPLFFQSQWQVIGPSV 974
+ FF + W +I V
Sbjct: 391 GFSASFFHAYWDIIEADV 408
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 171 bits (434), Expect = 1e-42
Identities = 106/311 (34%), Positives = 162/311 (52%), Gaps = 26/311 (8%)
Query: 682 LGFKGPPFTWKGRGVKE------RIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQL 735
+GFKG FTW+ RG+ E R+D L A + EA +
Sbjct: 1 MGFKGNRFTWR-RGLVESTFVAKRLDRVLFCAHARLKWQEALLCPA-------------- 45
Query: 736 GPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAFCEEAASWNRDVFGE 795
+++D+ +RPFRF AAWL+HE F L+ +SWD+ + + A + WN++VFG
Sbjct: 46 --QNVDARRRPFRFEAAWLSHEGFKELLTASWDTGLSTPV-ALNRLRWQLKKWNKEVFGN 102
Query: 796 IGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNH 855
I K +++ L+ + + L + + L + L KE+ +L +EE LW QKSR L
Sbjct: 103 IHVRKEKVVSDLKAVQDLLEVVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREKLLAL 162
Query: 856 GDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYH 915
GDRNT FFHT+T++ R+RN+IE L + +T+++ L +A+ ++ LY+
Sbjct: 163 GDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYSLEDVSVVRG 222
Query: 916 V--RNAFPSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPS 973
FP L RE + + P E+ VAV SMG KAPGPD P+F+Q W+ +G S
Sbjct: 223 TLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGES 282
Query: 974 VVKLAMTVYSN 984
V K M + +
Sbjct: 283 VSKFVMEFFES 293
>At1g10160 putative reverse transcriptase
Length = 1118
Score = 154 bits (389), Expect = 2e-37
Identities = 122/429 (28%), Positives = 187/429 (43%), Gaps = 34/429 (7%)
Query: 573 GGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQE--- 629
G IW++W V + Q + I + + +F V FVYG + R LW++
Sbjct: 67 GRIWIVWDPSIS-VLVFKRTDQIMFCSIKIPSLLQSFAVAFVYGRNSELDRRSLWEDILV 125
Query: 630 LQNIASQVTKPWVTVGDFNTYLQASD--KMGGGPPNLLSMAKFRDCLDACSLSDLGFKGP 687
L + PW+ +GDFN AS+ + NL M + CL LSDL +G
Sbjct: 126 LSRTSPLSVTPWLLLGDFNQIAAASEHYSINQSLLNLRGMEDLQCCLRDSQLSDLPSRGV 185
Query: 688 PFTWKGRG----VKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSS 743
FTW + ++D AL N W + FP A + P DH P +I + + S
Sbjct: 186 FFTWSNHQQDNPILRKLDRALANGEWFAVFPSALAVFDPPGDSDHAPCIILIDNQP-PPS 244
Query: 744 QRPFRFLAAWLTHEDFPRLVKSSWD------SHHDSW---IPASEAFCEEAASWNRDVFG 794
++ F++ + +H + + ++W+ SH S + ++ C + NR F
Sbjct: 245 KKSFKYFSFLSSHPSYLAALSTAWEENTLVGSHMFSLRQHLKVAKLCCR---TLNRLRFS 301
Query: 795 EIGRTKRRLMRRLEGINNHLRMSHSPYL---EKLQKRLWKEYQTVLIREELLWRQKSRVN 851
I + + + RLE I L S S L E + ++ W + L E +RQKSR+
Sbjct: 302 NIQQRTAQSLTRLEDIQVELLTSPSDTLFRREHVARKQWIFFAAAL---ESFFRQKSRIR 358
Query: 852 WLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPG 911
WL+ GD NT FFH + H+ N I+ L D G + + DQ++ M + + +L P
Sbjct: 359 WLHEGDANTRFFHRAVIAHQATNLIKFLRGDDGFRVENVDQIKGMLIAYYSHLLGIPSEN 418
Query: 912 -EPYHVRNAFPSLPRETLDEIMGPL----EAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQ 966
P+ V LP + L EI +FSM KAPGPD FF
Sbjct: 419 VTPFSVEKIKGLLPFRCDSFLASQLTTIPSEEEITQVLFSMPRNKAPGPDGFPVEFFIEA 478
Query: 967 WQVIGPSVV 975
W ++ SVV
Sbjct: 479 WAIVKSSVV 487
>At1g25430 hypothetical protein
Length = 1213
Score = 143 bits (361), Expect = 4e-34
Identities = 129/502 (25%), Positives = 213/502 (41%), Gaps = 29/502 (5%)
Query: 506 MLYNLFSWNIICAGAQGVPLLLKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFV 565
M NLF WNI K V ++ + E V+ K + L +
Sbjct: 1 MCTNLFCWNIRGFNNVSHRSGFKKWVKANKPIFGGVIETHVKQPKDRKFINAL-LPGWSF 59
Query: 566 VEAEGFS--GGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIR 623
VE FS G IWV+W V +++ Q + + + +V+ VY + + R
Sbjct: 60 VENYAFSDLGKIWVMWDPSV-QVVVVAKSLQMITCEVLLPGSPSWIIVSVVYAANEVASR 118
Query: 624 NFLWQELQN-IASQVT--KPWVTVGDFNTYLQASDKMGGGPPNL-LSMAKFRDCLDACSL 679
LW E+ N + S + +PW+ +GDFN L + N+ ++M FRDCL A L
Sbjct: 119 KELWIEIVNMVVSGIIGDRPWLVLGDFNQVLNPQEHSNPVSLNVDINMRDFRDCLLAAEL 178
Query: 680 SDLGFKGPPFTWKGRG----VKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQL 735
SDL +KG FTW + V ++ID L N W + FP + + DH + L
Sbjct: 179 SDLRYKGNTFTWWNKSHTTPVAKKIDRILVNDSWNALFPSSLGIFGSLDFSDHVSCGVVL 238
Query: 736 GPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHH---DSWIPASE---AFCEEAASWN 789
E ++RPF+F L + DF LV+ +W + + S S+ A + ++
Sbjct: 239 -EETSIKAKRPFKFFNYLLKNLDFLNLVRDNWFTLNVVGSSMFRVSKKLKALKKPIKDFS 297
Query: 790 RDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSR 849
R + E+ + + L G + +P + +++ + EE +RQKSR
Sbjct: 298 RLNYSELEKRTKEAHDFLIGCQDRTLADPTPINASFELEAERKWHILTAAEESFFRQKSR 357
Query: 850 VNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPG 909
++W GD NT +FH N I AL + G+L+ Q+ + + + F +L
Sbjct: 358 ISWFAEGDGNTKYFHRMADARNSSNSISALYDGNGKLVDSQEGILDLCASYFGSLLGD-- 415
Query: 910 PGEPYHVRN-------AFPSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLF 962
+PY + ++ P + E+ +IR A+FS+ K+ GPD F
Sbjct: 416 EVDPYLMEQNDMNLLLSYRCSPAQVC-ELESTFSNEDIRAALFSLPRNKSCGPDGFTAEF 474
Query: 963 FQSQWQVIGPSVVKLAMTVYSN 984
F W ++G V +S+
Sbjct: 475 FIDSWSIVGAEVTDAIKEFFSS 496
>At4g08830 putative protein
Length = 947
Score = 139 bits (351), Expect = 6e-33
Identities = 75/202 (37%), Positives = 119/202 (58%), Gaps = 20/202 (9%)
Query: 788 WNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQK 847
WNRDVFG++ + K LMR ++ + + L + S + + ++ L K + K
Sbjct: 49 WNRDVFGDVKKKKEELMREIQVVQDALDVHQSDDMLRKEEDLIKAFD------------K 96
Query: 848 SRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTS 907
SR W+ GDRNT++FHTTT++ R++N++E L+N+ G I D +L ++AV +Q LY+
Sbjct: 97 SREKWIALGDRNTNYFHTTTIIRRRQNRVERLHNEEGTWIFDAKELEALAVQYYQRLYSM 156
Query: 908 PGPGEPYHVRNAFP--SLPRETLDEIMG---PLEAAEIRVAVFSMGAMKAPGPDDLNPLF 962
+ V N P L R T +E++ P A E+ VA+ SMG KAPGPD P+F
Sbjct: 157 ---DDVLQVVNPLPQDGLERLTREEVLSLNKPFLATEVEVAIRSMGKYKAPGPDGYQPIF 213
Query: 963 FQSQWQVIGPSVVKLAMTVYSN 984
+QS W+V+G SV++ A+ +S+
Sbjct: 214 YQSCWEVVGNSVIRFALDFFSS 235
>At2g19100 putative non-LTR retroelement reverse transcriptase
Length = 1447
Score = 139 bits (351), Expect = 6e-33
Identities = 111/424 (26%), Positives = 184/424 (43%), Gaps = 26/424 (6%)
Query: 573 GGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQN 632
G IWV+W ++ S Q + + +N F +FVY S R LW ELQ+
Sbjct: 475 GRIWVVWRRNVRLSPFYKSE-QLITCSVKLENRDDEFFCSFVYASNFRDDRKVLWNELQD 533
Query: 633 IASQVT---KPWVTVGDFNTYLQASD--KMGGGPPNLLSMAKFRDCLDACSLSDLGFKGP 687
KPW+ GDFN L+ + K+ P + M FR ++ CSL+D+ GP
Sbjct: 534 HYDSPIIKKKPWIIFGDFNETLELEEHSKVEDNPVVSMGMRDFRSMVNYCSLTDMAHHGP 593
Query: 688 PFTWKGRG----VKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQL--GPEHMD 741
+TW + + +++D + N W SFP++ + DH I L GP +
Sbjct: 594 LYTWSNKREHDLIAKKLDRVMVNDVWTQSFPQSYSVFEAGGCLDHLRGRINLNDGPGSIV 653
Query: 742 SSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAF--CEEAASWN-------RDV 792
+RPF+F+ EDF V S W ++ S F ++ S ++
Sbjct: 654 RGKRPFKFVNVLTEMEDFKPTVDSYWKETEPIFLSTSSLFRFSKKLKSLKPLLRNLAKER 713
Query: 793 FGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNW 852
G + + R L + +P K + ++ V EE ++KS+++W
Sbjct: 714 LGNLVKKTREAYDTLCKKQESTLNNPTPNAMKEEVEAHDRWEHVAGLEEKFLKKKSKLHW 773
Query: 853 LNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQN-LYTSPGPG 911
L+ GD+N FH + +N I + G + D++++ A F+ L P
Sbjct: 774 LDGGDKNNKAFHRAVVTREAQNSISEIQCQDGSVTAKGDEIKAYAERFFREFLQLIPNEY 833
Query: 912 EPYHVRNAFPSLP---RETLDEIM-GPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQW 967
E + + LP ET E++ + A EI+ +FSM K+PGPD FF++ W
Sbjct: 834 EGVTMADLQDLLPFRCSETEHELLTRVVTAEEIKKVLFSMPNDKSPGPDGFTSEFFKATW 893
Query: 968 QVIG 971
+++G
Sbjct: 894 EILG 897
>At4g26360 putative protein
Length = 1141
Score = 131 bits (329), Expect = 2e-30
Identities = 119/421 (28%), Positives = 187/421 (44%), Gaps = 36/421 (8%)
Query: 563 FFVVEAEGFS--GGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNA 620
+F E GFS G IWVLW +V I++ Q + + N+ +++ VY +
Sbjct: 49 WFFDENYGFSDLGKIWVLWDPSV-EVVIVAKSLQMITCEVLFPNSRTWIVISVVYAANED 107
Query: 621 SIRNFLWQELQN-IASQVT--KPWVTVGDFNTYLQASDKMGGGPPNL-LSMAKFRDCLDA 676
R LW+E+ +AS VT +PW+ +GDFN L + N+ + FR+CL
Sbjct: 108 DKRKELWREITALVASPVTFNRPWILLGDFNQVLHPHEHSRHVSLNVDRRIRDFRECLLD 167
Query: 677 CSLSDLGFKGPPFTW----KGRGVKERIDWALGNARWISSFPEATVLHLPKLKYDHKP-- 730
LSDL +KG FTW K R V ++ID L N W + FP + L P DH
Sbjct: 168 AELSDLVYKGSSFTWWNKSKTRPVAKKIDRILVNESWSNLFPSSFGLFGPPDFSDHASCG 227
Query: 731 ILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAF--CEEAASW 788
++++L P ++RPF+F L + +F LV WD + + + S F ++ +
Sbjct: 228 VVLELDPI---KAKRPFKFFNFLLKNPEFLNLV---WDVWYSTNVVGSSMFRVSKKLKAL 281
Query: 789 NRDVFGEIGRTKRRLMRRLEGINNHLRMSHS-----PYLEKLQKRL--WKEYQTVLIREE 841
+ + L +R E + L + P LE L +++Q + EE
Sbjct: 282 KKPIKDFSRLNYSNLEKRTEEAHETLLSFQNLTLDNPSLENAAHELEAQRKWQILATAEE 341
Query: 842 LLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSF 901
+RQ+SRV W GD NT +FH + N I L +D G I Q + F
Sbjct: 342 SFFRQRSRVTWFAEGDGNTRYFHRMADSRKSVNTITTLVDDSGTQIDSQQGIADHCALYF 401
Query: 902 QNLYTSPGPGEPYHVRNAFPSL------PRETLDEIMGPLEAAEIRVAVFSMGAMKAPGP 955
+NL + +PY + +L P + ++ +I+ A F + + KA GP
Sbjct: 402 ENLLSD--DNDPYSLEQDDMNLLLTYRCPYSQVADLEAMFSDEDIKAAFFGLPSNKACGP 459
Query: 956 D 956
D
Sbjct: 460 D 460
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 130 bits (327), Expect = 4e-30
Identities = 115/465 (24%), Positives = 190/465 (40%), Gaps = 35/465 (7%)
Query: 541 LYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFS--GGIWVLWSKHWGDVEILSSHRQFVHL 598
L E RV+ N + L F+D+ ++ F+ G +WV+W ++ S Q +
Sbjct: 449 LIETRVKEENSQWLGSKL-FKDWSMLTNYEFNRRGRLWVVWRENVRFTPFYKSD-QLITC 506
Query: 599 RIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVT---KPWVTVGDFNTYLQASD 655
+ ++ F +FVY S A R LW +L++ KPW+ GDFN L +
Sbjct: 507 SVKLESQEEEFFYSFVYASNFAEERKILWNDLRDHMDSPIIRDKPWIIFGDFNEILDMDE 566
Query: 656 --KMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRG----VKERIDWALGNARW 709
+M P M F+ ++ CS SDL GP FTW + + +++D + N W
Sbjct: 567 HSRMEDHPAVTSGMRDFQSLVNYCSFSDLASHGPLFTWCNKRDNDPIWKKLDRVMVNEAW 626
Query: 710 ISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSS-----QRPFRFLAAWLTHEDFPRLVK 764
+P++ + DH I L +M+S +PF+F+ A E+F LV+
Sbjct: 627 KMVYPQSYNVFEAGGCSDHLRCRINL---NMNSGAQVRGNKPFKFVNAVADMEEFKPLVE 683
Query: 765 SSWDSHHDSWIPASEAFC---------EEAASWNRDVFGEIGRTKRRLMRRLEGINNHLR 815
+ W + S F + ++ G + + R L
Sbjct: 684 NFWRETEPIHMSTSSLFRFTKKLKALKPKLRGLAKEKMGNLVKRTREAYLSLCQAQQSNS 743
Query: 816 MSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNK 875
+ S +++ + + + EE +Q S+++WL GD+N FH +N
Sbjct: 744 QNPSQRAMEIESEAYVRWDRIASIEEKYLKQVSKLHWLKVGDKNNKTFHRAATARAAQNS 803
Query: 876 IEALNNDVGELITDQDQLRSMAVTSFQN-LYTSPGPGEPYHVRNAFPSLPRE----TLDE 930
I + + G T +D +++ FQ L P E V LP D
Sbjct: 804 IREIQKEDGSTATTKDDIKNETERFFQEFLQLIPNDYEGITVEKLTSLLPYHCSPAEKDM 863
Query: 931 IMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
+ + A EIR A+FSM K+PGPD F++ W +IG V
Sbjct: 864 LTASVSAKEIRGALFSMPNDKSPGPDGYTSEFYKRAWDIIGAEFV 908
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.141 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,973,990
Number of Sequences: 26719
Number of extensions: 1082081
Number of successful extensions: 3229
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 2967
Number of HSP's gapped (non-prelim): 130
length of query: 992
length of database: 11,318,596
effective HSP length: 109
effective length of query: 883
effective length of database: 8,406,225
effective search space: 7422696675
effective search space used: 7422696675
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0113.5


