

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0112.12
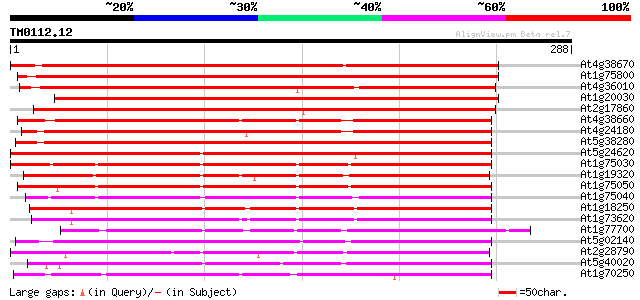
(288 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38670 thaumatin-like protein 384 e-107
At1g75800 thaumatin, putative 328 2e-90
At4g36010 thaumatin-like predicted GPI-anchored protein 322 2e-88
At1g20030 thaumatin-like protein, predicted GPI-anchored protein... 314 4e-86
At2g17860 putative thaumatin-like pathogenesis-related protein 309 1e-84
At4g38660 thaumatin-like protein 268 3e-72
At4g24180 thaumatin-like protein 266 1e-71
At5g38280 receptor serine/threonine kinase PR5K 261 4e-70
At5g24620 thaumatin-like protein 258 3e-69
At1g75030 thaumatin-like protein 241 3e-64
At1g19320 pathogenesis-related protein 5 precursor, putative 241 3e-64
At1g75050 thaumatin-like protein 238 3e-63
At1g75040 thaumatin-like protein 233 1e-61
At1g18250 putative protein 230 8e-61
At1g73620 thaumatin like protein 228 2e-60
At1g77700 thaumatin-like protein 216 1e-56
At5g02140 thaumatin-like protein 195 3e-50
At2g28790 thaumatin like protein 194 4e-50
At5g40020 thaumatin-like protein 189 1e-48
At1g70250 hypothetical protein 189 1e-48
>At4g38670 thaumatin-like protein
Length = 281
Score = 384 bits (985), Expect = e-107
Identities = 186/254 (73%), Positives = 207/254 (81%), Gaps = 7/254 (2%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
M R+LLF+IL + S LSEV+ SF+I N+CR+TIWPGLLSGANS PLPTTGF L G
Sbjct: 2 MNRILLFMILAFV---SSLSEVEPISFKITNRCRNTIWPGLLSGANSAPLPTTGFRLSRG 58
Query: 61 KSRTLHFPKAWSGRIWARTLCGHDRD-GKFSCVTADCGSDKVECIG-GAKPPATLAEFTL 118
KS+T+ P++WSGR+WARTLC DR G F C+T DCGS KVEC G GAKPPATLAEFTL
Sbjct: 59 KSKTVAIPESWSGRLWARTLCSQDRSSGSFVCLTGDCGSGKVECSGSGAKPPATLAEFTL 118
Query: 119 NGADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGG 178
NG GLDFYDVSLVDGYNLPMLI+ ++ GGC TGCLVDLNG CPRDLK+ G G
Sbjct: 119 NGTGGLDFYDVSLVDGYNLPMLILPKKIVIGGCGATGCLVDLNGACPRDLKLV-TRGNGN 177
Query: 179 SVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCAS 238
VAC SACEAFGDPRYCCS+AY+TPDTC PSVYSLFFKHACPRAYSYAYDDKTSTYTCA+
Sbjct: 178 GVACRSACEAFGDPRYCCSDAYATPDTCQPSVYSLFFKHACPRAYSYAYDDKTSTYTCAT 237
Query: 239 -ANYLIIFCPLPYT 251
A+Y IIFCP PYT
Sbjct: 238 GADYFIIFCPPPYT 251
>At1g75800 thaumatin, putative
Length = 330
Score = 328 bits (841), Expect = 2e-90
Identities = 157/250 (62%), Positives = 188/250 (74%), Gaps = 7/250 (2%)
Query: 5 LLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRT 64
L+FLI F+ F+S V+S SF +VNKC +T+WPGLLS A PPLPTTGF L+ G+ RT
Sbjct: 7 LIFLI----FSHLFVSGVRSTSFIMVNKCEYTVWPGLLSNAGVPPLPTTGFVLQKGEERT 62
Query: 65 LHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADG 123
+ P +W GR W RT C D DGKF+C+T DCGS +EC G GA PPATLAEFTL+G++G
Sbjct: 63 ISAPTSWGGRFWGRTQCSTDTDGKFTCLTGDCGSGTLECSGSGATPPATLAEFTLDGSNG 122
Query: 124 LDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTG-GSVAC 182
LDFYDVSLVDGYN+PML+ + G+ CS TGC+VDLNG CP +LKV ++G G S+ C
Sbjct: 123 LDFYDVSLVDGYNVPMLVAPQGGSGLNCSSTGCVVDLNGSCPSELKVTSLDGRGKQSMGC 182
Query: 183 MSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCA-SANY 241
SACEAF P YCCS A+ TPDTC PS YSL FK ACPRAYSYAYDD++ST+TCA S NY
Sbjct: 183 KSACEAFRTPEYCCSGAHGTPDTCKPSSYSLMFKTACPRAYSYAYDDQSSTFTCAESPNY 242
Query: 242 LIIFCPLPYT 251
+I FCP P T
Sbjct: 243 VITFCPTPNT 252
>At4g36010 thaumatin-like predicted GPI-anchored protein
Length = 301
Score = 322 bits (824), Expect = 2e-88
Identities = 157/251 (62%), Positives = 183/251 (72%), Gaps = 13/251 (5%)
Query: 6 LFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTL 65
LFL+L+ L F+ V S +F IVN+C +T+WPGLLSGA + PLPTTGF+L ++R +
Sbjct: 8 LFLLLI----LVFIYGVSSTTFTIVNQCSYTVWPGLLSGAGTSPLPTTGFSLNPTETRVI 63
Query: 66 HFPKAWSGRIWARTLCGHDRD-GKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADG 123
P AWSGRIW RTLC D G+F+C+T DCGS VEC G GA PPATLAEFTLNGA+G
Sbjct: 64 PIPAAWSGRIWGRTLCTQDATTGRFTCITGDCGSSTVECSGSGAAPPATLAEFTLNGANG 123
Query: 124 LDFYDVSLVDGYNLPMLIVARRG-----TRGGCSPTGCLVDLNGGCPRDLKVARVNGTGG 178
LDFYDVSLVDGYN+PM IV + G G C+ TGC+ +LNG CP LKVA G
Sbjct: 124 LDFYDVSLVDGYNIPMTIVPQGGGDAGGVAGNCTTTGCVAELNGPCPAQLKVATTGAEG- 182
Query: 179 SVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCAS 238
VAC SACEAFG P YCCS A+ TPDTC PS YS FFK+ACPRAYSYAYDD TST+TC
Sbjct: 183 -VACKSACEAFGTPEYCCSGAFGTPDTCKPSEYSQFFKNACPRAYSYAYDDGTSTFTCGG 241
Query: 239 ANYLIIFCPLP 249
A+Y+I FCP P
Sbjct: 242 ADYVITFCPSP 252
>At1g20030 thaumatin-like protein, predicted GPI-anchored protein
(by homology)
Length = 299
Score = 314 bits (804), Expect = 4e-86
Identities = 147/231 (63%), Positives = 173/231 (74%), Gaps = 3/231 (1%)
Query: 24 SASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSGRIWARTLCGH 83
S SF NKC +T+WPG+LS A PLPTTGF L G++RT++ P +W GR W RTLC
Sbjct: 2 SRSFTFSNKCDYTVWPGILSNAGVSPLPTTGFVLLKGETRTINAPSSWGGRFWGRTLCST 61
Query: 84 DRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGLDFYDVSLVDGYNLPMLIV 142
D DGKFSC T DCGS K+EC G GA PPATLAEFTL+G+ GLDFYDVSLVDGYN+ ML+V
Sbjct: 62 DSDGKFSCATGDCGSGKIECSGAGAAPPATLAEFTLDGSGGLDFYDVSLVDGYNVQMLVV 121
Query: 143 ARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGG-SVACMSACEAFGDPRYCCSEAYS 201
+ G+ CS TGC+VDLNG CP +L+V V+G ++AC SACEAF P YCCS A+
Sbjct: 122 PQGGSGQNCSSTGCVVDLNGSCPSELRVNSVDGKEAVAMACKSACEAFRQPEYCCSGAFG 181
Query: 202 TPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCA-SANYLIIFCPLPYT 251
+PDTC PS YS FK ACPRAYSYAYDDK+ST+TCA S NY+I FCP P T
Sbjct: 182 SPDTCKPSTYSRIFKSACPRAYSYAYDDKSSTFTCAKSPNYVITFCPSPNT 232
>At2g17860 putative thaumatin-like pathogenesis-related protein
Length = 253
Score = 309 bits (791), Expect = 1e-84
Identities = 148/241 (61%), Positives = 175/241 (72%), Gaps = 4/241 (1%)
Query: 13 LFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWS 72
L + F++ S +F IVN+C +T+WPGLLSGA + PLPTTGF+L S +SR + P AWS
Sbjct: 11 LAIMFFINGGSSTTFTIVNQCNYTVWPGLLSGAGTAPLPTTGFSLNSFESRLISIPAAWS 70
Query: 73 GRIWARTLCGHDR-DGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGLDFYDVS 130
GRIW RTLC + G F+CVT DCGS ++EC G GA PPATLAEFTLNGA+ LDFYDVS
Sbjct: 71 GRIWGRTLCNQNEITGIFTCVTGDCGSSQIECSGAGAIPPATLAEFTLNGAENLDFYDVS 130
Query: 131 LVDGYNLPMLIVARRGTRG--GCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEA 188
LVDGYN+PM IV G G C+ TGC DLNG CP LKV G+VAC SACEA
Sbjct: 131 LVDGYNIPMTIVPYGGAIGVGKCNATGCAADLNGLCPEQLKVTVEAAGTGAVACKSACEA 190
Query: 189 FGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFCPL 248
FG P +CC+ A+ TPDTC PS YS+FFK CP AYSYAYDD TST+TC+ A+Y+I FCP
Sbjct: 191 FGTPEFCCNGAFGTPDTCQPSEYSVFFKKTCPTAYSYAYDDGTSTFTCSGADYVITFCPS 250
Query: 249 P 249
P
Sbjct: 251 P 251
>At4g38660 thaumatin-like protein
Length = 345
Score = 268 bits (685), Expect = 3e-72
Identities = 135/244 (55%), Positives = 159/244 (64%), Gaps = 14/244 (5%)
Query: 5 LLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRT 64
L F +LL T S+ ++F N+C +T+WPG+LS A SP L TTGF L G SR+
Sbjct: 15 LSFTVLLLASTGSY-----GSTFTFANRCGYTVWPGILSNAGSPTLSTTGFELPKGTSRS 69
Query: 65 LHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADG 123
L P WSGR WART C D G +C T DCGS+ VEC G GA PP TLAEFTL G G
Sbjct: 70 LQAPTGWSGRFWARTGCKFDSSGSGTCKTGDCGSNAVECAGLGAAPPVTLAEFTL-GTGG 128
Query: 124 LDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACM 183
DFYDVSLVDGYN+PM++ G+ G C+ TGC DLN CP +L+ G AC
Sbjct: 129 DDFYDVSLVDGYNIPMIVEVAGGS-GQCASTGCTTDLNIQCPAELRF------GDGDACK 181
Query: 184 SACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLI 243
SAC AF P YCCS AY+TP +C PSVYS FK ACPR+YSYAYDD TST+TCA +Y +
Sbjct: 182 SACLAFRSPEYCCSGAYATPSSCRPSVYSEMFKAACPRSYSYAYDDATSTFTCAGGDYTV 241
Query: 244 IFCP 247
FCP
Sbjct: 242 TFCP 245
>At4g24180 thaumatin-like protein
Length = 255
Score = 266 bits (680), Expect = 1e-71
Identities = 133/248 (53%), Positives = 163/248 (65%), Gaps = 16/248 (6%)
Query: 7 FLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLH 66
+ I+LS F A+ IVN+C T+WPG+LS + S + TTGF L G SR+
Sbjct: 13 YFIILSFL---FFHGSDGATITIVNRCSFTVWPGILSNSGSGDIGTTGFELVPGGSRSFQ 69
Query: 67 FPKAWSGRIWARTLCGHDRD-GKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNG---- 120
P +WSGR WART C + D G+ +C+T DCGS++VEC G GAKPPATLAEFT+
Sbjct: 70 APASWSGRFWARTGCNFNSDTGQGTCLTGDCGSNQVECNGAGAKPPATLAEFTIGSGPAD 129
Query: 121 -ADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGS 179
A DFYDVSLVDGYN+PML+ A G+ G C TGC+ DLN CP +L+ G
Sbjct: 130 PARKQDFYDVSLVDGYNVPMLVEASGGSEGTCLTTGCVTDLNQKCPTELRF------GSG 183
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASA 239
AC SACEAFG P YCCS AY++P C PS+YS FK ACPR+YSYA+DD TST+TC A
Sbjct: 184 SACKSACEAFGSPEYCCSGAYASPTECKPSMYSEIFKSACPRSYSYAFDDATSTFTCTDA 243
Query: 240 NYLIIFCP 247
+Y I FCP
Sbjct: 244 DYTITFCP 251
>At5g38280 receptor serine/threonine kinase PR5K
Length = 665
Score = 261 bits (666), Expect = 4e-70
Identities = 123/247 (49%), Positives = 166/247 (66%), Gaps = 6/247 (2%)
Query: 4 LLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSR 63
L L +L+S F F+S V S +F I NKC +T+WPG L+ + LPT GF+L+ G+SR
Sbjct: 7 LSLMFLLVSHF---FVSGVMSRNFTIENKCDYTVWPGFLTMTTAVSLPTNGFSLKKGESR 63
Query: 64 TLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECI-GGAKPPATLAEFTLNGAD 122
++ P +WSGR+W R+ C G FSC T DCGS K+EC GA+PP TL +FTL+ D
Sbjct: 64 VINVPPSWSGRLWGRSFCSTSSTGNFSCATGDCGSGKIECSDSGARPPTTLIDFTLDATD 123
Query: 123 GLDFYDVSLVDGYNLPMLIVARR-GTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVA 181
G DFYDVS+VDGYNLP+++V +R G+ CS GC+V+LN CP +LKV + +A
Sbjct: 124 GQDFYDVSVVDGYNLPLVVVPQRLGSGRTCSNVGCVVNLNKTCPSELKVMGSSNKEHPIA 183
Query: 182 CMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCA-SAN 240
CM+AC+ FG P +CC Y P C P++YS FK+ CP AYSYAYD++ +T+ C+ S N
Sbjct: 184 CMNACQKFGLPEFCCYGEYGKPAKCQPTLYSTNFKNECPLAYSYAYDNENNTFRCSNSPN 243
Query: 241 YLIIFCP 247
Y+I FCP
Sbjct: 244 YVITFCP 250
>At5g24620 thaumatin-like protein
Length = 420
Score = 258 bits (658), Expect = 3e-69
Identities = 130/252 (51%), Positives = 166/252 (65%), Gaps = 6/252 (2%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
MGR + + + ++ S S++F I N C TIWPG L+G+ + PLPTTGF L+ G
Sbjct: 1 MGRSPMVPMAVLVYVSLLFSVSYSSTFVITNNCPFTIWPGTLAGSGTQPLPTTGFRLDVG 60
Query: 61 KSRTLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTL- 118
+S + WSGRIWART C D +G C+T DCG K+EC G GA PP +L E TL
Sbjct: 61 QSVRIPTALGWSGRIWARTGCNFDANGAGKCMTGDCGG-KLECAGNGAAPPTSLFEITLG 119
Query: 119 NGADGLDFYDVSLVDGYNLPMLIVARRGTR-GGCSPTGCLVDLNGGCPRDLKVARVNGT- 176
+G+D DFYDVSLVDGYNLP++ + G G C+ TGC+ D+N CP++L+V
Sbjct: 120 HGSDDKDFYDVSLVDGYNLPIVALPTGGGLVGACNATGCVADINISCPKELQVMGEEEAE 179
Query: 177 -GGSVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYT 235
GG VAC SACEAFG +YCCS ++ P TC PS+YS FK ACPRAYSYA+DD TST+T
Sbjct: 180 RGGVVACKSACEAFGLDQYCCSGQFANPTTCRPSLYSSIFKRACPRAYSYAFDDGTSTFT 239
Query: 236 CASANYLIIFCP 247
C ++ Y IIFCP
Sbjct: 240 CKASEYAIIFCP 251
>At1g75030 thaumatin-like protein
Length = 246
Score = 241 bits (616), Expect = 3e-64
Identities = 123/248 (49%), Positives = 158/248 (63%), Gaps = 6/248 (2%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
M ++ IL +F S +++ + F + N C +T+WPG LSG NS L GF L G
Sbjct: 1 MAKISSIHILFFVFITSGIAD-SATVFTLQNSCAYTVWPGTLSG-NSITLGDGGFPLTPG 58
Query: 61 KSRTLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNG 120
S L P WSGR WART C D G +CVT DCG ++C GG PPATLAEFT+
Sbjct: 59 ASVQLTAPTGWSGRFWARTGCNFDASGHGTCVTGDCGG-VLKCTGGGVPPATLAEFTVGS 117
Query: 121 AD-GLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGS 179
++ G+DFYDVSLVDGYN+ M I + G G C GC+ D+N CP +L++ N +G
Sbjct: 118 SNAGMDFYDVSLVDGYNVKMGIKPQGGF-GNCKYAGCVSDINEICPSELRIMDPN-SGSV 175
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASA 239
AC SAC AF P +CC+ A++TP TC+P+ YS FK+ACP AYSYAYDD +ST+TC +
Sbjct: 176 AACKSACAAFSSPEFCCTGAHATPQTCSPTYYSSMFKNACPSAYSYAYDDASSTFTCTGS 235
Query: 240 NYLIIFCP 247
NYLI FCP
Sbjct: 236 NYLITFCP 243
>At1g19320 pathogenesis-related protein 5 precursor, putative
Length = 247
Score = 241 bits (615), Expect = 3e-64
Identities = 127/245 (51%), Positives = 151/245 (60%), Gaps = 13/245 (5%)
Query: 8 LILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHF 67
L+ +S + V +F + N C TIWPG+L+ AN L GFAL SG S T
Sbjct: 9 LLFISFIIATCTISVSGTTFTLTNHCGSTIWPGILT-ANGAQLGDGGFALASGSSVTFTV 67
Query: 68 PKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGL-- 124
WSGR WART C D G C T DCGS K++C G G PPATLAEFT+ G+ G
Sbjct: 68 SPGWSGRFWARTYCNFDASGSGKCGTGDCGS-KLKCAGAGGAPPATLAEFTI-GSSGKKN 125
Query: 125 ---DFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVA 181
DFYDVSLVDGYN+ M I + G+ G C GC+ D+N CP++L+V G G A
Sbjct: 126 AVQDFYDVSLVDGYNVQMKITPQGGS-GDCKTAGCVSDVNAICPKELQVT---GPSGVAA 181
Query: 182 CMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANY 241
C SACEAF P YCC+ AYSTP TC P+ YS FK ACP AYSYAYDD +ST+TC +ANY
Sbjct: 182 CKSACEAFNKPEYCCTGAYSTPATCPPTNYSKIFKQACPSAYSYAYDDASSTFTCTNANY 241
Query: 242 LIIFC 246
I FC
Sbjct: 242 EISFC 246
>At1g75050 thaumatin-like protein
Length = 257
Score = 238 bits (607), Expect = 3e-63
Identities = 122/250 (48%), Positives = 154/250 (60%), Gaps = 11/250 (4%)
Query: 5 LLFLILLSLFTLSFLSEVQ-------SASFEIVNKCRHTIWPGLLSGANSPPLPTTGFAL 57
+LFL+ +++F + E+ + F + N C +T+WPG+LSG N L GF L
Sbjct: 9 ILFLVFITIFIVVVCFEIAGGIAVSAATVFTLQNSCPYTVWPGILSG-NDNTLGDGGFPL 67
Query: 58 ESGKSRTLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFT 117
G S L P WSGR WART C D G +C T DCG ++C GG PP TLAEFT
Sbjct: 68 TPGASVQLTAPAGWSGRFWARTGCNFDASGHGNCGTGDCGG-VLKCNGGGVPPVTLAEFT 126
Query: 118 LNGADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTG 177
L G G DFYDVSLVDGYN+ M I + G+ G C GC+ D+N CP +L++ + TG
Sbjct: 127 LVGDGGKDFYDVSLVDGYNVEMGIKPQGGS-GDCHYAGCVADVNAVCPNELRLMDPH-TG 184
Query: 178 GSVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCA 237
AC SAC AF +CC+ A++TP TC+P+ YS FK ACP AYSYAYDD TST+TC
Sbjct: 185 IIAACKSACAAFNSEEFCCTGAHATPQTCSPTHYSAMFKSACPGAYSYAYDDATSTFTCT 244
Query: 238 SANYLIIFCP 247
+NYLI FCP
Sbjct: 245 GSNYLISFCP 254
>At1g75040 thaumatin-like protein
Length = 239
Score = 233 bits (593), Expect = 1e-61
Identities = 124/239 (51%), Positives = 142/239 (58%), Gaps = 8/239 (3%)
Query: 9 ILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFP 68
IL +F S ++ V + F + N C T+W G L+G P L GF L G SR L P
Sbjct: 9 ILFLVFITSGIA-VMATDFTLRNNCPTTVWAGTLAG-QGPKLGDGGFELTPGASRQLTAP 66
Query: 69 KAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYD 128
WSGR WART C D G CVT DCG + C GG PP TLAEFTL G G DFYD
Sbjct: 67 AGWSGRFWARTGCNFDASGNGRCVTGDCGG--LRCNGGGVPPVTLAEFTLVGDGGKDFYD 124
Query: 129 VSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEA 188
VSLVDGYN+ + I G+ G C GC+ DLN CP LKV N VAC SACE
Sbjct: 125 VSLVDGYNVKLGIRPSGGS-GDCKYAGCVSDLNAACPDMLKVMDQNNV---VACKSACER 180
Query: 189 FGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFCP 247
F +YCC A P+TC P+ YS FK+ACP AYSYAYDD+TST+TC ANY I FCP
Sbjct: 181 FNTDQYCCRGANDKPETCPPTDYSRIFKNACPDAYSYAYDDETSTFTCTGANYEITFCP 239
>At1g18250 putative protein
Length = 243
Score = 230 bits (586), Expect = 8e-61
Identities = 120/241 (49%), Positives = 147/241 (60%), Gaps = 9/241 (3%)
Query: 11 LSLFTLSFLSEVQSASFEIV---NKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHF 67
++LF +FL + AS V NKC+H +WPG+ A L GF L + K+ +L
Sbjct: 4 INLFLFAFLLLLSHASASTVIFYNKCKHPVWPGIQPSAGQNLLAGGGFKLPANKAHSLQL 63
Query: 68 PKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGLDF 126
P WSGR W R C DR G+ C T DCG + C G G +PPATLAE TL LDF
Sbjct: 64 PPLWSGRFWGRHGCTFDRSGRGHCATGDCGGS-LSCNGAGGEPPATLAEITLGPE--LDF 120
Query: 127 YDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSAC 186
YDVSLVDGYNL M I+ +G+ G CS GC+ DLN CP L+V NG VAC SAC
Sbjct: 121 YDVSLVDGYNLAMSIMPVKGS-GQCSYAGCVSDLNQMCPVGLQVRSRNGKR-VVACKSAC 178
Query: 187 EAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFC 246
AF P+YCC+ + P +C P+ YS FK ACP+AYSYAYDD TS TC+ ANY++ FC
Sbjct: 179 SAFNSPQYCCTGLFGNPQSCKPTAYSKIFKVACPKAYSYAYDDPTSIATCSKANYIVTFC 238
Query: 247 P 247
P
Sbjct: 239 P 239
>At1g73620 thaumatin like protein
Length = 264
Score = 228 bits (582), Expect = 2e-60
Identities = 121/239 (50%), Positives = 144/239 (59%), Gaps = 7/239 (2%)
Query: 12 SLFTLSFLSEVQSASFEIV---NKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFP 68
SLF L L V AS V NKC +T+WPG+ + L GF L ++ TL P
Sbjct: 26 SLFLLPLLLLVSQASASTVIFYNKCTYTVWPGIQPSSGQSLLAGGGFKLSPNRAYTLQLP 85
Query: 69 KAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYD 128
WSGR W R C DR G+ C T DCG + G PPATLAE TL G D +DFYD
Sbjct: 86 PLWSGRFWGRHGCSFDRSGRGRCATGDCGGSFLCNGAGGVPPATLAEITL-GHD-MDFYD 143
Query: 129 VSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEA 188
VSLVDGYNL M I+ +GT G C+ GC+ DLN CP L+V +GT VAC SAC A
Sbjct: 144 VSLVDGYNLAMSIMPVKGT-GKCTYAGCVSDLNRMCPVGLQVRSRDGTQ-VVACKSACSA 201
Query: 189 FGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFCP 247
F PRYCC+ + P +C P+ YS FK ACP+AYSYAYDD TS TC+ ANY++ FCP
Sbjct: 202 FNSPRYCCTGLFGNPQSCKPTAYSKIFKVACPKAYSYAYDDPTSIATCSKANYVVTFCP 260
>At1g77700 thaumatin-like protein
Length = 356
Score = 216 bits (550), Expect = 1e-56
Identities = 109/241 (45%), Positives = 138/241 (57%), Gaps = 12/241 (4%)
Query: 27 FEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSGRIWARTLCGHDRD 86
F I+N C TIWP + G N GF L+ G+S H P WSGRIW RT C D
Sbjct: 91 FTIINSCDQTIWPAITPGEN---FNGGGFELKPGQSIVFHAPVGWSGRIWGRTGCKFDST 147
Query: 87 GKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYDVSLVDGYNLPMLIVARRG 146
G +C T CGS ++C KPPA+LAEFTL LDFYDVSLVDG+NLPM + G
Sbjct: 148 GTGTCETGSCGST-LKCSASGKPPASLAEFTLAA---LDFYDVSLVDGFNLPMSVTPMNG 203
Query: 147 TRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDPRYCCSEAYSTPDTC 206
+G CS GC+ DL CP++L V G ++C SAC+ F YCC Y P C
Sbjct: 204 -KGNCSVAGCVADLRPHCPQELAV---KSNGKVISCRSACDVFDRDEYCCRGVYGNPVVC 259
Query: 207 APSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFCPLPYTRYNYSIIILYDFVGF 266
P+ YS FK ACP AYSYAYDD TS TC +++Y+I FC + + + + +D+ +
Sbjct: 260 QPTYYSKIFKQACPTAYSYAYDDPTSIMTCTASDYVISFCSSRFVLFTF-LTTSHDYFNY 318
Query: 267 D 267
D
Sbjct: 319 D 319
>At5g02140 thaumatin-like protein
Length = 294
Score = 195 bits (495), Expect = 3e-50
Identities = 103/246 (41%), Positives = 135/246 (54%), Gaps = 13/246 (5%)
Query: 4 LLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSR 63
L L + +S FT A IVN C+ +IWPG+L G GF + SG+
Sbjct: 7 LFLLFLTISAFT-------DGAQLIIVNNCQESIWPGILGGGGQITPRNGGFHMGSGEET 59
Query: 64 TLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGAD 122
+ P WSGRIW R C +++GK SC T DC + C G G PPAT+ E TL +
Sbjct: 60 IIDVPDKWSGRIWGRQGCTFNQNGKGSCQTGDCNDGSINCQGTGGVPPATVVEMTLGSSS 119
Query: 123 G-LDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVA 181
L FYDVSLVDG+NLP+ + G G C C V+LN CP L+V R G V
Sbjct: 120 SPLHFYDVSLVDGFNLPVSMKPIGGGVG-CGVAACEVNLNICCPSALEVKR---DGKVVG 175
Query: 182 CMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANY 241
C SAC A +YCC+ Y+ P C P++++ FK CP+AYSYA+DD +S C ++ Y
Sbjct: 176 CKSACLAMQSAKYCCTGEYANPQACKPTLFANLFKAICPKAYSYAFDDSSSLNKCRASRY 235
Query: 242 LIIFCP 247
+I FCP
Sbjct: 236 VITFCP 241
>At2g28790 thaumatin like protein
Length = 249
Score = 194 bits (494), Expect = 4e-50
Identities = 112/253 (44%), Positives = 147/253 (57%), Gaps = 12/253 (4%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASF--EIVNKCRHTIWPGLLSGANSPPLPTTGFALE 58
M + L L L +SF S V ++ +VN C T+WP + A P L GFAL
Sbjct: 1 MAKTSLPLAASFLLLISFSSAVDTSRLFLTVVNNCPFTVWPAIQPNAGHPVLEKGGFALP 60
Query: 59 SGKSRTLHFPKA-WSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEF 116
+ R+ + P WSGRIWART C H +GKFSC+T DCG+ ++EC G G PPA+LA+F
Sbjct: 61 TFTHRSFNVPTTHWSGRIWARTWCAH-YNGKFSCLTGDCGN-RLECNGLGGAPPASLAQF 118
Query: 117 TLNGADGLDF--YDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVN 174
L+ DF Y VSLVDGYN+PM + G G C GC DL CP L+V +
Sbjct: 119 DLHHGGHHDFSSYGVSLVDGYNVPMTVTPHEG-HGVCPVVGCREDLIKTCPAHLQVR--S 175
Query: 175 GTGGSVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTY 234
+G VAC S CEAF CC Y++P+TC S +SLFFKHACP ++++A+D + +
Sbjct: 176 HSGHVVACKSGCEAFHTDELCCRGHYNSPNTCKASSHSLFFKHACPSSFTFAHDSPSLMH 235
Query: 235 TCASANYL-IIFC 246
CAS L +IFC
Sbjct: 236 DCASPRELKVIFC 248
>At5g40020 thaumatin-like protein
Length = 256
Score = 189 bits (481), Expect = 1e-48
Identities = 106/245 (43%), Positives = 136/245 (55%), Gaps = 11/245 (4%)
Query: 10 LLSLFTLS---FLSEVQS--ASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRT 64
LL LF +S +V S +F + NKC IWP + + P L + GF L G +
Sbjct: 8 LLQLFIISSCIVYGKVTSHEVTFYVQNKCSFPIWPAVAPNSGHPVLASGGFYLPCGGIKR 67
Query: 65 LHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGG-AKPPATLAEFTLNGADG 123
+ P WSGRIWART C + K +C T DC +EC G KPPATL + +
Sbjct: 68 IDAPLGWSGRIWARTGCDFTSNSKQACETGDCDGH-LECNGLIGKPPATLIQIAVQADKS 126
Query: 124 L-DFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVAC 182
+FYDVSLVDGYNLP+ +V + C+ GC DL CP +L+V +N G VAC
Sbjct: 127 KPNFYDVSLVDGYNLPV-VVNSKPVSSKCTILGCHKDLKTTCPEELQV--LNEEGRVVAC 183
Query: 183 MSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYL 242
SAC AF + R+CC AY TP+ C + YS+ FK ACP YSYAY+ TC++ YL
Sbjct: 184 KSACLAFDNDRFCCRNAYGTPEKCKRNTYSMLFKEACPNYYSYAYETPPPLVTCSAKEYL 243
Query: 243 IIFCP 247
I FCP
Sbjct: 244 ITFCP 248
>At1g70250 hypothetical protein
Length = 676
Score = 189 bits (480), Expect = 1e-48
Identities = 101/251 (40%), Positives = 140/251 (55%), Gaps = 15/251 (5%)
Query: 3 RLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKS 62
RL +L+ F L S SF I NKC++TIWP S L TTGF LE G++
Sbjct: 14 RLKSLQYILNFFLLYV---AMSRSFTIENKCQYTIWPATYGYRRS--LETTGFVLEKGET 68
Query: 63 RTLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGAD 122
RT+ P +W GR W RTLC + G FSC T DC S K++C+G P T+ EF L +
Sbjct: 69 RTIKAPSSWIGRFWGRTLCSTNSTGGFSCATGDCTSGKIKCLGIPIDPTTVVEFNL-ASY 127
Query: 123 GLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVN-GTGGSVA 181
G+D+Y V++ +GYNLP+L+ C C++D+N CP +L V G+ +A
Sbjct: 128 GVDYYVVNVFNGYNLPLLVTPE---NKNCRSIECVIDMNETCPSELMVNSSGLGSHHPIA 184
Query: 182 CMSACEAFGDPRYCC----SEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCA 237
CM+ C+ + P CC S P C ++YS F + CP AYSYAYD S++TC
Sbjct: 185 CMTTCQRYQLPELCCIGLSSGVVVPPGICKRTIYSRTFNNVCPSAYSYAYDVDNSSFTCP 244
Query: 238 S-ANYLIIFCP 247
+ +N++I FCP
Sbjct: 245 NFSNFVITFCP 255
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,270,747
Number of Sequences: 26719
Number of extensions: 332793
Number of successful extensions: 825
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 710
Number of HSP's gapped (non-prelim): 32
length of query: 288
length of database: 11,318,596
effective HSP length: 98
effective length of query: 190
effective length of database: 8,700,134
effective search space: 1653025460
effective search space used: 1653025460
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0112.12


