

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0112.9
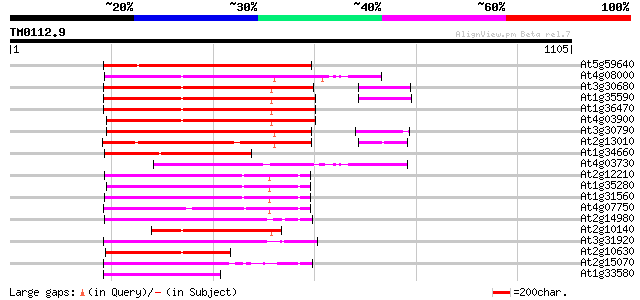
(1105 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59640 serine/threonine-specific protein kinase - like 449 e-126
At4g08000 putative transposon protein 418 e-117
At3g30680 hypothetical protein 415 e-116
At1g35590 CACTA-element, putative 413 e-115
At1g36470 hypothetical protein 406 e-113
At4g03900 putative transposon protein 404 e-112
At3g30790 hypothetical protein 402 e-112
At2g13010 pseudogene 388 e-108
At1g34660 hypothetical protein 336 5e-92
At4g03730 putative transposon protein 313 3e-85
At2g12210 putative TNP2-like transposon protein 308 1e-83
At1g35280 hypothetical protein 300 4e-81
At1g31560 putative protein 297 3e-80
At4g07750 putative transposon protein 279 7e-75
At2g14980 pseudogene 278 2e-74
At2g10140 putative protein 275 8e-74
At3g31920 hypothetical protein 256 5e-68
At2g10630 En/Spm transposon hypothetical protein 1 255 9e-68
At2g15070 En/Spm-like transposon protein 246 7e-65
At1g33580 En/Spm-like transposon protein, putative 192 7e-49
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 449 bits (1156), Expect = e-126
Identities = 214/413 (51%), Positives = 276/413 (66%), Gaps = 5/413 (1%)
Query: 186 KFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSC--SPEK 243
+ M + LP GN PK+ Y KK + LGL KID C CM+++KE E + C +
Sbjct: 104 QIMQEYLPEGNNSPKSYYEIKKLMRSLGLPYQKIDVCQDNCMIFWKETEKEEYCLFCKKD 163
Query: 244 RYKEVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDA 303
RY+ +K G +K +P ++M+Y P+ RL+RLY S TA HMRWH EH + HPSD
Sbjct: 164 RYRPTQKIG-QKSIPYRQMFYLPIADRLKRLYQSHNTAKHMRWHAEHLASDGEMGHPSDG 222
Query: 304 EAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRRE 363
EAW F + FA +PRNV LGLC+DGF PF +SG YS WPVI+TPYNLPP MCM++E
Sbjct: 223 EAWKHFHKVHPDFASKPRNVYLGLCTDGFNPFGMSGHNYSLWPVILTPYNLPPEMCMKQE 282
Query: 364 FMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTIND 423
FMFLTI++PGP+ PK+S+D++L+PLI+ELK LW +GV YD+S KQNFL+K +MWTI+D
Sbjct: 283 FMFLTILVPGPNHPKRSLDIFLQPLIEELKDLWVNGVEAYDISTKQNFLLKVVLMWTISD 342
Query: 424 FPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFT 483
FPAYGMLSGW T G+LACP C +++ AF L RK WFDCHR FL +H YR K F
Sbjct: 343 FPAYGMLSGWTTHGRLACPYCSDQTGAFWLKNGRKTCWFDCHRCFLPVNHSYRGNKKDFK 402
Query: 484 KGKGETSTKPRRLNGEEVWERIKHLPSKEE--SKEGPSQGYGVKHHWTSRSIFWELPYWK 541
KGK +KP L GEE++ + LP + G +GYG H+W +SI WEL YWK
Sbjct: 403 KGKVVEDSKPEILTGEELYNEVCCLPKTVDCGGNHGRLEGYGKTHNWHKQSILWELSYWK 462
Query: 542 TLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEV 594
L L HN+DVMH E+NV N + T+++ +GKTKD KSR DL+E C R L +
Sbjct: 463 DLKLRHNLDVMHIEKNVLDNFIKTLLNVQGKTKDNIKSRLDLQENCNRKDLHL 515
Score = 32.7 bits (73), Expect = 1.2
Identities = 16/39 (41%), Positives = 21/39 (53%)
Query: 751 IFNHAGRGYGKCITTTLDGMEVKSAHLYILLNCPEVTPY 789
IF+ AGR GK L + AH YIL NC ++ P+
Sbjct: 620 IFSQAGRDSGKESEIWLQDKDYHIAHRYILRNCDQLRPF 658
>At4g08000 putative transposon protein
Length = 609
Score = 418 bits (1075), Expect = e-117
Identities = 231/558 (41%), Positives = 316/558 (56%), Gaps = 39/558 (6%)
Query: 188 MSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--PEKRY 245
+ + LP GN + Y +K + LGL ID C CML++KEDE C +R+
Sbjct: 75 LEEFLPEGNQATSSHYQTEKLMRNLGLPYHTIDVCSNNCMLFWKEDEQEDHCRFCGAQRW 134
Query: 246 KEVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAEA 305
K + + K VP +MWY P+ RL+R+Y S +TA+ MRWH EH + ++HPSDA
Sbjct: 135 KPKDDRRRTK-VPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAE 193
Query: 306 WMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREFM 365
W F E FA+EPRNV LGLC+DGF PF +S + +S WPVI+TPYNLPP MCM +++
Sbjct: 194 WRYFQELHPRFAEEPRNVYLGLCTDGFNPFGMS-RNHSLWPVILTPYNLPPSMCMNTKYL 252
Query: 366 FLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDFP 425
FLTI+ P+ P+ S+DV+L+PLI+ELK LW GV YDVSL QNF +KA ++WTI+DFP
Sbjct: 253 FLTILNVEPNHPRASLDVFLQPLIEELKELWSTGVDAYDVSLSQNFNLKAVLLWTISDFP 312
Query: 426 AYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFTKG 485
AY MLSGW+T GK +CP+CME +K+F L RK WFDCHR+FL H R+ K F KG
Sbjct: 313 AYSMLSGWITHGKFSCPICMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKG 372
Query: 486 KGETST-KPRRLNGEEV-WERIKHL-PSKEESKEGPSQ-----GYGVKHHWTSRSIFWEL 537
+ +S P+ L GE+V +ER+ + P+K + G GYG +H+W SI WEL
Sbjct: 373 RDSSSEYPPKSLTGEQVYYERLASVNPTKTKDVGGNGHEKKMPGYGKEHNWHKESILWEL 432
Query: 538 PYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEVQQS 597
YW L L HNIDVMHTE+N +I+ T+M K K+KD SR D+E +C R L +
Sbjct: 433 SYWNDLNLRHNIDVMHTEKNFLDDIMNTLMSVKDKSKDNIMSRLDIEIFCSRSDLHIDSQ 492
Query: 598 ENGNFFKPK*IAHYGGKH----PNNFVQIRANISPKFL*LNGTPPNSFTV*SKSRRSCPL 653
G FF+ N V I N+ F PP+ F V P
Sbjct: 493 GIGAFFRDLCSRTLQKSRVQILKQNIVLIICNLEKIF------PPSFFDV----MEHLP- 541
Query: 654 *MDVSV*EVSS*IHLISLKFINSFPLRSFFHFLNRFIRTLKQKVTNKAHVEGSICKAYLL 713
IHL + + + RF + K K NK + GSI ++Y+
Sbjct: 542 ------------IHLPYEPELGGPVQYRWMYPFERFFKKFKGKAKNKRYAAGSIVESYIN 589
Query: 714 EEMSTFASYYYPPDVPSR 731
+E++ F+ YY+ + ++
Sbjct: 590 DEIAYFSEYYFADHIQTK 607
>At3g30680 hypothetical protein
Length = 1116
Score = 415 bits (1066), Expect = e-116
Identities = 209/422 (49%), Positives = 273/422 (64%), Gaps = 12/422 (2%)
Query: 186 KFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--PEK 243
+ +D LP GN + Y +K + LGL ID C CML++KEDE C +
Sbjct: 219 EMFTDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCKNNCMLFWKEDEKEDQCRFCGAQ 278
Query: 244 RYKEVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDA 303
R+K + + K VP +MWY P+ +L+R+Y S +TA+ MRWH EH + ++HPSDA
Sbjct: 279 RWKPKDDRRRTK-VPYSRMWYLPIADQLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDA 337
Query: 304 EAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRRE 363
W F E FA+EPRNV LGLC+DGF PF +S + +S WPVI+TPYNLPP MCM E
Sbjct: 338 AEWRYFQELHPRFAEEPRNVYLGLCTDGFNPFSMS-RNHSLWPVILTPYNLPPRMCMNTE 396
Query: 364 FMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTIND 423
++FLTI+ GP+ P+ S+DV+L+PLI+ELK LW GV YDVSL QNF +KA ++WTI+D
Sbjct: 397 YLFLTILNSGPNHPRASLDVFLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISD 456
Query: 424 FPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFT 483
FPAY MLSGW T GKL+CPVCME +K+F L RK WFDCHR+FL H R+ K F
Sbjct: 457 FPAYSMLSGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFL 516
Query: 484 KGKGETST-KPRRLNGEEV-WERIKHL-PSKEE-----SKEGPSQGYGVKHHWTSRSIFW 535
KG+ +S P L GE+V +ER+ + P K + E +GYG +H+W SI W
Sbjct: 517 KGRDASSEYPPESLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILW 576
Query: 536 ELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEVQ 595
EL YWK L L HNIDVMHTE+N NI+ T++ KGK+KD SR D+E+YC R GL +
Sbjct: 577 ELSYWKDLNLRHNIDVMHTEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHID 636
Query: 596 QS 597
+
Sbjct: 637 ST 638
Score = 58.2 bits (139), Expect = 3e-08
Identities = 31/104 (29%), Positives = 53/104 (50%), Gaps = 2/104 (1%)
Query: 688 RFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSRRTRVPRNDGECSSQNP 747
RF + LK K NK + GSI ++Y+ +E++ F+ +Y+ + ++ N+GE +
Sbjct: 742 RFFKKLKGKAKNKRYAAGSIVESYINDEIAYFSEHYFADHIQTKSRLTRFNEGEVPVYHV 801
Query: 748 P--LSIFNHAGRGYGKCITTTLDGMEVKSAHLYILLNCPEVTPY 789
P +IF GR G L + ++AH Y+L NC P+
Sbjct: 802 PGVPNIFMQVGRPSGAMHVDWLSEKDYQNAHAYVLRNCDYFKPF 845
>At1g35590 CACTA-element, putative
Length = 1180
Score = 413 bits (1061), Expect = e-115
Identities = 209/427 (48%), Positives = 273/427 (62%), Gaps = 12/427 (2%)
Query: 186 KFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--PEK 243
+ +D LP GN + Y +K + LGL ID C CML++KEDE C +
Sbjct: 219 EMFTDFLPEGNQATTSHYQTEKLMHNLGLPYHTIDVCKNNCMLFWKEDEKEDQCRFCGAQ 278
Query: 244 RYKEVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDA 303
R+K + + K VP +MWY P+ RL+R+Y +TA+ MRWH EH + ++HPSDA
Sbjct: 279 RWKPKDDRRRTK-VPYSRMWYLPIADRLKRMYQCHKTAAAMRWHAEHQSKEGEMNHPSDA 337
Query: 304 EAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRRE 363
W F E FA+EPRNV LGLC+DGF PF +S + +S WPVI+TPYNLPP MCM E
Sbjct: 338 AEWRYFQELHPRFAEEPRNVYLGLCTDGFNPFGMS-RNHSLWPVILTPYNLPPGMCMNTE 396
Query: 364 FMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTIND 423
++FLTI+ GP+ P+ S+DV+L+PLI+ELK LW GV Y+VSL QNF +KA ++WTI+D
Sbjct: 397 YLFLTILNYGPNHPRASLDVFLQPLIEELKELWCTGVDAYEVSLSQNFNLKAVLLWTISD 456
Query: 424 FPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFT 483
FPAY MLSGW T GKL+CPVCME +K+F L RK WFDCHR+FL H R+ K F
Sbjct: 457 FPAYNMLSGWTTHGKLSCPVCMESTKSFYLPIGRKTCWFDCHRRFLPHGHPSRRNKKDFL 516
Query: 484 KGKGETST-KPRRLNGEEV-WERIKHL-PSKEE-----SKEGPSQGYGVKHHWTSRSIFW 535
KG+ +S P L GE+V +ER+ + P K + E +GYG +H+W SI W
Sbjct: 517 KGRDASSEYPPESLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILW 576
Query: 536 ELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEVQ 595
EL YWK L L HNIDVMHTE+N NI+ T++ KGK+KD SR D+E+YC R GL +
Sbjct: 577 ELSYWKDLNLRHNIDVMHTEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHID 636
Query: 596 QSENGNF 602
+ F
Sbjct: 637 STGKAPF 643
Score = 50.4 bits (119), Expect = 5e-06
Identities = 29/105 (27%), Positives = 51/105 (47%), Gaps = 2/105 (1%)
Query: 688 RFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSRRTRVPRNDGECSSQNP 747
RF + LK K NK + SI K+Y+ +E++ F+ +Y+ + ++ ++GE +
Sbjct: 813 RFFKKLKGKAKNKRYAASSIVKSYINDEIAYFSEHYFTDHIQTKSRLTRFDEGEVHVYHV 872
Query: 748 P--LSIFNHAGRGYGKCITTTLDGMEVKSAHLYILLNCPEVTPYI 790
P +IF GR G L + ++AH Y+L N Y+
Sbjct: 873 PGVPNIFMQVGRPSGAMHVEWLSEKDYQNAHAYVLRNYSMFEDYL 917
>At1g36470 hypothetical protein
Length = 753
Score = 406 bits (1044), Expect = e-113
Identities = 210/428 (49%), Positives = 269/428 (62%), Gaps = 12/428 (2%)
Query: 185 VKFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--PE 242
V+ D LP GN + Y +K + LGL ID C CML +KEDE C
Sbjct: 50 VQSSRDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLSWKEDEKEDQCRFCGA 109
Query: 243 KRYKEVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSD 302
KR+K + + K VP +MWY P+ RL+R+Y S +TA+ MRWH EH + ++H SD
Sbjct: 110 KRWKPKDDRRRTK-VPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHLSD 168
Query: 303 AEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRR 362
A W F FA+EPRNV LGLC+DGF PF +S + +S WPVI+TPYNLPP MCM
Sbjct: 169 AAEWRYFQGLHPEFAEEPRNVYLGLCTDGFNPFGMS-RNHSLWPVILTPYNLPPGMCMNT 227
Query: 363 EFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTIN 422
E++FLTI+ GP+ P+ S+DV+L+PLI+ELK W GV YDVSL QNF +KA ++WTI+
Sbjct: 228 EYLFLTILNSGPNHPRASLDVFLQPLIEELKEFWSTGVDAYDVSLSQNFNLKAVLLWTIS 287
Query: 423 DFPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKF 482
DFPAY ML GW T GKL+CPVCME +K+F L RK WFDCHR+FL H R+ K F
Sbjct: 288 DFPAYNMLLGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNRKDF 347
Query: 483 TKGKGETST-KPRRLNGEEV-WERIKHL-PSKEE-----SKEGPSQGYGVKHHWTSRSIF 534
KG+ +S P L GE+V +ER+ + P K + E +GYG +H+W SI
Sbjct: 348 LKGRDASSEYPPESLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESIL 407
Query: 535 WELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEV 594
WEL YWK L L HNIDVMHTE+N NI+ T+M KGK+KD SR D+E++C R GL +
Sbjct: 408 WELSYWKDLNLRHNIDVMHTEKNFLDNIMNTLMRVKGKSKDNIMSRLDIEKFCSRPGLHI 467
Query: 595 QQSENGNF 602
S F
Sbjct: 468 DSSGKAPF 475
>At4g03900 putative transposon protein
Length = 817
Score = 404 bits (1039), Expect = e-112
Identities = 207/421 (49%), Positives = 268/421 (63%), Gaps = 12/421 (2%)
Query: 192 LPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--PEKRYKEVR 249
LP GN + Y +K + LGL ID CML++KEDE C +R+K
Sbjct: 225 LPEGNQATTSHYQTEKLMRNLGLPYHTIDVYKNNCMLFWKEDEKEDQCRFCGAQRWKPKD 284
Query: 250 KQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAEAWMKF 309
+ + K VP +MWY P+ RL+R+Y S +TA+ MRWH EH + ++HPSDA W F
Sbjct: 285 DRRRTK-VPYSRMWYLPIADRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYF 343
Query: 310 DEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREFMFLTI 369
E FA+EP NV LGL +DGF PF +S + +S WPVI+TPYNLPP MCM E++FLTI
Sbjct: 344 QELHPRFAEEPHNVYLGLYTDGFNPFGMS-RNHSLWPVILTPYNLPPGMCMNTEYLFLTI 402
Query: 370 MIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDFPAYGM 429
+ GP+ P+ S+DV+L+PLI+ELK LW GV YDVSL QNF +KA ++WTI+DFPAY M
Sbjct: 403 LNSGPNHPRASLDVFLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSM 462
Query: 430 LSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFTKGKGET 489
LSGW T GKL+CPVCME +K+F L RK WFDCHR+FL H R+ K F KG+ +
Sbjct: 463 LSGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDAS 522
Query: 490 ST-KPRRLNGEEV-WERIKHL-PSKEE-----SKEGPSQGYGVKHHWTSRSIFWELPYWK 541
S P L GE+V +ER+ + P K + E +GYG +H+W SI WEL YWK
Sbjct: 523 SEYPPESLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWK 582
Query: 542 TLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEVQQSENGN 601
L L HNIDVMHTE+N NI+ T++ KGK+KD SR D+E+YC R GL + +
Sbjct: 583 DLNLRHNIDVMHTEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHIDSTGKAP 642
Query: 602 F 602
F
Sbjct: 643 F 643
Score = 36.6 bits (83), Expect = 0.081
Identities = 14/44 (31%), Positives = 28/44 (62%)
Query: 688 RFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSR 731
RF + LK K NK + GSI ++Y+ +E++ F+ +Y+ + ++
Sbjct: 772 RFFKKLKGKAKNKRYAAGSIVESYINDEIAYFSEHYFADHIQTK 815
>At3g30790 hypothetical protein
Length = 953
Score = 402 bits (1034), Expect = e-112
Identities = 196/411 (47%), Positives = 267/411 (64%), Gaps = 9/411 (2%)
Query: 192 LPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSPEKRYKEVRKQ 251
LP N ++ + +K V LGL ID C CM+++ +D LQ C + +
Sbjct: 126 LPPDNISAESYFEIQKLVLSLGLPSEMIDVCIDYCMIFWGDDVNLQECRFCGKPRYQTTS 185
Query: 252 GKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAEAWMKFDE 311
G+ + VP ++MWY P+ RL+RLY S +TA+ MRWH +H + HPSD +AW F
Sbjct: 186 GRTR-VPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTTADGEIKHPSDTKAWKHFQT 244
Query: 312 AWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREFMFLTIMI 371
+ FA E RNV LGLC+DGF+PF +SG++YS W +I+TPYNLPP M M+ EF+FL+I++
Sbjct: 245 VYPDFASECRNVYLGLCTDGFSPFVLSGRQYSLWHIILTPYNLPPDMYMQSEFLFLSILV 304
Query: 372 PGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDFPAYGMLS 431
PGP PK+++DV+L+PLI ELK LW +GV T+D S KQNF M+A +MWTI+DFPAYGMLS
Sbjct: 305 PGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAVLMWTISDFPAYGMLS 364
Query: 432 GWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFTKGKGETST 491
GW T G+L+CP CM R+ AF L RK WFD HR+FL H YR+ K F K K
Sbjct: 365 GWTTHGRLSCPYCMGRTDAFQLKKGRKPCWFDYHRRFLPLHHPYRRNKKLFRKNKIVRLP 424
Query: 492 KPRRLNGEEVWERIKHLPSKEESKEGPS--------QGYGVKHHWTSRSIFWELPYWKTL 543
P ++G +++++I + ++E K G + YG H+W +SIFW+LPYWK L
Sbjct: 425 PPSYVSGTDLFKQIDYYGAQETCKRGGNWLTTANMPDEYGSTHNWHKQSIFWQLPYWKDL 484
Query: 544 LLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEV 594
L H +DVMH E+N F NI+ T+++ +GKTKD+ KSR DLEE C R L V
Sbjct: 485 FLRHCLDVMHIEKNFFDNIMNTLLNVQGKTKDSLKSRLDLEEICSRPELHV 535
Score = 55.8 bits (133), Expect = 1e-07
Identities = 38/109 (34%), Positives = 53/109 (47%), Gaps = 11/109 (10%)
Query: 682 FFHFLNRFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSRRTRVPRND-- 739
+ + +F+++LK K N A VEGSI L EE S F SYY+ P V +++TR R D
Sbjct: 683 WMYLFEQFMKSLKGKAKNLARVEGSIVAGSLTEETSHFTSYYFSPSVRTKKTRPRRYDDG 742
Query: 740 GECSSQNPP--LSIFNHAGRGYGKCITTTLDGMEVKSAHLYILLNCPEV 786
G + N P F GR GK L +++ A CPE+
Sbjct: 743 GVAPTYNVPDVTDTFAQIGRLAGKLKEVWLFDSQIREA-------CPEL 784
>At2g13010 pseudogene
Length = 1040
Score = 388 bits (997), Expect = e-108
Identities = 194/421 (46%), Positives = 264/421 (62%), Gaps = 24/421 (5%)
Query: 184 WVKFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--P 241
W + + + LP N ++ Y +K V+ GL ID C CM+++ +D LQ C
Sbjct: 368 WAELIKEYLPPDNISAESYYEIQKLVSSHGLPSEMIDVCIDYCMIFWGDDVNLQECRFCG 427
Query: 242 EKRYKEVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPS 301
+ RY+ G VP ++MWY P+ RL+RLY S +TA+ MRWH +H ++HPS
Sbjct: 428 KPRYQTT---GGRTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTTADGEITHPS 484
Query: 302 DAEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMR 361
DA+AW F + FA+E RNV LGLC+DGF+PF + G++YS WPVI+TPY LPP MCM+
Sbjct: 485 DAKAWKHFQTVYPDFANECRNVYLGLCTDGFSPFGLCGRQYSLWPVILTPYKLPPDMCMQ 544
Query: 362 REFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTI 421
EFMFL+I++PGP PK+++DV+L+PLI ELK LW +GV T+D S KQNF M+A +MWTI
Sbjct: 545 SEFMFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAMLMWTI 604
Query: 422 NDFPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKK 481
+DFPAY MLSGW G+L P RK WF+CHR+FL H YR+ K
Sbjct: 605 SDFPAYRMLSGWTMHGRLYVP-----------TKDRKPCWFNCHRRFLPLHHPYRRNKKL 653
Query: 482 FTKGKGETSTKPRRLNGEEVWERIKHLPSKEESKEGPS--------QGYGVKHHWTSRSI 533
F K K P ++G +++E+I + ++E K G + YG H+W +SI
Sbjct: 654 FRKNKIVRLLPPSYVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDEYGSTHNWYKQSI 713
Query: 534 FWELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLE 593
FW+LPYWK LLL HN+ VMH E+N F NI+ T+++ +GKTKDT KSR DL E C + L
Sbjct: 714 FWQLPYWKDLLLRHNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRLDLAEICSKPELH 773
Query: 594 V 594
V
Sbjct: 774 V 774
Score = 55.5 bits (132), Expect = 2e-07
Identities = 41/101 (40%), Positives = 53/101 (51%), Gaps = 7/101 (6%)
Query: 688 RFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSRRTRVPR---NDGECSS 744
RF+++LK K N A VEGSI L EE S F SYY+ P V +++TR PR + G +
Sbjct: 928 RFMKSLKGKAKNLARVEGSIVAGSLTEETSHFTSYYFSPSVRTKKTR-PRCYDDGGVAPT 986
Query: 745 QNPP--LSIFNHAGRGYGKCITTTLDGMEV-KSAHLYILLN 782
N P F GR GK D + ++AH YIL N
Sbjct: 987 YNVPDVPDTFAQIGRLAGKLKEVWWDDHTLSRAAHNYILRN 1027
>At1g34660 hypothetical protein
Length = 501
Score = 336 bits (861), Expect = 5e-92
Identities = 152/289 (52%), Positives = 208/289 (71%), Gaps = 2/289 (0%)
Query: 187 FMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSPEKRYK 246
F+ D LP N P + Y +K V+ LGL ID C CM+++++ L +C+ ++ +
Sbjct: 190 FVKDLLPEPNLAPGSYYEIQKLVSSLGLPYQMIDVCVDNCMIFWRDTVNLDACTFCQKPR 249
Query: 247 EVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAEAW 306
GK + VP ++MWY P+ RL+RLY S +TA MRWH EH + + +SHPSDAEAW
Sbjct: 250 FQVTSGKSR-VPFKRMWYLPITDRLKRLYQSERTAGPMRWHAEHSSDGN-ISHPSDAEAW 307
Query: 307 MKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREFMF 366
F+ + FA+E RNV LGLC+DGF PF SG+KYS WPVI+TPYNLPP +CM+REF+F
Sbjct: 308 KHFNSMYKDFANEHRNVYLGLCTDGFNPFGKSGRKYSLWPVILTPYNLPPSLCMKREFLF 367
Query: 367 LTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDFPA 426
L+I++PGP P++S+DV+L+PLI EL++LW +GV+ YDVSLK NF+M+A +MWTI+DFPA
Sbjct: 368 LSILVPGPEHPRRSLDVFLQPLIHELQLLWSEGVVAYDVSLKNNFVMRAVLMWTISDFPA 427
Query: 427 YGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEY 475
YGMLSGW T G+L+CP C + AF L + +K WFDCHR+FL ++H Y
Sbjct: 428 YGMLSGWTTHGRLSCPYCQDTIDAFQLKHGKKSCWFDCHRRFLPKNHSY 476
>At4g03730 putative transposon protein
Length = 696
Score = 313 bits (803), Expect = 3e-85
Identities = 186/503 (36%), Positives = 253/503 (49%), Gaps = 52/503 (10%)
Query: 284 MRWHHEHYKEPDFLSHPSDAEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYS 343
MRWH EH + +PSD AW F+E + FA E RN+ LGL +DGF ++G+ +S
Sbjct: 3 MRWHAEHMSPEGEMHNPSDGAAWKNFNEVYPNFAAESRNIYLGLSTDGFNLVSMNGEAHS 62
Query: 344 CWPVIVTPYNLPPWMCMRREFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTY 403
W VIVTPYNLP MC++REF FL++++PGP PKK++D+YL+PLI EL LW DG Y
Sbjct: 63 VWHVIVTPYNLPSGMCIKREFFFLSVLVPGPKHPKKNLDIYLQPLIKELVSLWIDGEEAY 122
Query: 404 DVSLKQNFLMKAAVMWTINDFPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFD 463
D S K+ F M+ A+ W I+DF YGMLS WMT G L+CP C++ + +F L RK SWFD
Sbjct: 123 DKSKKKKFTMRVALTWAISDFSTYGMLSVWMTHGLLSCPYCLDNTISFWLPNERKHSWFD 182
Query: 464 CHRQFLSEDHEYRKCPKKFTKGKGETSTKPRRLN--GEEVWERIKHLPSKEESKEGPSQG 521
CH FL H Y + + F GK P ++ G E E G
Sbjct: 183 CHGMFLPLGHMYMQNVQAFRGGKEVRDDPPFTVDCGGNE-----------HEKFAQTIDG 231
Query: 522 YGVKHHWTSRSIFWELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRK 581
YG+ H+W RSIFWELPYW+ L+L HN+D MH ++N F N++ TV+ KTKD KSR
Sbjct: 232 YGIHHNWVKRSIFWELPYWENLILRHNLDFMHIKKNFFDNLINTVLKVLEKTKDKIKSRM 291
Query: 582 DLEEYCLRGGLEVQQSENGNFFKPK*IAHYGGKHPNNFVQIRANISPKFL*LNGTPPNSF 641
DL C R GL + + I G N +I PP+ F
Sbjct: 292 DLATLCNRSGLHLTRDGT--------ILQILGLKLCNLEKI-------------FPPSFF 330
Query: 642 TV*SKSRRSCPL*MDVSV*EVSS*IHLISLKFINSFPLRSFFHFLNRFIRTLKQKVTNKA 701
V P HL ++ + + R++ LK+KV N A
Sbjct: 331 DV----MEHLP-------------DHLPDEALLSGPVQYRWMYLFERYMYHLKRKVKNCA 373
Query: 702 HVEGSICKAYLLEEM-STFASYYYPPDVPSRRTRVPRNDGECSSQNPPLSIFNHAGRGYG 760
H+ GSI + EE+ +T A+Y+ P++ V S+F H GR G
Sbjct: 374 HIAGSIVAQCVNEEIANTSANYFGHPEIKDDDVPVSGEIRFTYFYPDVSSLFYHEGRVSG 433
Query: 761 KCITTTLDGMEVKSAHLYILLNC 783
K + + ++LLNC
Sbjct: 434 KLTNGWFNDNDYTVLQTFLLLNC 456
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 308 bits (788), Expect = 1e-83
Identities = 167/437 (38%), Positives = 245/437 (55%), Gaps = 35/437 (8%)
Query: 188 MSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSP--EKRY 245
+ + LP N + +LY KK + + KI C C L+ K + L++C R+
Sbjct: 173 LPEMLPADNVLHTSLYEVKKFLRSFDMGYEKIHACVNDCCLFRKRYKNLENCPKCNASRW 232
Query: 246 KEVRKQGK-EKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAE 304
K + G+ +K VPQ+ + YFP++PR +R++ S + A +RWH+ + L HP D+
Sbjct: 233 KSNMQTGEAKKGVPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHYSNKSTDGKLRHPVDSV 292
Query: 305 AWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREF 364
W K ++ + FA E RN+RLGL +DGF PF++ +YSCWPV++ YNLPP +CM++E
Sbjct: 293 TWDKMNDKYPEFAIEERNLRLGLSTDGFNPFNMKNTRYSCWPVLLVNYNLPPDLCMKKEN 352
Query: 365 MFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDF 424
+ L+++IPGP P SIDVYL PLI++L LW+ G +TYD K F +KA ++WTI+DF
Sbjct: 353 IMLSLLIPGPQQPGNSIDVYLEPLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTISDF 412
Query: 425 PAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFTK 484
PAYG L+G GK+ CP+C + + + L YSRK F CHR+ L+ H YR K +
Sbjct: 413 PAYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRK-HVFMCHRKGLAPTHSYRN-KKAWFD 470
Query: 485 GKGETSTKPRRLNGEEVWERIKHLP-----------------------------SKEESK 515
GK E K R L G EV + +K+ S+ E
Sbjct: 471 GKIEHGRKARILTGREVSQTLKNFKNDFGNAKESGRKRKRNDCIESVTDSDDESSESEED 530
Query: 516 EGPSQGYGVKHHWTSRSIFWELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKD 575
E W RSIF++L YW+ L + HN+DVMH ERNV +I+ T++ GK+KD
Sbjct: 531 EELEVDEDELSRWKKRSIFFKLSYWEDLPVRHNLDVMHVERNVGASIVSTLLHC-GKSKD 589
Query: 576 TFKSRKDLEEYCLRGGL 592
+RKDL++ +R L
Sbjct: 590 GLNARKDLQDLGVRKDL 606
>At1g35280 hypothetical protein
Length = 557
Score = 300 bits (767), Expect = 4e-81
Identities = 165/433 (38%), Positives = 241/433 (55%), Gaps = 35/433 (8%)
Query: 192 LPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSP--EKRYKEVR 249
LP N + +LY KK + + KI C C L+ K + L++C R+K
Sbjct: 2 LPADNVLHTSLYEVKKFLKSFDMGYEKIHACVNDCCLFRKRYKNLENCPKCNASRWKSNM 61
Query: 250 KQGK-EKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAEAWMK 308
+ G+ +K VPQ+ + YFP++PR +R++ S + A +RWH+ + L HP D+ W K
Sbjct: 62 QTGEAKKGVPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHYSNKSIDGKLRHPVDSVTWDK 121
Query: 309 FDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREFMFLT 368
++ + FA E RN+RLGL +DGF PF++ +YSCWPV++ YNLP +CM++E + L+
Sbjct: 122 MNDKYPEFAAEERNLRLGLSTDGFNPFNMKNTRYSCWPVLLVNYNLPLDLCMKKENIMLS 181
Query: 369 IMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDFPAYG 428
++IPGP P SIDVYL PLI++L LW+ G +TYD K F +KA ++WTI+DF AYG
Sbjct: 182 LLIPGPQQPGNSIDVYLEPLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTISDFSAYG 241
Query: 429 MLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFTKGKGE 488
L+G GK+ CP+C + + + L YSRK F CHR+ L+ H YR K + GK E
Sbjct: 242 NLAGCKVKGKMGCPLCGKGTDSMWLKYSRK-HVFMCHRKGLAPTHSYRN-KKAWFDGKIE 299
Query: 489 TSTKPRRLNGEEVWERIKHLP-----------------------------SKEESKEGPS 519
K R L G EV + +K+ S+ E E
Sbjct: 300 HGRKARILTGREVSQTLKNFKNDFRNAKESGRKRKRNDCIESVTDSDDESSESEEDEELE 359
Query: 520 QGYGVKHHWTSRSIFWELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKS 579
W RSIF++L YW+ L + HN+DVMH ERNV +I+ T++ GK+KD +
Sbjct: 360 VDKDELSRWKKRSIFFKLSYWEDLPVRHNLDVMHVERNVGASIVSTLLHC-GKSKDGLNA 418
Query: 580 RKDLEEYCLRGGL 592
RKDL++ +R L
Sbjct: 419 RKDLQDLGVRKDL 431
>At1g31560 putative protein
Length = 1431
Score = 297 bits (760), Expect = 3e-80
Identities = 163/437 (37%), Positives = 237/437 (53%), Gaps = 35/437 (8%)
Query: 188 MSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSP--EKRY 245
+ + LP N + +LY KK + + KI C C L+ K + L++C R+
Sbjct: 104 LPEMLPADNVLRTSLYEVKKFLKSFDMGYEKIHACVNDCCLFRKRYKNLENCPKCNASRW 163
Query: 246 KEVRKQGK-EKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAE 304
K + G+ +K VPQ+ + YF ++PR ++++ S + A +RWH+ + HP D+
Sbjct: 164 KNYMQTGEAKKGVPQKVLRYFLIIPRFKKMFRSKEMAKDLRWHYSNKSTDGKHRHPVDSV 223
Query: 305 AWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREF 364
W K + + FA E RN+RLGL +DGF PF++ +YSCWPV++ YNLPP CM+ E
Sbjct: 224 TWDKMIDKYPEFAAEERNMRLGLSTDGFNPFNMKNTRYSCWPVLLVNYNLPPDQCMKNEN 283
Query: 365 MFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDF 424
+ L+++IPGP P SIDVYL P I++L LW+ G +TYD K F +KA + WTI+DF
Sbjct: 284 IMLSLLIPGPQQPGNSIDVYLEPFIEDLNHLWKKGELTYDAFSKTTFTLKAMLFWTISDF 343
Query: 425 PAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFTK 484
PAYG L+G GK+ CP+C + + + L YSRK F CHR+ L+ H Y+ K +
Sbjct: 344 PAYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRK-HVFMCHRKGLAPTHSYKN-KKAWFD 401
Query: 485 GKGETSTKPRRLNGEEVWERIKHLP-----------------------------SKEESK 515
GK E K R L G EV +K+ S+ E
Sbjct: 402 GKVEHGRKARILTGREVSHNLKNFKNDFGNAKESGRKRKRNDCIESVTDSDNESSESEED 461
Query: 516 EGPSQGYGVKHHWTSRSIFWELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKD 575
E W RSIF++L YW+ L + HN+DVMH ERNV +I+ T++ GK+KD
Sbjct: 462 EEVEVDEDELSRWKKRSIFFKLSYWENLPVRHNLDVMHVERNVGASIVSTLLHC-GKSKD 520
Query: 576 TFKSRKDLEEYCLRGGL 592
SRKDL++ +R L
Sbjct: 521 GLNSRKDLQDLGVRKDL 537
Score = 30.8 bits (68), Expect = 4.5
Identities = 25/103 (24%), Positives = 44/103 (42%), Gaps = 8/103 (7%)
Query: 688 RFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSRRTRVPRNDGECSSQNP 747
R+++ LK V N + I ++YL EE F S + ++T E +++
Sbjct: 593 RYMKVLKDFVRNSVKPDVCIAESYLAEECMQFCSDFL------KKTTSVEEKPERNTEYE 646
Query: 748 PLSIFNHAGRGYGKCITTTLDGMEVKSAHLYILLNCPEVTPYI 790
SI GR + L ++ AHL ++ N + PY+
Sbjct: 647 NNSILE--GRPISAGTSFMLTEVDKNIAHLAVIQNMAALDPYV 687
>At4g07750 putative transposon protein
Length = 569
Score = 279 bits (713), Expect = 7e-75
Identities = 155/440 (35%), Positives = 236/440 (53%), Gaps = 45/440 (10%)
Query: 185 VKFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSP--E 242
++ + + LP N + +LY KK + + KI C C L+ K + L SC
Sbjct: 111 LEILPEMLPKDNVLHTSLYKVKKFLKSFDMGYQKIHACVNDCCLFRKNYKKLDSCPKCNA 170
Query: 243 KRYKEVRKQGK-EKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPS 301
R+K G+ +K VPQ+ + YFP++PRL+ ++ S + +RWH + + L HP
Sbjct: 171 SRWKTNLHIGEIKKGVPQKVLRYFPIIPRLKGMFRSKEMDKDLRWHFSNKSSDEKLCHPV 230
Query: 302 DAEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMR 361
D W + + + +FA E RN+R GL +DGF PF++ +YSCWP + CM+
Sbjct: 231 DLVTWDQMNAKYPSFAGEERNLRHGLSTDGFNPFNMKNTRYSCWPDL----------CMK 280
Query: 362 REFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTI 421
+E + LT++IPGP P +IDVYL PLI++L LW++G +TYD K F +KA ++WTI
Sbjct: 281 KENIMLTLLIPGPQQPGNNIDVYLEPLIEDLCHLWDNGELTYDAYSKSTFDLKAMLLWTI 340
Query: 422 NDFPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKK 481
+DFPAYG L+G GK+ CP+C + + + L +SRK + CH + L H +R K
Sbjct: 341 SDFPAYGNLAGCKVKGKMGCPLCGKNTDSMWLKFSRKHVYM-CHIKGLPPTHHFRM-KKS 398
Query: 482 FTKGKGETSTKPRRLNGEEVWERIKHLP-----------------------------SKE 512
+ GK E K R L+G+E+++ +K+ S+
Sbjct: 399 WFDGKAENGRKRRILSGQEIYQNLKNFKNNFGTLKQSASKRKRTVNIEADSDSDDQSSES 458
Query: 513 ESKEGPSQGYGVKHHWTSRSIFWELPYWKTLLLPHNIDVMHTERNVFLNILFTVMDTKGK 572
E E W +SIF++LPYW+ LL+ HN+DVMH ERNV +I+ T++ K
Sbjct: 459 EEDEQDQVDEEELSRWKKKSIFFKLPYWQELLVRHNLDVMHVERNVAASIISTLLHC-WK 517
Query: 573 TKDTFKSRKDLEEYCLRGGL 592
+KD +RKDLE +R L
Sbjct: 518 SKDGLNARKDLEVLGIRKDL 537
>At2g14980 pseudogene
Length = 665
Score = 278 bits (710), Expect = 2e-74
Identities = 158/411 (38%), Positives = 231/411 (55%), Gaps = 20/411 (4%)
Query: 188 MSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSP--EKRY 245
+ + LP N + +LY KK + + KI C C L+ K+ + LQ+C R+
Sbjct: 66 LPEMLPEDNVLHTSLYEVKKFLKSFDMGYQKIHACVNDCCLFRKKYKKLQNCLKCNASRW 125
Query: 246 KEVRKQGK-EKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAE 304
K G+ +K VPQ+ + YF +VPRL+R++ S + A +RWH + L HP D+
Sbjct: 126 KTNMHTGEVKKGVPQKVLRYFSIVPRLKRMFRSEEMARDLRWHFHNKSTDGKLCHPVDSV 185
Query: 305 AWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREF 364
W + + + FA E RN+RLGL ++GF PF++ +YSCWPV++ YNLPP +CM++E
Sbjct: 186 TWDQMNAKYPLFASEERNLRLGLSTNGFNPFNMKNTRYSCWPVLLVNYNLPPDLCMKKEN 245
Query: 365 MFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDF 424
+ LT++IPG P SIDVYL PLID+L LW++G TYD K +F +KA ++WTI+DF
Sbjct: 246 IMLTLLIPGSQQPGNSIDVYLEPLIDDLCHLWDNGEWTYDAFSKSSFTLKAMLLWTISDF 305
Query: 425 PAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKKFTK 484
PAYG L+G GK+AC +C + +++ L +SRK + HR L H +R K +
Sbjct: 306 PAYGNLAGCKVKGKMACLLCGKHTESMWLKFSRKHVYMG-HRMGLPPSHRFRN-KKSWFD 363
Query: 485 GKGETSTKPRRLNGEEVWERIKHLPSKEESKEGPSQGYGVKHHWTSRSIFWELPYWKTLL 544
GK E K R L+G E+ +K+ +G T + E L
Sbjct: 364 GKAEHRRKSRILSGLEISHNLKNF----------QNNFGNFKQSTRKRKQPE----SDLP 409
Query: 545 LPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEVQ 595
+ HN+DVMH ERNV +I+ T+M GK+KD +RKDLE +R L Q
Sbjct: 410 VRHNLDVMHVERNVVASIVSTLMHC-GKSKDGVNARKDLEVLGIRKDLHPQ 459
Score = 42.0 bits (97), Expect = 0.002
Identities = 31/104 (29%), Positives = 48/104 (45%), Gaps = 8/104 (7%)
Query: 688 RFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSRRTRVPRNDGECSSQNP 747
R+++ LK V N A EG I + YL EE F S + ++T + + +++
Sbjct: 570 RYMKVLKDFVRNTARPEGCIAECYLAEECIQFCSEFL------KKTTKVQEKADRNTEYE 623
Query: 748 PLSIFNHAGRGYGKCITTTLDGMEVKSAHLYILLNCPEVTPYIK 791
SI GR +L ME K AHL ++ N V PY++
Sbjct: 624 NNSILG--GRPISAGTFISLTEMERKIAHLAVIQNMAVVEPYVE 665
>At2g10140 putative protein
Length = 663
Score = 275 bits (704), Expect = 8e-74
Identities = 139/264 (52%), Positives = 175/264 (65%), Gaps = 9/264 (3%)
Query: 279 QTASHMRWHHEHYKEPDFLSHPSDAEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDIS 338
+TA+ MRWH EH + ++HPSDA W F E FA EPRNV LGLC DGF PF IS
Sbjct: 244 KTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHTWFAKEPRNVYLGLCIDGFNPFGIS 303
Query: 339 GKKYSCWPVIVTPYNLPPWMCMRREFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWED 398
+ +S WPVI+T YNLPP MCM +++FLTI+ GP+ P+ S+DV+L+PLI+ELK LW
Sbjct: 304 -RNHSLWPVILTSYNLPPGMCMNTKYLFLTILNSGPNHPRASLDVFLQPLIEELKELWCT 362
Query: 399 GVMTYDVSLKQNFLMKAAVMWTINDFPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRK 458
GV TYDVSL QNF +KA ++W I+DFPAY MLSGW T GKL+CPVCME +K+F L RK
Sbjct: 363 GVDTYDVSLSQNFNLKAVLLWAISDFPAYSMLSGWTTHGKLSCPVCMESTKSFYLPNGRK 422
Query: 459 VSWFDCHRQFLSEDHEYRKCPKKFTKGK-GETSTKPRRLNGEEV-WERIKHL-PSKEE-- 513
WFDCHR+FL H R+ K F KG+ P L GE+V +ER+ + P K +
Sbjct: 423 TCWFDCHRRFLPHSHPSRRNKKDFLKGRDASREYPPGSLTGEQVYYERLASVNPPKTKDV 482
Query: 514 ---SKEGPSQGYGVKHHWTSRSIF 534
E +GYG +H+W SIF
Sbjct: 483 GGNGHEKKMRGYGKEHNWDKESIF 506
>At3g31920 hypothetical protein
Length = 725
Score = 256 bits (654), Expect = 5e-68
Identities = 150/426 (35%), Positives = 225/426 (52%), Gaps = 32/426 (7%)
Query: 185 VKFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCSP--E 242
+ + D LP N +P + KK + G I C C+LY KE E+ +C
Sbjct: 178 LSLVHDILPGENVLPTSTNEIKKFLKMFGFGYDIIHACQNDCILYRKEYELRDTCPRCNA 237
Query: 243 KRYKEVRKQGKEKF-VPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPS 301
R++ + G+EK +P + + YFP+ R +R++ S + A RWH + + + H
Sbjct: 238 SRWERDKHTGEEKKGIPAKVLGYFPIKDRFKRMFRSKKMAEDQRWHFSNASDDGTMRHHV 297
Query: 302 DAEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMR 361
D+ AW + ++ W FA EPRN+RLGL +DG PF I KYS WP+++ YN+ P MCM+
Sbjct: 298 DSLAWAQVNDKWPEFAAEPRNLRLGLSADGMNPFSIQNTKYSTWPILLVNYNMAPTMCMK 357
Query: 362 REFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTI 421
E + LT++I GP+AP +IDVYL PLID+LK LW +G+ YD K++F ++A +W+I
Sbjct: 358 AENIMLTLLIHGPTAPSNNIDVYLAPLIDDLKDLWSEGIHVYDSFKKESFTLRAMFLWSI 417
Query: 422 NDFPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKK 481
+D+PA G L+G GK AC VC + + L +SRK + +R+ L H YR+ +
Sbjct: 418 SDYPALGTLAGCKVKGKQACNVCGKDTPFRWLKFSRKHVYLR-NRRRLRPGHPYRR-RRG 475
Query: 482 FTKGKGETSTKPRRLNGEEVWERIKHLPSKEESKEGPSQGYGVKHHWTSRSIFWELPYWK 541
+ E T R GEE++E +K L + I +L W+
Sbjct: 476 WFDNTVEEGTTSRIQTGEEIFEALKAL----------------------KMILGDL--WR 511
Query: 542 TLLLP--HNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEVQQSEN 599
+P HNIDVMH E+NV I+ +M + K+KD +RKDLE+ R L Q
Sbjct: 512 ETDMPVRHNIDVMHVEKNVSDAIVSMLMQSV-KSKDGLNARKDLEDMGTRSNLHPQLRVK 570
Query: 600 GNFFKP 605
+ P
Sbjct: 571 RTYLPP 576
>At2g10630 En/Spm transposon hypothetical protein 1
Length = 515
Score = 255 bits (652), Expect = 9e-68
Identities = 126/249 (50%), Positives = 164/249 (65%), Gaps = 4/249 (1%)
Query: 189 SDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--PEKRYK 246
+D LP N + Y +K + LGL ID C CML++KE+E C KR+K
Sbjct: 216 TDFLPEENQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLFWKEEEKEDQCRFCGAKRWK 275
Query: 247 EVRKQGKEKFVPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPSDAEAW 306
+ + K VP MWY P+ RL+R+Y S +TA+ MRWH EH + ++HPSDA W
Sbjct: 276 PKDDRRRTK-VPYSCMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEW 334
Query: 307 MKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMRREFMF 366
F FA++PRNV LGLC+DGF PF +S + +S WPVI+TPYNLPP MCM E++F
Sbjct: 335 RYFQGLHPQFAEDPRNVYLGLCTDGFNPFGMS-RNHSLWPVILTPYNLPPGMCMNTEYLF 393
Query: 367 LTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTINDFPA 426
LTI+ GP+ P+ S+DV+L+PLI+ELK LW GV YDVSL QNF +KA ++WTI+DFPA
Sbjct: 394 LTILNSGPNHPRASLDVFLQPLIEELKELWSTGVDAYDVSLSQNFNLKAVLLWTISDFPA 453
Query: 427 YGMLSGWMT 435
Y MLSGW T
Sbjct: 454 YSMLSGWTT 462
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 246 bits (627), Expect = 7e-65
Identities = 142/414 (34%), Positives = 206/414 (49%), Gaps = 62/414 (14%)
Query: 185 VKFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSC--SPE 242
+ + D LP N +P + KK + G I CP C+LY KE E+ +C
Sbjct: 103 LSLVHDILPGENVLPTSTNEIKKFLKMFGFGYDIIHACPNDCILYKKEYELRDTCLGCSA 162
Query: 243 KRYKEVRKQGKEKF-VPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPS 301
R+K + G+EK +P + +WYF + R +R++ S + ++WH + + HP
Sbjct: 163 SRWKRDKHTGEEKKGIPAKVLWYFSIKDRFKRMFRSKKMVEDLQWHFSNASVDGTMRHPI 222
Query: 302 DAEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMR 361
D+ AW + ++ W FA E RN+RLGL +D PF I KYS WPV++ YN+ P MCM+
Sbjct: 223 DSLAWAQVNDKWPQFAAESRNLRLGLSTDKMNPFSIQNTKYSTWPVLLVNYNMAPTMCMK 282
Query: 362 REFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMKAAVMWTI 421
+ + LT++I GP AP +IDVYL PLI++LK L DG+ YD K+NF ++A ++W+I
Sbjct: 283 AKNIMLTLLIHGPKAPSNNIDVYLAPLINDLKDLSSDGIQVYDSFFKENFTLRAMLLWSI 342
Query: 422 NDFPAYGMLSGWMTMGKLACPVCMERSKAFSLAYSRKVSWFDCHRQFLSEDHEYRKCPKK 481
ND+P G L+G C + F R R L ED EY
Sbjct: 343 NDYPTLGTLAG-----------CKDFKNDFGRPLERNNK---RKRSDLCEDDEY------ 382
Query: 482 FTKGKGETSTKPRRLNGEEVWERIKHLPSKEESKEGPSQGYGVKHHWTSRSIFWELPYWK 541
GE+ + W RSI +ELPYWK
Sbjct: 383 ----------------GEDTHQ----------------------WRWKKRSILFELPYWK 404
Query: 542 TLLLPHNIDVMHTERNVFLNILFTVMDTKGKTKDTFKSRKDLEEYCLRGGLEVQ 595
L + HNIDVMH E+NV I+ T+M + K+KD +RK+LE+ +R L +Q
Sbjct: 405 DLPVRHNIDVMHVEKNVSDVIVSTLMQS-AKSKDGLNARKNLEDMGIRSNLHIQ 457
Score = 42.7 bits (99), Expect = 0.001
Identities = 32/106 (30%), Positives = 46/106 (43%), Gaps = 7/106 (6%)
Query: 686 LNRFIRTLKQKVTNKAHVEGSICKAYLLEEMSTFASYYYPPDVPSRRTRVPRNDGECSSQ 745
L R+++TLK V N A +E + + YL E F + + V RN+G S+
Sbjct: 574 LERYMKTLKAYVKNFATLEACMAEGYLAGECMAFCLKFLQNSLSVPEV-VNRNEGVDSNS 632
Query: 746 NPPLSIFNHAGRGYGKCITTTLDGMEVKSAHLYILLNCPEVTPYIK 791
GR K TL E AH Y+L+N + PY+K
Sbjct: 633 ------LVLEGRPLHKATQVTLTSKERDIAHRYVLMNMAVMDPYVK 672
>At1g33580 En/Spm-like transposon protein, putative
Length = 428
Score = 192 bits (489), Expect = 7e-49
Identities = 93/233 (39%), Positives = 137/233 (57%), Gaps = 3/233 (1%)
Query: 185 VKFMSDALPNGNCMPKNLYHAKKTVAELGLACVKIDCCPGGCMLYYKEDEMLQSCS--PE 242
+ + D LP N + + KK + G I CP C+LY KE E+ +C
Sbjct: 185 LSLVHDILPGENVLLTSTNEIKKFLKMFGFGYDIIHACPNDCILYRKEYELRDTCPRCSA 244
Query: 243 KRYKEVRKQGKEKF-VPQQKMWYFPLVPRLQRLYSSMQTASHMRWHHEHYKEPDFLSHPS 301
R++ + G+EK +P + + YFP+ R +R++ S + A +RWH + + + HP
Sbjct: 245 FRWERDKHTGEEKKGIPAKVLRYFPIKDRFKRMFRSKKMAEDLRWHFSNASDDGTMRHPV 304
Query: 302 DAEAWMKFDEAWGTFADEPRNVRLGLCSDGFAPFDISGKKYSCWPVIVTPYNLPPWMCMR 361
D+ AW + + W FA EPRN+RLGL +DG PF I KYS WPV++ YN+ P MCM+
Sbjct: 305 DSLAWAQVNYKWPQFAAEPRNLRLGLSTDGMNPFSIQNTKYSTWPVLLVNYNMAPTMCMK 364
Query: 362 REFMFLTIMIPGPSAPKKSIDVYLRPLIDELKMLWEDGVMTYDVSLKQNFLMK 414
E + LT++IPGP+ P +IDVYL PLID+LK LW DG+ YD LK++F ++
Sbjct: 365 AENIMLTLLIPGPTTPNNNIDVYLAPLIDDLKDLWSDGIQVYDSFLKESFTLR 417
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.348 0.152 0.547
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,094,068
Number of Sequences: 26719
Number of extensions: 1011863
Number of successful extensions: 4063
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 3931
Number of HSP's gapped (non-prelim): 59
length of query: 1105
length of database: 11,318,596
effective HSP length: 110
effective length of query: 995
effective length of database: 8,379,506
effective search space: 8337608470
effective search space used: 8337608470
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0112.9


