

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0112.15
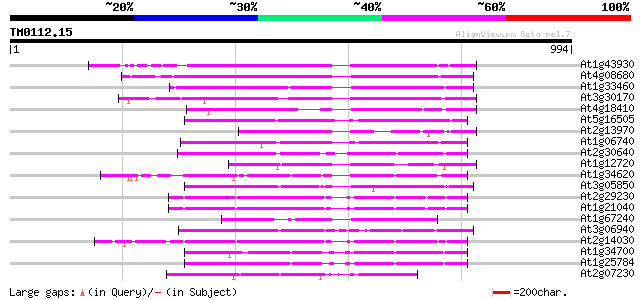
(994 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g43930 hypothetical protein 256 3e-68
At4g08680 putative MuDR-A-like transposon protein 249 5e-66
At1g33460 mutator transposase MUDRA, putative 240 3e-63
At3g30170 unknown protein 226 4e-59
At4g18410 MuDR transposable element - like protein 190 4e-48
At5g16505 mutator-like transposase-like protein (MQK4.25) 183 4e-46
At2g13970 Mutator-like transposase 181 2e-45
At1g06740 mudrA-like protein 181 2e-45
At2g30640 Mutator-like transposase 179 7e-45
At1g12720 hypothetical protein 177 3e-44
At1g34620 Mutator-like protein 171 2e-42
At3g05850 unknown protein 170 4e-42
At2g29230 Mutator-like transposase 169 5e-42
At1g21040 hypothetical protein 169 5e-42
At1g67240 hypothetical protein 164 3e-40
At3g06940 putative mudrA protein 160 3e-39
At2g14030 Mutator-like transposase 153 4e-37
At1g34700 hypothetical protein 138 2e-32
At1g25784 mutator-like transposase, putative 136 7e-32
At2g07230 Mutator-like transposase 130 3e-30
>At1g43930 hypothetical protein
Length = 946
Score = 256 bits (655), Expect = 3e-68
Identities = 184/693 (26%), Positives = 317/693 (45%), Gaps = 74/693 (10%)
Query: 140 EVPNSAPVNDALNSSSALVHEVPNSAPVNDALNSSSALVHEVPNSAPVNEPLNESEDESY 199
EV + V ++ L +V + V +N+ EV + V +N E
Sbjct: 104 EVNDGGGVEVEVDDGGGLERDVNDGGIVEAEVNNGGNAEAEVNDGGGVEAEVNNGE---- 159
Query: 200 IPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGAR 259
S E EG + +++ D E D+ AE+ +D D D+ + R
Sbjct: 160 ---SVEEEGNG-GLDMEEEDCE---DFEAEFGAEAREEENESDGD-----SGDDIWDDER 207
Query: 260 IEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDL 319
I +P SD ++GD+ D D ++ S RL + TF++ +
Sbjct: 208 IPDPLVHSD---DEGDNDDEDGAPQTDDPEESLRLAK----------------TFNNPED 248
Query: 320 FKQAVKDYALQQKKDLTFKKDDKKRCVVICV---AGCPFYLRLSKTDSRSYWLLVSLEAD 376
FK AV Y+L+ + D+ + R C C + S + + +
Sbjct: 249 FKIAVLRYSLKTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSYDKKKQKMQVKVYKDG 308
Query: 377 HTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKA 436
H+C RS ++ K + +A F +R P IT +++E + + + + + + +AK
Sbjct: 309 HSCVRSGYSKMLKQRTIAWLFRERLRKNPKITKQEMVDEIKREYKLTVTEDQCSKAKTMV 368
Query: 437 VEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAK-GPV--FERIYVCFAATKTA 493
+ Q + +A + Y A++ RSNPG+T I++ G G + F R+++CF + K +
Sbjct: 369 MRERQATHQEHFARIWDYQAKIFRSNPGTTFEIETILGPTIGSLQRFLRLFICFKSQKES 428
Query: 494 FATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLV 553
+ TCRP+IG+DG FLK G LLAA G+DG+N++ PIA+AVVE E +W WFV +L
Sbjct: 429 WKQTCRPIIGIDGAFLKWDIKGHLLAATGRDGDNRIVPIAWAVVEIENDDNWDWFVRMLS 488
Query: 554 NDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELV 613
LD+ + + A ISD+Q GLV + ++
Sbjct: 489 RTLDLQDGRNVAIISDKQ-------------------------------SGLVKAIHSVI 517
Query: 614 GDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMM 673
EHR C +H+ N+++ E+++ W+ R+ T E+ A M +K N +AF+ ++
Sbjct: 518 PQAEHRQCARHIMENWKRNSHDMELQRLFWKIVRSYTEGEFGAHMRALKSYNASAFELLL 577
Query: 674 NLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQK 733
P W+R+ + + C+ +NN+ E+FNR I E R KP++ +LE ++ R K+
Sbjct: 578 KTLPVTWSRAFFKIGSCCNDNLNNLSESFNRTIREARRKPLLDMLEDIRRQCMVRNEKRY 637
Query: 734 QLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCA 793
+ R RT ++E S + WM +V + ++V++ + +C
Sbjct: 638 VIADRLRTRFTKRAHAEIEKMIAGS-QVCQRWMARHNKHEIKVG-PVDSFTVDMNNNTCG 695
Query: 794 CRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
C +W ++GIPC HA + + R+ EDYV Y
Sbjct: 696 CMKWQMTGIPCIHAASVIIGKRQKVEDYVSDWY 728
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 249 bits (636), Expect = 5e-66
Identities = 173/628 (27%), Positives = 301/628 (47%), Gaps = 50/628 (7%)
Query: 199 YIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGA 258
Y G EY G +E S + EE ++ ++ D + P + +V +G
Sbjct: 12 YSDGDVEYNGGNESES--EERIEEEIVRSKKNTKKNKKEKKKKDEEDPLERFDFEVDDGV 69
Query: 259 RIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKD 318
++FEDE D+ S DE+E L R + ++ KF +G TF +
Sbjct: 70 --------ANFEDEIPIGRDIIPESSDEDE--DPILVRDRRIRRAMGDKFVIGSTFFTGI 119
Query: 319 LFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAG--CPFYLRLSKTDSRSYWLLVSLEAD 376
FK+A+ + L+ ++ + +K++ C G C + + S R +++ +
Sbjct: 120 EFKEAILEVTLKHGHNVVQDRWEKEKISFKCGMGGKCKWRVYCSYDPDRQLFVVKTSCTW 179
Query: 377 HTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKA 436
H+C + + K+ +AR F+ +R + E + RW ++ + R ++ A
Sbjct: 180 HSCSPNGKCQILKSPVIARLFLDKLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLA 239
Query: 437 VEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGP-VFERIYVCFAATKTAFA 495
++M++ E +Q+A++R Y E+ NPGS I + + KG VF R YVCF +T +A
Sbjct: 240 LKMLKKEYEEQFAHIRGYVEEIHSQNPGSVAFIDTYRNEKGEDVFNRFYVCFNILRTQWA 299
Query: 496 TTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVND 555
+CRP+IGLDG FLK + G LL AVG D NNQ++PIA+AVV++E +W WFV + D
Sbjct: 300 GSCRPIIGLDGTFLKVVVKGVLLTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKD 359
Query: 556 LDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGD 615
L++ + R+ +SD+ K GL+ + + + +
Sbjct: 360 LNLEDGSRFVILSDRSK-------------------------------GLLSAVKQELPN 388
Query: 616 VEHRLCIKHLYGNFRKKYPGAEM-KQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMN 674
EHR+C+KH+ N +K + +M K +W+ A + E+ + ++ +EA + D++N
Sbjct: 389 AEHRMCVKHIVENLKKNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDEALYNDVLN 448
Query: 675 LPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQ 734
P W+R Y + C+ NN E+FN I + R K +I +LE ++ RIVK+ +
Sbjct: 449 EEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGMTRIVKRNK 508
Query: 735 LMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCAC 794
LR + L K ++D+ + + +FEV D + V++ C+C
Sbjct: 509 KSLRHEGRFTKYALKMLALEKTDADR---SKVYRCTHGVFEVYIDENGHRVDIPKTQCSC 565
Query: 795 RRWDLSGIPCAHAVASMWFGRRVPEDYV 822
+W +SGIPC H+ +M E+Y+
Sbjct: 566 GKWQISGIPCEHSYGAMIEAGLDAENYI 593
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 240 bits (613), Expect = 3e-63
Identities = 153/546 (28%), Positives = 273/546 (49%), Gaps = 43/546 (7%)
Query: 283 SDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDK 342
SD+EEE + R + ++ + + +G TF + FK+ V YA++ + + + +K
Sbjct: 153 SDNEEEDHV--ITRDRKIRLASDDRLAIGRTFFTGFEFKEVVLHYAMKHRINAKQNRWEK 210
Query: 343 KRCVVICV--AGCPFYLRLSKTDSRSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPI 400
+ C C +Y+ S + R W+L + DH+C + + K + + R FM
Sbjct: 211 DKISFRCAQRKECEWYVYASYSHERQLWVLKTKCLDHSCTSNGKCKLLKRRVIGRLFMDK 270
Query: 401 IRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLR 460
+R PN + + +W ++ + ++ A++ ++ E Q+A+LR Y AE+L
Sbjct: 271 LRLQPNFMPLDIQRHIKEQWKLVSTIGQVQDGRLLALKWLKEEYAQQFAHLRGYVAEILS 330
Query: 461 SNPGSTVIIKSTKGA-KGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLA 519
+N GST I+ + + A + VF RIYVCF A K AF CRPLIG+DG FLK G LL
Sbjct: 331 TNKGSTAIVDTIRDANENDVFNRIYVCFGAMKNAF-YFCRPLIGIDGTFLKHAVKGCLLT 389
Query: 520 AVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SC 579
A+ D NNQ++P+A+A V+ E +W WF++ L +DL++ + + ISD+ K
Sbjct: 390 AIAHDANNQIYPVAWATVQFENAENWLWFLNQLKHDLELKDGSGYVVISDRCK------- 442
Query: 580 IH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAE-M 638
G++ + + + EHR C+KH+ N +K++ + +
Sbjct: 443 ------------------------GIISAVKNALPNAEHRPCVKHIVENLKKRHGSLDLL 478
Query: 639 KQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNM 698
K+ +W A + + ++ A + M+ + ++D+M P W RS + + C+ NN
Sbjct: 479 KKFVWNLAWSYSDTQYKANLNEMRAYIMSLYEDVMKEEPNTWCRSWFRIGSYCEDVDNNA 538
Query: 699 CEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNS 758
E+FN I++ R K ++ ++E ++ RI K+K + +W+ + + + L++ +
Sbjct: 539 TESFNATIVKARAKALVPMMETIRRQAMARISKRKAKIGKWKKKISEYVSEILKEEWELA 598
Query: 759 DKWFANWMGDEYMSLFEVSRDNEKYSVNL--ESRSCACRRWDLSGIPCAHAVASMWFGRR 816
K E FEV D +V+L E C+C +W ++GIPC HA A++ +
Sbjct: 599 VKCEVTKGTHE---KFEVWFDGNSNAVSLKTEEWDCSCCKWQVTGIPCEHAYAAINDVGK 655
Query: 817 VPEDYV 822
ED+V
Sbjct: 656 DVEDFV 661
>At3g30170 unknown protein
Length = 995
Score = 226 bits (577), Expect = 4e-59
Identities = 167/649 (25%), Positives = 292/649 (44%), Gaps = 80/649 (12%)
Query: 193 ESEDESYIPGSDEYEG----ESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFV 248
E D+ + G D+ G + E + DD + + + E N+ A+ P F
Sbjct: 116 EGNDDEAVGGGDDATGCGGNDDEALGYDDDATSDGGNDDDDGGVEEDANLNLAEEFPEF- 174
Query: 249 QGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSP--SDDEEEGRSKRLPRFKAVQDGEEV 306
A +EE + D D+ + + P SDDE+E R++ + + + EEV
Sbjct: 175 ---------AGVEEDCSDDDPPDDVWEEDKIPDPFSSDDEDESRTREVHGHRDAAN-EEV 224
Query: 307 KFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKK----RCVVICVAG--CPFYLRLS 360
++ T+++ D FK AV Y+L+ + D+ + + + +C + G CP+ + S
Sbjct: 225 LLELKKTYNTPDDFKLAVLRYSLKTRYDIKLFRSEARIVAAKCSYVDKDGVQCPWKVYCS 284
Query: 361 KTDSRSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRW 420
+ + + +H C RS ++ K +A F +R P +T + E +
Sbjct: 285 YEKTMHKMQIRTYVNNHICVRSGHSKMLKRSSIAYLFEERLRVNPKLTKYEMAAEILREY 344
Query: 421 GILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVF 480
+ + + +AK K V+ +A + Y AE+++ NP + I++T GA
Sbjct: 345 NLEVTPDQCAKAKTKVVKARNASHEAHFARVWDYQAEVIKQNPWTEFEIETTAGA----- 399
Query: 481 ERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAE 540
RP IGLDG FLK G LLAAVG+DG+N++ PIA+AVVE E
Sbjct: 400 ------------VIGAKQRPFIGLDGAFLKWDIKGHLLAAVGRDGDNKIVPIAWAVVEIE 447
Query: 541 TKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSC 600
+W WF+ L L + + A +SD+Q
Sbjct: 448 NDDNWDWFLRHLSASLGLCQMANIAILSDKQ----------------------------- 478
Query: 601 VMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEV 660
GLV + ++ EHR C KH+ N+++ E+++ W+ AR+ T+ E+ M
Sbjct: 479 --SGLVKAIHTILPHAEHRQCSKHIMDNWKRDSHDLELQRLFWKIARSYTVEEFNNHMME 536
Query: 661 MKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLER 720
+++ N A++ + P W+R+ + T C+ +NN+ E+FNR I + R KPI+ +LE
Sbjct: 537 LQQYNRQAYESLQLTSPVTWSRAFFRIGTCCNDNLNNLSESFNRTIRQARRKPILDMLED 596
Query: 721 LKFYLNNRIVKQKQLMLRWRTALCPMIQQKLED---SKRNSDKWFANWMGDEYMSLFEVS 777
++ R K+ + + +T +++E R +++ A +L E+
Sbjct: 597 IRRQCMVRSAKRFLIAEKLQTRFTKRAHEEIEKMIVGLRQCERYMAR------ENLHEIY 650
Query: 778 RDNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
+N Y V++E ++C CR+ + GIPC HA + + EDYV Y
Sbjct: 651 VNNVSYFVDMEQKTCDCRKCQMVGIPCIHATCVIIGKKEKVEDYVSDYY 699
>At4g18410 MuDR transposable element - like protein
Length = 633
Score = 190 bits (482), Expect = 4e-48
Identities = 143/529 (27%), Positives = 237/529 (44%), Gaps = 89/529 (16%)
Query: 313 TFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV--------AGCPFYLRLSKTDS 364
+F S FK+AV DYAL+ +L + DK + ICV A C + + + S
Sbjct: 72 SFFSGVAFKEAVIDYALRTGHNLKQYRYDKDKIGFICVGCDGKEGGAKCEWKVYAAILPS 131
Query: 365 RSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARL-RWGIL 423
+ W + H+C + K ++AR F+ IR +P + A IEE L +W I
Sbjct: 132 DNMWKIRKFIDTHSCIPNGECEMFKVPHIARLFVDKIRDSPEFYLPAKIEEIILEQWKIS 191
Query: 424 LGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGP-VFER 482
+ + + A+ KA++ I+ E DQ++ LR YAAE+L SN S V ++ +G +F R
Sbjct: 192 VTRNQCQSARKKALQWIEKEYDDQFSRLRDYAAEILESNKDSHVEVECLVSDEGKDMFNR 251
Query: 483 IYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETK 542
YVCF + + + +CRPLIG+DGCFLK
Sbjct: 252 FYVCFDSLRRTWKESCRPLIGIDGCFLKN------------------------------- 280
Query: 543 ASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVM 602
LL D D+ + + + ISD+QK
Sbjct: 281 -------KLLKEDFDLSDGEGFIMISDRQK------------------------------ 303
Query: 603 QGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEM-KQALWRAARASTLPEWYAAMEVM 661
GL+ + + +EHR+C++H+YGN +K Y M K LW A + E+ +E +
Sbjct: 304 -GLIKAVQLELPKIEHRMCVQHIYGNLKKTYGSKTMIKPLLWNLAWSYNETEYKQHLEKI 362
Query: 662 KELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERL 721
+ + ++ +M P+ W R+ + C+ NN E+FN ++ + R+K + +LE +
Sbjct: 363 RCYDTKVYESVMKTNPRSWVRAFQKIGSFCEDVDNNSVESFNGSLNKAREKQFVAMLETI 422
Query: 722 KFYLNNRIVKQKQLMLRWRTALC-PMIQQKL--EDSKRNSDKWFANWMGDEYMSLFEVSR 778
+ RI K + + T +C P + + L E ++ K G ++EV
Sbjct: 423 RRMAMVRIAK-RSVESHTHTGVCTPYVMKFLAGEHKVASTAKVAPGTNG-----MYEVRH 476
Query: 779 DNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSYR 827
+ + V+L + +C C +W + GIPC HA + + PED+V +R
Sbjct: 477 GGDTHRVDLAAYTCTCIKWQICGIPCEHAYGVILHKKLQPEDFVCQWFR 525
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 183 bits (465), Expect = 4e-46
Identities = 123/509 (24%), Positives = 232/509 (45%), Gaps = 46/509 (9%)
Query: 310 VGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCPFYLRLSKTDSRSYW 368
VG F + ++ +KD A+ DL K D+ R + C GCP+ + +K +
Sbjct: 27 VGKEFPDVETCRRTLKDLAIALHFDLRIVKSDRSRFIAKCSKEGCPWRIHAAKCPGVQTF 86
Query: 369 LLVSLEADHTC--CRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGK 426
+ +L ++HTC R ++QA ++AR IR P ++++ R G+ +
Sbjct: 87 TVRTLNSEHTCEGVRDLHHQQASVGWVARSVEARIRDNPQYKPKEILQDIRDEHGVAVSY 146
Query: 427 WKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGP--VFERIY 484
+A+R K +++ + G + Y L Y ++ NPGS + A GP F+R++
Sbjct: 147 MQAWRGKERSMAALHGTYEEGYRFLPAYCEQIKLVNPGSFASVS----ALGPENCFQRLF 202
Query: 485 VCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKAS 544
+ + A + F ++CRPL+ LD LKG Y G +L A D ++ +FP+A A+V+ E+ +
Sbjct: 203 IAYRACISGFFSSCRPLLELDRAHLKGKYLGAILCAAAVDADDGLFPLAIAIVDNESDEN 262
Query: 545 WSWFVDLLVNDLDM--VERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVM 602
WSWF+ L L M + +S++Q ++ +
Sbjct: 263 WSWFLSELRKLLGMNTDSMPKLTILSERQSAVVE------------------------AV 298
Query: 603 QGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMK 662
+ PT H C++++ NFR + ++ W A A T E+ M
Sbjct: 299 ETHFPT-------AFHGFCLRYVSENFRDTFKNTKLVNIFWSAVYALTPAEFETKSNEMI 351
Query: 663 ELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLK 722
E+++ Q P +W AY + + + E LE + PII ++E ++
Sbjct: 352 EISQDVVQWFELYLPHLWA-VAYFQGVRYGHFGLGITEVLYNWALECHELPIIQMMEHIR 410
Query: 723 FYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEK 782
+++ +++L + W + L P ++++ ++ ++ + + + R N
Sbjct: 411 HQISSWFDNRRELSMGWNSILVPSAERRITEAVADARCYQVLRANEVEFEIVSTERTN-- 468
Query: 783 YSVNLESRSCACRRWDLSGIPCAHAVASM 811
V++ +R C+CRRW + G+PCAHA A++
Sbjct: 469 -IVDIRTRDCSCRRWQIYGLPCAHAAAAL 496
>At2g13970 Mutator-like transposase
Length = 597
Score = 181 bits (459), Expect = 2e-45
Identities = 118/430 (27%), Positives = 203/430 (46%), Gaps = 76/430 (17%)
Query: 405 PNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPG 464
P IT A+++E + + + + + + +AK + + + ++ + Y AE+ R+NPG
Sbjct: 19 PKITKKAMVDEIQREYKLTVTEDQCSKAKTIVMRERKATHQEHFSRIWDYQAEIFRTNPG 78
Query: 465 STVIIKSTKGAK-GPV--FERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAV 521
+ I++ G G + F R+++CF + K ++ TCRP+IG+DG FLK G LLAA
Sbjct: 79 TKFEIETIPGPTIGSLQRFYRLFICFKSQKDSWKQTCRPIIGIDGAFLKWDIKGHLLAAT 138
Query: 522 GKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH 581
G+DG+N++ PIA+AVVE + +W WFV L LD+ + + A ISD+Q
Sbjct: 139 GRDGDNRIVPIAWAVVEIQNDDNWDWFVRQLSESLDLQDGRSVAIISDKQ---------- 188
Query: 582 *SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQA 641
GL + ++ EHR +H+ N++K E+++
Sbjct: 189 ---------------------SGLGKAIHTVIPQAEHRQYARHIMDNWKKNSHDMELQRL 227
Query: 642 LWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEA 701
W+ P W+R+ + + C+ +NN+ E+
Sbjct: 228 FWKT------------------------------NPLTWSRAFFRIGSCCNDNLNNLSES 257
Query: 702 FNRAILEYRDKPIITLLERLKFYLNNRIVKQKQLMLRWR-----TALCPMIQQKLEDSKR 756
FNR I R KP++ +LE ++ R +K+ ++ +R T M +K+ D
Sbjct: 258 FNRTIRNARRKPLLDMLEDIRRQCMVR--NEKRFVIAYRLKSRFTKRAHMEIEKMIDGAA 315
Query: 757 NSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRR 816
D+W A E V D + +SV++ R+C CR+W ++GIPC HA + + R+
Sbjct: 316 VCDRWMARHNRIE----VRVGLD-DSFSVDMNDRTCGCRKWQMTGIPCIHAASVIIGNRQ 370
Query: 817 VPEDYVHSSY 826
ED+V Y
Sbjct: 371 KVEDFVSDWY 380
>At1g06740 mudrA-like protein
Length = 726
Score = 181 bits (459), Expect = 2e-45
Identities = 129/520 (24%), Positives = 234/520 (44%), Gaps = 51/520 (9%)
Query: 303 GEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCPFYLRLSK 361
G + + VG FS ++A+K+ A+ + ++ K DK R C + GCP+ + +K
Sbjct: 151 GTDHEMVVGMEFSDAYACRRAIKNAAISLRFEMRTIKSDKTRFTAKCNSKGCPWRIHCAK 210
Query: 362 TDSRSYWLLVSLEADHTC--CRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLR 419
+ + + ++ HTC ++QA +++A ++ P+ ++EE
Sbjct: 211 VSNAPTFTIRTIHGSHTCGGISHLGHQQASVQWVADVVAEKLKENPHFKPKEILEEIYRV 270
Query: 420 WGILLGKWKAYRAKVKAVEMIQGETL-----DQYANLRKYAAELLRSNPGSTVIIKSTKG 474
GI L +A+R K + + ++G TL ++Y L +Y E+ RSNPGS ++
Sbjct: 271 HGISLSYKQAWRGKERIMATLRGSTLRGSFEEEYRLLPQYCDEIRRSNPGSVAVVHVN-- 328
Query: 475 AKGPV---FERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFP 531
P+ F+ +++ F A+ + F CRPLI LD LK Y G LL A G DG+ +FP
Sbjct: 329 ---PIDGCFQHLFISFQASISGFLNACRPLIALDSTVLKSKYPGTLLLATGFDGDGAVFP 385
Query: 532 IAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I 591
+AFA+V E +W F+ L LD K S ++
Sbjct: 386 LAFAIVNEENDDNWHRFLSELRKILDENMPKLTILSSGERP------------------- 426
Query: 592 *YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTL 651
V+ G+ H C+ +L F++++ + + W AA T+
Sbjct: 427 ---------VVDGVEANFPAAF----HGFCLHYLTERFQREFQSSVLVDLFWEAAHCLTV 473
Query: 652 PEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRD 711
E+ + + +++++ A + N P W S + L N + E+ + + +
Sbjct: 474 LEFKSKINKIEQISPEASLWIQNKSPARWASSYFEGTRFGQLTANVITESLSNWVEDTSG 533
Query: 712 KPIITLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYM 771
PII +E + +L N + ++++ L W L P ++++ + S A+ +
Sbjct: 534 LPIIQTMECIHRHLINMLKERRETSLHWSNVLVPSAEKQMLAAIEQSR---AHRVYRANE 590
Query: 772 SLFEVSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVASM 811
+ FEV VN+E+ SC C RW + G+PC+HAV ++
Sbjct: 591 AEFEVMTCEGNVVVNIENCSCLCGRWQVYGLPCSHAVGAL 630
>At2g30640 Mutator-like transposase
Length = 754
Score = 179 bits (454), Expect = 7e-45
Identities = 125/520 (24%), Positives = 235/520 (45%), Gaps = 44/520 (8%)
Query: 298 KAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCPFY 356
+++Q E VG FS ++A++D A+ + ++ K DK R C + GCP+
Sbjct: 170 RSLQTAPEHNLIVGMEFSDVLACRRALRDAAIALRFEMQTVKSDKTRFTAKCNSEGCPWR 229
Query: 357 LRLSKTDSRSYWLLVSLEADHTC--CRSASNRQAKTKYLARKFMPIIRHTPNITVNALIE 414
+ +K + + ++ HTC + QA +++A ++ P+ ++E
Sbjct: 230 IHCAKLPGLPTFTIRTIHGSHTCGGISHLGHHQASVQWVADAVTERLKVNPHCKPKEILE 289
Query: 415 EARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKG 474
+ GI L +A+R K + + ++G + Y L +Y ++ R+NPGS ++ +
Sbjct: 290 QIHQVHGITLTYKQAWRGKERIMAAVRGSYEEDYRLLPRYCDQIRRTNPGSVAVVHGS-- 347
Query: 475 AKGPV---FERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFP 531
PV F++ ++ F A+ F CRPLIGLD LK Y G LL A G DG +FP
Sbjct: 348 ---PVDGSFQQFFISFQASICGFLNACRPLIGLDRTVLKSKYVGTLLLATGFDGEGAVFP 404
Query: 532 IAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I 591
+AFA++ E +SW WF+ E ++ ++ + L
Sbjct: 405 LAFAIISEENDSSWQWFLS---------ELRQLLEVNSENMPKLT--------------- 440
Query: 592 *YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTL 651
+ S Q +V + H LC+ L + R ++ + + +W AA+ T
Sbjct: 441 -----ILSSRDQSIVDGVDTNFPTAFHGLCVHCLTESVRTQFNNSILVNLVWEAAKCLTD 495
Query: 652 PEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRD 711
E+ M + +++ A + N+ W + T+ N+ E+ N + +
Sbjct: 496 FEFEGKMGEIAQISPEAASWIRNIQHSQWATYCF-EGTRFGHLTANVSESLNSWVQDASG 554
Query: 712 KPIITLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYM 771
PII +LE ++ L ++++ ++W L P ++ + ++ + + +
Sbjct: 555 LPIIQMLESIRRQLMTLFNERRETSMQWSGMLVPSAERHVLEAIEECRLYPVHKANE--- 611
Query: 772 SLFEVSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVASM 811
+ FEV K+ V++ R+C CR W+L G+PC+HAVA++
Sbjct: 612 AQFEVMTSEGKWIVDIRCRTCYCRGWELYGLPCSHAVAAL 651
>At1g12720 hypothetical protein
Length = 585
Score = 177 bits (449), Expect = 3e-44
Identities = 120/446 (26%), Positives = 201/446 (44%), Gaps = 71/446 (15%)
Query: 389 KTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQY 448
K +A F +R P +T ++ E + + + + + +AK K ++ +
Sbjct: 3 KRSTIAALFEERLRVNPKMTKYEMVAEIKREYKLEVTPDQCAKAKTKVLKARNASHDTHF 62
Query: 449 ANLRKYAAELLRSNPGSTVIIKSTK----GAKGPVFERIYVCFAATKTAFATTCRPLIGL 504
+ + Y AE+L NP S I++T G+K F R+Y+CF + K ++ CRP+IG+
Sbjct: 63 SRIWDYQAEVLNRNPNSDFDIETTARTFIGSKQRFF-RLYICFNSQKVSWKQHCRPVIGI 121
Query: 505 DGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERKRW 564
DG FLK G LLAAVG+DG+N++ P+A+AVVE E +W WF+ L L + E
Sbjct: 122 DGAFLKWDIKGHLLAAVGRDGDNRIVPLAWAVVEIENDDNWDWFLKKLSESLGLCEMVNL 181
Query: 565 AFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKH 624
A ISD+Q GLV + ++ EHR C KH
Sbjct: 182 ALISDKQ-------------------------------SGLVKAIHNVLPQAEHRQCSKH 210
Query: 625 LYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSA 684
+ N+++ E+++ W+ +R+ T+ E+ M +K N A+ + P WT
Sbjct: 211 IMDNWKRDSHDMELQRLFWKISRSYTIEEFNTHMANLKSYNPQAYASLQLTSPMTWT--- 267
Query: 685 YSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQLMLRWRTALC 744
I + R KP++ +LE ++ R K+ + R ++
Sbjct: 268 ---------------------IRQARRKPLLDMLEDIRRQCMVRTAKRFIIAERLKSRFT 306
Query: 745 PMIQQKLEDSKRNSDKWFANWMGDE----YMSLFEVSRDNEKYSVNLESRSCACRRWDLS 800
P ++E K A G E +L E+ ++ Y V+++ ++C CR+W++
Sbjct: 307 PRAHAEIE-------KMIAGSAGCERHLARNNLHEIYVNDVGYFVDMDKKTCGCRKWEMV 359
Query: 801 GIPCAHAVASMWFGRRVPEDYVHSSY 826
GIPC H + + EDYV Y
Sbjct: 360 GIPCVHTPCVIIGRKEKVEDYVSDYY 385
>At1g34620 Mutator-like protein
Length = 1034
Score = 171 bits (433), Expect = 2e-42
Identities = 168/677 (24%), Positives = 292/677 (42%), Gaps = 106/677 (15%)
Query: 162 PNSAPVNDALNSSSALVHEVPNSAPVNEPLNESEDESYIPGSDEYEG-----ESEDVS-- 214
P+ A V D S + VP + P N P + + I G+D VS
Sbjct: 238 PSRATVTDTSTDSDDDLLIVPFTLPPNPPAITIPENADIEGADSSRAAVGTPHGNHVSPI 297
Query: 215 ---VDDSDYEEN----WDWAEVLPAESLINIATADRDPPFVQGSVDVGNGARIEEPATFS 267
V ++D + W A+ E+L I + D P + G + P F+
Sbjct: 298 GNAVAETDLQRQNILFWGRAQ----EALNTILSDFSDDPILFGR---------DAPPVFN 344
Query: 268 DFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDY 327
D + E DS+ D + + K VG F SK K + +
Sbjct: 345 DGKGEGVDSAFFDVKYEGD--------------------KLFVGRVFKSKSDCKIKIAIH 384
Query: 328 ALQQKKDLTFKKDDKKRCVVICVAG-CPFYLRLSKTDSRSYWLLVSLEADHTCC---RSA 383
A+ +K + K V+ C++ CP+ + SK D+ + + HTC R
Sbjct: 385 AINRKFHFRTARSTPKFMVLKCISKTCPWRVYASKVDTSDSFQVRQANQRHTCTIDQRRR 444
Query: 384 SNRQAKTKYLAR----KFMPIIRHTPNITV--NALIEEARLRWGILLGKWKAYRAKVKAV 437
+R A T+ + +F+ I R PN V L+++ + + + WKA+RA+ A+
Sbjct: 445 YHRLATTQVIGELMQSRFLGIKRG-PNAAVIRKFLLDD----YHVSISYWKAWRAREVAM 499
Query: 438 EMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPV-FERIYVCFAATKTAFAT 496
E G YA + YA L ++NPGS + GP F+ ++ FAA+ +A
Sbjct: 500 EKSLGSMAGSYALIPAYAGLLQQANPGSLCFTEYDDDPTGPRRFKYQFIAFAASIKGYAF 559
Query: 497 TCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDL 556
R +I +DG +KG YGG L++A +DGN Q+FP+AF +V +E +++ WF L +
Sbjct: 560 M-RKVIVVDGTSMKGRYGGCLISACCQDGNFQIFPLAFGIVNSENDSAYEWFFQRL--SI 616
Query: 557 DMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDV 616
+ FISD+ + + L+++
Sbjct: 617 IAPDNPDLMFISDRH-------------------------------ESIYTGLSKVYTQA 645
Query: 617 EHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLP 676
H C HL+ N R Y + + + AA+A + ++ +++LN +++L
Sbjct: 646 NHAACTVHLWRNIRHLYKPKSLCRLMSEAAQAFHVTDFNRIFLKIQKLNPGCAAYLVDLG 705
Query: 677 PKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQLM 736
WTR +S+ + ++ +N+CE++N I E R+ P+I +LE ++ L + ++
Sbjct: 706 FSEWTR-VHSKGRRFNIMDSNICESWNNVIREAREYPLICMLEYIRTTLMDWFATRRAQA 764
Query: 737 LRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEV---SRDNEKYSVNLESRSCA 793
T L P +Q+++E++ ++ A M + + FE R E + V L+ +C+
Sbjct: 765 EDCPTTLAPRVQERVEENYQS-----AMSMSVKPICNFEFQVQERTGECFIVKLDESTCS 819
Query: 794 CRRWDLSGIPCAHAVAS 810
C + GIPCAHA+A+
Sbjct: 820 CLEFQGLGIPCAHAIAA 836
>At3g05850 unknown protein
Length = 777
Score = 170 bits (430), Expect = 4e-42
Identities = 136/523 (26%), Positives = 233/523 (44%), Gaps = 51/523 (9%)
Query: 310 VGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCPFYLRLSKTDSRSYW 368
VG F + F++A++ YA+ + +KK+D R V C A GCP+ + S+ +
Sbjct: 206 VGQRFKNVGEFREALRKYAIANQFGFRYKKNDSHRVTVKCKAEGCPWRIHASRLSTTQLI 265
Query: 369 LLVSLEADHTC--CRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGK 426
+ + HTC + Q ++A ++ PN ++ + + +GI L
Sbjct: 266 CIKKMNPTHTCEGAGGINGLQTSRSWVASIIKEKLKVFPNYKPKDIVSDIKEEYGIQLNY 325
Query: 427 WKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVC 486
++A+R K A E +QG D Y L + +++ +NPGS + + + F R++V
Sbjct: 326 FQAWRGKEIAREQLQGSYKDGYKQLPLFCEKIMETNPGSLATFTTKEDSS---FHRVFVS 382
Query: 487 FAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWS 546
F A+ F CRPL+ LD LK Y G LLAA DG++++FP+AFAVV+AET +W
Sbjct: 383 FHASVHGFLEACRPLVFLDSMQLKSKYQGTLLAATSVDGDDEVFPLAFAVVDAETDDNWE 442
Query: 547 WFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLV 606
WF+ L + L + F++D+QK L
Sbjct: 443 WFL-LQLRSL-LSTPCYITFVADRQK-------------------------------NLQ 469
Query: 607 PTLAELVGDVEHRLCIKHLYGNFRKKYPGA---EMKQAL----WRAARASTLPEWYAAME 659
++ ++ H C+++L K G E+K+ + + AA A + +E
Sbjct: 470 ESIPKVFEKSFHAYCLRYLTDELIKDLKGPFSHEIKRLIVDDFYSAAYAPRADSFERHVE 529
Query: 660 VMKELNEAAFQ-DMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLL 718
+K L+ A+ + P W +AY R + + ++ E F + D PI ++
Sbjct: 530 NIKGLSPEAYDWIVQKSQPDHWA-NAYFRGARYNHMTSHSGEPFFSWASDANDLPITQMV 588
Query: 719 ERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSR 778
+ ++ + I ++ L P ++ KLE + D +LF+V R
Sbjct: 589 DVIRGKIMGLIHVRRISANEANGNLTPSMEVKLEKESLRAQTVHVAPSADN--NLFQV-R 645
Query: 779 DNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDY 821
VN+ C+C+ W L+G+PC HAVA + + R P DY
Sbjct: 646 GETYELVNMAECDCSCKGWQLTGLPCHHAVAVINYYGRNPYDY 688
>At2g29230 Mutator-like transposase
Length = 915
Score = 169 bits (429), Expect = 5e-42
Identities = 149/544 (27%), Positives = 252/544 (45%), Gaps = 58/544 (10%)
Query: 281 SPSDDE--EEGR-SKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTF 337
+P+ DE EEGR L V +G+E+ VG F +K + +AL ++
Sbjct: 316 APTLDEQGEEGRVGMELAIRDVVYEGDELF--VGRVFKNKQDCNVKLAVHALNRRFHFRR 373
Query: 338 KKDDKKRCVVICVAG-CPFYLRLSKTDSRSYWLLVSLEADHTCC---RSASNRQAKTKYL 393
+ KK + C++ CP+ + + K + + + S +HTC RS +R A T+ +
Sbjct: 374 DRSCKKLMTLTCISETCPWRVYIVKLEDSDNYQIRSANLEHTCTVEERSNYHRAATTRVI 433
Query: 394 ARKFMPIIRHTPNITVNALIEEARLR---WGILLGKWKAYRAKVKAVEMIQGETLDQYAN 450
++ N I+ R+ + + + WKA++++ A++ QG + +
Sbjct: 434 GSIIQS--KYAGNSRGPRAIDLQRILLTDYSVRISYWKAWKSREIAMDSAQGSAANSFTL 491
Query: 451 LRKYAAELLRSNPGSTVIIKSTKGAKGPV-FERIYVCFAATKTAFATTCRPLIGLDGCFL 509
L Y L +NPGS V +K+ KG F+ +++ FAA+ F+ R ++ +DG L
Sbjct: 492 LPAYLHVLREANPGSIVDLKTEVDGKGNHRFKYMFLAFAASIQGFSCMKRVIV-IDGAHL 550
Query: 510 KGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISD 569
KG YGG LL A G+D N Q+FPIAF VV++E +W WF +L + + F+SD
Sbjct: 551 KGKYGGCLLTASGQDANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIP--DGDNLTFVSD 608
Query: 570 QQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNF 629
+ +S + G L + +H CI HL N
Sbjct: 609 R---------------------------HSSIYTG----LRRVYPKAKHGACIVHLQRNI 637
Query: 630 RKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDT 689
Y + + RAARA + E++ + L+ A + + ++ WTR AY
Sbjct: 638 ATSYKKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTR-AYFLGK 696
Query: 690 QCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQ 749
+ ++ +N+ E+ N + E R+ PII+LLE ++ L + +++ RT P+ +
Sbjct: 697 RYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAA---RTEASPLPPK 753
Query: 750 KLEDSKRNSDK--WFANWMGDEYMSLFEVSRDNEK-YSVNLESRSCACRRWDLSGIPCAH 806
E RN +K FA D Y +E+ + Y V L R+C+CR +DL +PC H
Sbjct: 754 MREVVHRNFEKSVRFAVHRLDRYD--YEIREEGASVYHVKLMERTCSCRAFDLLHLPCPH 811
Query: 807 AVAS 810
A+A+
Sbjct: 812 AIAA 815
>At1g21040 hypothetical protein
Length = 904
Score = 169 bits (429), Expect = 5e-42
Identities = 149/544 (27%), Positives = 252/544 (45%), Gaps = 58/544 (10%)
Query: 281 SPSDDE--EEGR-SKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTF 337
+P+ DE EEGR L V +G+E+ VG F +K + +AL ++
Sbjct: 316 APTLDEQGEEGRVGMELAIRDVVYEGDELF--VGRVFKNKQDCNVKLAVHALNRRFHFRR 373
Query: 338 KKDDKKRCVVICVAG-CPFYLRLSKTDSRSYWLLVSLEADHTCC---RSASNRQAKTKYL 393
+ KK + C++ CP+ + + K + + + S +HTC RS +R A T+ +
Sbjct: 374 DRSCKKLMTLTCISETCPWRVYIVKLEDSDNYQIRSANLEHTCTVEERSNYHRAATTRVI 433
Query: 394 ARKFMPIIRHTPNITVNALIEEARLR---WGILLGKWKAYRAKVKAVEMIQGETLDQYAN 450
++ N I+ R+ + + + WKA++++ A++ QG + +
Sbjct: 434 GSIIQS--KYAGNSRGPRAIDLQRILLTDYSVRISYWKAWKSREIAMDSAQGSAANSFTL 491
Query: 451 LRKYAAELLRSNPGSTVIIKSTKGAKGPV-FERIYVCFAATKTAFATTCRPLIGLDGCFL 509
L Y L +NPGS V +K+ KG F+ +++ FAA+ F+ R ++ +DG L
Sbjct: 492 LPAYLHVLREANPGSIVDLKTEVDGKGNHRFKYMFLAFAASIQGFSCMKRVIV-IDGAHL 550
Query: 510 KGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISD 569
KG YGG LL A G+D N Q+FPIAF VV++E +W WF +L + + F+SD
Sbjct: 551 KGKYGGCLLTASGQDANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIP--DGDNLTFVSD 608
Query: 570 QQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNF 629
+ +S + G L + +H CI HL N
Sbjct: 609 R---------------------------HSSIYTG----LRRVYPKAKHGACIVHLQRNI 637
Query: 630 RKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDT 689
Y + + RAARA + E++ + L+ A + + ++ WTR AY
Sbjct: 638 ATSYKKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTR-AYFLGE 696
Query: 690 QCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQ 749
+ ++ +N+ E+ N + E R+ PII+LLE ++ L + +++ RT P+ +
Sbjct: 697 RYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAA---RTEASPLPPK 753
Query: 750 KLEDSKRNSDK--WFANWMGDEYMSLFEVSRDNEK-YSVNLESRSCACRRWDLSGIPCAH 806
E RN +K FA D Y +E+ + Y V L R+C+CR +DL +PC H
Sbjct: 754 MREVVHRNFEKSVRFAVHRLDRYD--YEIREEGASVYHVKLMERTCSCRAFDLLHLPCPH 811
Query: 807 AVAS 810
A+A+
Sbjct: 812 AIAA 815
>At1g67240 hypothetical protein
Length = 851
Score = 164 bits (414), Expect = 3e-40
Identities = 107/383 (27%), Positives = 174/383 (44%), Gaps = 59/383 (15%)
Query: 376 DHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVK 435
+H S R K +AR F R P + + +E +R+ I + KW +A+
Sbjct: 339 EHEHAISGKARMLKQGVIARLFRDEARRRPTLRWTDIKDEIMMRYTISVSKWICQKARRL 398
Query: 436 AVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFA 495
A +M+ +Q+A L Y AEL RSN I
Sbjct: 399 AFDMVIQTQREQFAKLWDYEAELQRSNKDIHTEI----------------------VTIP 436
Query: 496 TTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVND 555
C DG FLK G++LAAVG+D +N+++PIA+A+V E SW+WFV+ L D
Sbjct: 437 QDC------DGAFLKWELKGEILAAVGRDADNRIYPIAWAIVRVEDNDSWAWFVEHLKTD 490
Query: 556 LDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGD 615
L + ISD++K GL+ +A+L+
Sbjct: 491 LGLGLGSLLTVISDKKK-------------------------------GLINVVADLLPQ 519
Query: 616 VEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNL 675
EHR C +H+Y N+RK Y + W A +ST ++ M+ ++ + A D++
Sbjct: 520 AEHRHCARHIYANWRKVYSDYSHESYFWAIAYSSTNGDYRWNMDALRLYDPQAHDDLLKT 579
Query: 676 PPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQL 735
P+ W R+ +S ++C+ NN+CE+FNR I + R+ P+I +LE ++ RI K
Sbjct: 580 DPRTWCRAFFSTHSRCEDVSNNLCESFNRTIRDARNLPVINMLEEVRRTSMKRIAKLCGK 639
Query: 736 MLRWRTALCPMIQQKLEDSKRNS 758
++ T P I + LE +++++
Sbjct: 640 TMKCETRFPPKIMEILEGNRKSA 662
>At3g06940 putative mudrA protein
Length = 749
Score = 160 bits (405), Expect = 3e-39
Identities = 127/527 (24%), Positives = 237/527 (44%), Gaps = 36/527 (6%)
Query: 299 AVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVA-GCPFYL 357
A+Q E V V F++ F+ A+ Y++ +KK+D R V C A GCP+ +
Sbjct: 171 AIQQWENVITGVDQRFNTFLEFRDALHKYSVAHGFAYKYKKNDSHRVSVKCKAQGCPWRI 230
Query: 358 RLSKTDSRSYWLLVSLEADHTCCRSA--SNRQAKTKYLARKFMPIIRHTPNITVNALIEE 415
S+ + + + HTC R+ +A ++ ++ P+ + E+
Sbjct: 231 TASRLSTTQLICIKKMNPRHTCERAVIKPGYRATRGWVRTILKEKLKAFPDYKPKDIAED 290
Query: 416 ARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGA 475
+ +GI L +A+RAK A E +QG + Y+ L ++ +NPGS + + +
Sbjct: 291 IKKEYGIQLNYSQAWRAKEIAREQLQGSYKEAYSQLPLICEKIKETNPGSIATFMTKEDS 350
Query: 476 KGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFA 535
F R+++ F A+ + F RPL+ LD L Y G +L A D + +FP+AFA
Sbjct: 351 S---FHRLFISFYASISGFKQGSRPLLFLDNAILNSKYQGVMLVATASDAEDGIFPVAFA 407
Query: 536 VVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLS 595
+V++ET+ +W WF++ L L E + F++D Q + N +I ++ + +
Sbjct: 408 IVDSETEENWLWFLEQLKTALS--ESRIITFVADFQNGLKN------AIAQVFEKDAHHA 459
Query: 596 YLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWY 655
Y C+ Q +L G H ++Y M + A A+T +Y
Sbjct: 460 Y---CLGQLAEKLNVDLKGQFSHE----------ARRY----MLNDFYSVAYATTPVGYY 502
Query: 656 AAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPII 715
A+E +K ++ A+ ++ P W + + + + +N + F + E + PI
Sbjct: 503 LALENIKSISPDAYNWVIESEPHHWANALFQGERYNKMN-SNFGKDFYSWVSEAHEFPIT 561
Query: 716 TLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFE 775
+++ + + I ++ W T L P ++KL+ + + + SLF+
Sbjct: 562 QMIDEFRAKMMQSIYTRQVKSREWVTTLTPSNEEKLQ---KEIELAQLLQVSSPEGSLFQ 618
Query: 776 VSRDNEKYS-VNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDY 821
V+ S V++ C C+ W L+G+PC+HA+A + + P +Y
Sbjct: 619 VNGGESSVSIVDINQCDCDCKTWRLTGLPCSHAIAVIGCIEKSPYEY 665
>At2g14030 Mutator-like transposase
Length = 874
Score = 153 bits (387), Expect = 4e-37
Identities = 165/686 (24%), Positives = 294/686 (42%), Gaps = 81/686 (11%)
Query: 151 LNSSSALVHEVPNSAPVNDALNSSSALVHEVPNSAPVNEPLNESEDESYIPG-------- 202
+ SS+ VH P A V AL + + +E +E ED + +
Sbjct: 170 MGDSSSNVHAQPAGATVEPALTLETNQSMDT-----ASEKADEEEDSNLVEADFLANLGL 224
Query: 203 ------SDEYEGESEDVSVDDSDYEENWDWAEVLPAESLIN-IATADRDPPFVQGSVDVG 255
+D Y +S +D E ++D E L + AT +P V + V
Sbjct: 225 PSINLATDNYGFDSGSEGYND---ENDYDGGETLKESNFFGPSATILTEPLGVHLDLQVP 281
Query: 256 NGARIEEPA-TFSDFEDEDGDSSDLDSPSDDEEEGRS-KRLPRFKAVQDGEEVKFQVGHT 313
+E+ F+ F + + D + EEGR L V +G+E+ VG
Sbjct: 282 LQQALEDMCINFASFNRDAAPTLD-----EQGEEGRDGMELAIRDVVYEGDELF--VGRV 334
Query: 314 FSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAG-CPFYLRLSKTDSRSYWLLVS 372
F +K + +AL + + KK + C++ C + + + K + + + S
Sbjct: 335 FKNKQDCNVKLAVHALNSRFHFRRDRSYKKLMTLTCISELCLWRVYIVKLEDSDNYQIRS 394
Query: 373 LEADHTCC---RSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLR---WGILLGK 426
+HTC RS +R A T+ + ++ N ++ R+ + + +
Sbjct: 395 ATLEHTCTVEERSNYHRGATTRVIRSIIKS--KYDGNTRGPRAVDLQRILLTDYSVRISY 452
Query: 427 WKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPV-FERIYV 485
WKA++++ A+E +QG + ++ L Y L +NP S V +K+ AKG F+ +++
Sbjct: 453 WKAWKSREIALESVQGSATNSFSLLTAYIHVLQEANPSSIVDLKTEIDAKGNYRFKYLFL 512
Query: 486 CFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASW 545
FAA+ F+ R ++ + G LKG YGG LL A +D N Q++P+AF VV+++ +W
Sbjct: 513 AFAASIQGFSCMKRVIV-IGGAHLKGKYGGCLLTASAQDANFQVYPLAFGVVDSKNDDAW 571
Query: 546 SWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGL 605
WF +L + + + F+SD+ +S + G
Sbjct: 572 EWFFRVLSTAIP--DGEILTFVSDR---------------------------HSSIYTG- 601
Query: 606 VPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELN 665
L ++ H CI HL N Y + + RAARA + E+ L+
Sbjct: 602 ---LRKVYPKARHGACIVHLQRNIATSYKKKHLLFHVSRAARAYRICEFQTYFNEGIRLD 658
Query: 666 EAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYL 725
A + + + WTR AY + ++ +N+ E+ N + E R+ PI +LLE ++ L
Sbjct: 659 PACARYLELVGFCHWTR-AYFLGERYNVMTSNVVESLNPVLKEARELPIFSLLEFIRTTL 717
Query: 726 NNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEK-YS 784
+ +++ +AL P +++ + + S FA D Y ++V + +
Sbjct: 718 ISWFAMRRKAARSKTSALLPKMREVVHQNFEKS-VTFAVHRIDRYD--YKVRGEGSSVFH 774
Query: 785 VNLESRSCACRRWDLSGIPCAHAVAS 810
V L R+C+CR +DL +PC HA+A+
Sbjct: 775 VKLMERTCSCRAFDLLHLPCPHAIAA 800
>At1g34700 hypothetical protein
Length = 990
Score = 138 bits (347), Expect = 2e-32
Identities = 123/515 (23%), Positives = 221/515 (42%), Gaps = 59/515 (11%)
Query: 310 VGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV-AGCPFYLRLSKTDSRSYW 368
VG F ++ + +Q + A++Q K K ++CV CP+ L +
Sbjct: 379 VGSVFKNRKILQQTMSLQAIKQCFCFKQPKSCPKTLKMVCVDETCPWQLTARVVKDSESF 438
Query: 369 LLVSLEADHTC---CRSASNRQA---------KTKYLARKFMPIIRHTPNITVNALIEEA 416
+ S HTC R N+ A +++Y + + P P + +N L
Sbjct: 439 KITSYATTHTCNIDSRKNYNKHANYKLLGEVVRSRYSSTQGGPRAVDLPQLLLNDL---- 494
Query: 417 RLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAK 476
+ + A+RAK AVE ++G+ + Y L Y L +NPG+ + T
Sbjct: 495 ----NVRISYSTAWRAKEVAVENVRGDEIANYRFLPTYLYLLQLANPGTITHLHYTPEDD 550
Query: 477 GPV-FERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFA 535
G F+ ++V A+ R ++ +DG L G Y G LL A +DGN Q+FPIAF
Sbjct: 551 GKQRFKYVFVSLGASIKCLIYM-RKVVVVDGTQLVGPYKGCLLIACAQDGNFQIFPIAFG 609
Query: 536 VVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLS 595
VV+ ET ASW+WF + L I D +M+ +S
Sbjct: 610 VVDGETDASWAWFFEKLAE-----------IIPDSDDLMI------------------VS 640
Query: 596 YLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWY 655
+S + +G L+ + H C HL N Y + + AA+A + ++
Sbjct: 641 DRHSSIYKG----LSVVYPRAHHGACAVHLERNLSTYYGKFGVSALFFSAAKAYRVKDFE 696
Query: 656 AAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPII 715
E+++E + + + ++ + WTR A+ R + ++ +N E+ N + + + PI+
Sbjct: 697 KYFELLREKSAKCGKYLEDIGFEHWTR-AHCRGERYNIMSSNNFESMNHVLTKAKTYPIV 755
Query: 716 TLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFE 775
++E ++ L +++ + R ++++ P + ++ S K+ G S
Sbjct: 756 YMIEFIRDVLMRWFASRRKKVARCKSSVTPEVDERFLHELPASGKYVVKMSGP--WSYQV 813
Query: 776 VSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVAS 810
S+ E + V L+ +C C R+ IPC HA+A+
Sbjct: 814 TSKSGEHFHVVLDQCTCTCLRYTKLRIPCEHALAA 848
>At1g25784 mutator-like transposase, putative
Length = 1127
Score = 136 bits (342), Expect = 7e-32
Identities = 121/507 (23%), Positives = 216/507 (41%), Gaps = 43/507 (8%)
Query: 310 VGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV-AGCPFYLRLSKTDSRSYW 368
VG F ++ + +Q + A++Q K K ++CV CP+ L +
Sbjct: 379 VGSVFKNRKVLQQTMSLQAIKQCFCFKQPKSCPKTLKMVCVDETCPWQLTARVVKDSESF 438
Query: 369 LLVSLEADHTC---CRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLR-WGILL 424
+ S HTC R N+ A K L T + + L+ + +
Sbjct: 439 KITSYATTHTCNIDSRKYYNKHANYKLLGEVVRSRYSSTQGGPRAVDLPQLLLKDLNVRI 498
Query: 425 GKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPV-FERI 483
A+RAK AVE ++G+ + Y L Y L +NPG+ + T G F+ +
Sbjct: 499 SYSTAWRAKEVAVENVRGDEIANYRFLPTYLYLLQLANPGTITHLHYTPEDDGKQRFKYV 558
Query: 484 YVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKA 543
+V A+ R ++ +DG L G Y G LL A +DGN Q+FPIAF VV+ ET A
Sbjct: 559 FVSLGASIKGLIYM-RKVVVVDGTQLVGPYKGCLLIACAQDGNFQIFPIAFGVVDGETDA 617
Query: 544 SWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQ 603
SW+WF + L + D +M+ +S +S + +
Sbjct: 618 SWAWFFEKLAE-----------IVPDSDDLMI------------------VSDRHSSIYK 648
Query: 604 GLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKE 663
G L+ + H C HL N Y + + AA+A + ++ E+++E
Sbjct: 649 G----LSVVYPRAHHGACAVHLERNLSTYYGKFGVSALFFSAAKAYRVRDFEKYFELLRE 704
Query: 664 LNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKF 723
+ + + ++ + WTR A+ R + ++ +N E+ N + + + PI+ ++E ++
Sbjct: 705 KSAKCAKYLEDIGFEHWTR-AHCRGERYNIMSSNNSESMNHVLTKAKTYPIVYMIEFIRD 763
Query: 724 YLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKY 783
L +++ + R ++++ P + ++ S K+ G S S+ E +
Sbjct: 764 VLMRWFASRRKKVARCKSSVTPEVDERFLQELPASGKYAVKMSGP--WSYQVTSKSGEHF 821
Query: 784 SVNLESRSCACRRWDLSGIPCAHAVAS 810
V L+ +C C R+ IPC HA+A+
Sbjct: 822 HVVLDQCTCTCLRYTKLRIPCEHALAA 848
>At2g07230 Mutator-like transposase
Length = 590
Score = 130 bits (328), Expect = 3e-30
Identities = 120/462 (25%), Positives = 206/462 (43%), Gaps = 60/462 (12%)
Query: 278 DLDSP---SDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKD 334
D D+P D EEG + L R D E +G F +K+ + +A+++K +
Sbjct: 20 DRDAPPYFDDPGEEGNTYYLHRALKDADYEGDNIFIGRLFKNKEDCATKLAIHAIRRKFN 79
Query: 335 LTFKKDDKKRCVVICVAG-CPFYLRLSKTDSRSYWLLVSLEADHTC---CRSASNRQAKT 390
K + +CV+ CP+ + +K + + + HTC R ++QA T
Sbjct: 80 FITAKSCPNIVLAVCVSHTCPWRVYATKLEDSERFEIKCATQQHTCSVDARGDFHKQAST 139
Query: 391 KYLAR----KFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLD 446
+ + K++ + + + N L + R + + + WKA+RA+ +++ G +
Sbjct: 140 AVIGQLMRTKYLGVGKGPRS---NELRKMLRDEFSLNVSYWKAWRAREISMDNAMGSAMG 196
Query: 447 QYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFA--TTCRPLIGL 504
YA ++ Y L+ +NP S V + + K KG ER F A A R +I +
Sbjct: 197 SYALVQPYFKLLMETNPNSLVAMDTEKDKKG--LERFRYLFFALDAAVKGYAYMRKVIVI 254
Query: 505 DGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFV----DLLVNDLDMVE 560
DG L+G YGG L+AA +D N Q+FPIAF +V +E +W+WF+ D + ND D+V
Sbjct: 255 DGTHLRGRYGGCLIAASAQDANFQVFPIAFGIVNSENDDAWTWFMERLTDAIPNDPDLV- 313
Query: 561 RKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRL 620
F+SD+ H SI Y S M+ + P + H
Sbjct: 314 -----FVSDR----------HSSI--------YAS------MRKVYPMSS-------HAA 337
Query: 621 CIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMW 680
C+ HL N + + + AAR+ ++ ++ ++ A + + + W
Sbjct: 338 CVVHLKRNIVSIFKSEGLSFLVASAARSYRPSDFNRIFAEVRAMHPACADYLEGIGFEHW 397
Query: 681 TRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLK 722
TRS + D + +N+ E+ N + RD P+I+LLE L+
Sbjct: 398 TRSHFVGD-RYFFMTSNIAESLNNVLTMARDYPVISLLETLR 438
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,406,017
Number of Sequences: 26719
Number of extensions: 1006600
Number of successful extensions: 5469
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 92
Number of HSP's successfully gapped in prelim test: 109
Number of HSP's that attempted gapping in prelim test: 4798
Number of HSP's gapped (non-prelim): 489
length of query: 994
length of database: 11,318,596
effective HSP length: 109
effective length of query: 885
effective length of database: 8,406,225
effective search space: 7439509125
effective search space used: 7439509125
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0112.15


