

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0109.6
(545 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
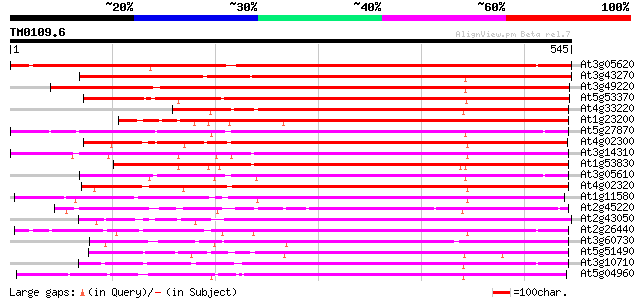
Score E
Sequences producing significant alignments: (bits) Value
At3g05620 putative pectinesterase 685 0.0
At3g43270 pectinesterase like protein 485 e-137
At3g49220 pectinesterase - like protein 427 e-119
At5g53370 pectinesterase 424 e-119
At4g33220 pectinesterase like protein 419 e-117
At1g23200 putative pectinesterase 407 e-114
At5g27870 pectin methyl-esterase - like protein 400 e-112
At4g02300 putative pectinesterase 395 e-110
At3g14310 pectin methylesterase like protein 393 e-109
At1g53830 unknown protein 390 e-109
At3g05610 putative pectinesterase 390 e-108
At4g02320 pectinesterase - like protein 389 e-108
At1g11580 unknown protein 389 e-108
At2g45220 pectinesterase like protein 387 e-108
At2g43050 putative pectinesterase 377 e-105
At2g26440 putative pectinesterase 377 e-105
At3g60730 pectinesterase - like protein 372 e-103
At5g51490 pectinesterase 369 e-102
At3g10710 putative pectinesterase 365 e-101
At5g04960 pectinesterase 363 e-100
>At3g05620 putative pectinesterase
Length = 543
Score = 685 bits (1767), Expect = 0.0
Identities = 346/556 (62%), Positives = 420/556 (75%), Gaps = 24/556 (4%)
Query: 1 MALVNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNE 60
M + AL ++M++ + SS + I + + ++L+A+AC I+ C+ NI
Sbjct: 1 MGITTALLLVMLMSVHTSS---YETTILKPYKEDNFRSLVAKACQFIDAHELCVSNIWTH 57
Query: 61 LTKIGPP-SPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVS 119
+ + G +P SVL AA++ ++A+ A++ + S+ +REQ+AI DCKELV FSV+
Sbjct: 58 VKESGHGLNPHSVLRAAVKEAHDKAKLAMERIPTVMMLSIRSREQVAIEDCKELVGFSVT 117
Query: 120 ELAWSLGEMRRIRAGD---------TSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLE 170
ELAWS+ EM ++ G + GNL+ WLSAA+SNQDTCLEGFEGT+R+ E
Sbjct: 118 ELAWSMLEMNKLHGGGGIDLDDGSHDAAAAGGNLKTWLSAAMSNQDTCLEGFEGTERKYE 177
Query: 171 SYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQEL 230
I GSL QVTQL+SNVL +YTQL+ALPF+ RN + S PEW+TE D+ L
Sbjct: 178 ELIKGSLRQVTQLVSNVLDMYTQLNALPFKASRNESVIAS---------PEWLTETDESL 228
Query: 231 L-RSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKM 289
+ R P + VVA+DG G YRTI EA+N AP+HS +RYVIYVKKG+Y+EN+D+K+K
Sbjct: 229 MMRHDPSVMHPNTVVAIDGKGKYRTINEAINEAPNHSTKRYVIYVKKGVYKENIDLKKKK 288
Query: 290 TNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVA 349
TNIMLVGDGIGQTIIT +RNFMQG TTFRTATVAVSG+GFIA+D++FRNTAGP N QAVA
Sbjct: 289 TNIMLVGDGIGQTIITGDRNFMQGLTTFRTATVAVSGRGFIAKDITFRNTAGPQNRQAVA 348
Query: 350 LRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRA 409
LRVDSDQSAFYRCS+EG+QDTLYAHSLRQFYR+CEIYGTIDFIFGNGAAVLQNCKIYTR
Sbjct: 349 LRVDSDQSAFYRCSVEGYQDTLYAHSLRQFYRDCEIYGTIDFIFGNGAAVLQNCKIYTRV 408
Query: 410 PLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQPTYLGRPWKQYSRTVYINTYMSGMVQ 469
PLPLQKVT+TAQGRKSP+Q+TGF IQ+S++LATQPTYLGRPWK YSRTVY+NTYMS +VQ
Sbjct: 409 PLPLQKVTITAQGRKSPNQNTGFVIQNSYVLATQPTYLGRPWKLYSRTVYMNTYMSQLVQ 468
Query: 470 PRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGD 529
PRGWLEWFG+FAL TLWYGEY N GPG +GRVKWPGYHIM D TA FTV FI G
Sbjct: 469 PRGWLEWFGNFALDTLWYGEYNNIGPGWRSSGRVKWPGYHIM-DKRTALSFTVGSFIDGR 527
Query: 530 SWLPGTGVKFTAGLTN 545
WLP TGV FTAGL N
Sbjct: 528 RWLPATGVTFTAGLAN 543
>At3g43270 pectinesterase like protein
Length = 527
Score = 485 bits (1249), Expect = e-137
Identities = 248/489 (50%), Positives = 339/489 (68%), Gaps = 16/489 (3%)
Query: 69 PTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEM 128
P + A +T ++ A+ ++KF + +R AI DC +L+D + EL+W +
Sbjct: 43 PPLEFAEAAKTVVDAITKAVAIVSKFDKKAGKSRVSNAIVDCVDLLDSAAEELSWIISAS 102
Query: 129 RRIRAGDTSVQYEGN-LEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISNV 187
+ D S G+ L W+SAALSNQDTCL+GFEGT+ ++ ++G L++V + N+
Sbjct: 103 QSPNGKDNSTGDVGSDLRTWISAALSNQDTCLDGFEGTNGIIKKIVAGGLSKVGTTVRNL 162
Query: 188 LSLYTQLHALPFRP-PRNNTHKTSESLDLD-SEFPEWMTEGDQELLRSKPHRSRADAVVA 245
L T +H+ P +P P+ +T S+FP W+ GD++LL++ + + ADAVVA
Sbjct: 163 L---TMVHSPPSKPKPKPIKAQTMTKAHSGFSKFPSWVKPGDRKLLQTD-NITVADAVVA 218
Query: 246 LDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIIT 305
DG+G++ TI++AV AAP +S +RYVI+VK+G+Y ENV++K+K NIM+VGDGI T+IT
Sbjct: 219 ADGTGNFTTISDAVLAAPDYSTKRYVIHVKRGVYVENVEIKKKKWNIMMVGDGIDATVIT 278
Query: 306 SNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIE 365
NR+F+ GWTTFR+AT AVSG+GFIARD++F+NTAGP HQAVA+R D+D FYRC++
Sbjct: 279 GNRSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVAIRSDTDLGVFYRCAMR 338
Query: 366 GFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKS 425
G+QDTLYAHS+RQF+REC I GT+DFIFG+ AV Q+C+I + LP QK ++TAQGRK
Sbjct: 339 GYQDTLYAHSMRQFFRECIITGTVDFIFGDATAVFQSCQIKAKQGLPNQKNSITAQGRKD 398
Query: 426 PHQSTGFTIQDSFILA---------TQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEW 476
P++ TGFTIQ S I A T TYLGRPWK YSRTV++ YMS + P GWLEW
Sbjct: 399 PNEPTGFTIQFSNIAADTDLLLNLNTTATYLGRPWKLYSRTVFMQNYMSDAINPVGWLEW 458
Query: 477 FGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTG 536
G+FAL TL+YGEY N GPGASL RVKWPGYH++ +A A+ FTV + I G+ WLP TG
Sbjct: 459 NGNFALDTLYYGEYMNSGPGASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGNLWLPSTG 518
Query: 537 VKFTAGLTN 545
+ F AGL +
Sbjct: 519 ITFIAGLVS 527
>At3g49220 pectinesterase - like protein
Length = 598
Score = 427 bits (1097), Expect = e-119
Identities = 224/517 (43%), Positives = 314/517 (60%), Gaps = 18/517 (3%)
Query: 40 IAQACMDIENQNSCLQNIHNELTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSV 99
I++AC C+ ++ + + S ++ + TL+ A+ + S +
Sbjct: 87 ISKACELTRFPELCVDSLMDFPGSLAASSSKDLIHVTVNMTLHHFSHALYSSASLSFVDM 146
Query: 100 SNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCL 159
R + A C EL+D SV L+ +L + A V WLSAAL+N DTC
Sbjct: 147 PPRARSAYDSCVELLDDSVDALSRALSSVVSSSAKPQDVT------TWLSAALTNHDTCT 200
Query: 160 EGFEGTDRR-LESYISGSLTQVTQLISNVLSLYTQLH-ALPFRPPRNNTHKTSESLDLDS 217
EGF+G D ++ +++ +L +++L+SN L++++ H F + + +
Sbjct: 201 EGFDGVDDGGVKDHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEEREE 260
Query: 218 EFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKG 277
+FP WM ++E+L + +AD +V+ DG+G +TI+EA+ AP +S RR +IYVK G
Sbjct: 261 KFPRWMRPKEREILEMPVSQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKAG 320
Query: 278 LYREN-VDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSF 336
Y EN + + RK N+M VGDG G+T+I+ ++ TTF TA+ A +G GFIARD++F
Sbjct: 321 RYEENNLKVGRKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDITF 380
Query: 337 RNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNG 396
N AGP HQAVALR+ +D + YRC+I G+QDTLY HS RQF+REC+IYGT+DFIFGN
Sbjct: 381 ENWAGPAKHQAVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGNA 440
Query: 397 AAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILA---------TQPTYL 447
A VLQNC IY R P+ QK T+TAQ RK P+Q+TG +I S +LA + TYL
Sbjct: 441 AVVLQNCSIYARKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTYL 500
Query: 448 GRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPG 507
GRPWK +SRTVY+ +Y+ G V RGWLEW FAL TL+YGEY N GPG+ L RV WPG
Sbjct: 501 GRPWKLFSRTVYMMSYIGGHVHTRGWLEWNTTFALDTLYYGEYLNSGPGSGLGQRVSWPG 560
Query: 508 YHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGLT 544
Y ++ A A+ FTV FI+G SWLP TGV F AGL+
Sbjct: 561 YRVINSTAEANRFTVAEFIYGSSWLPSTGVSFLAGLS 597
>At5g53370 pectinesterase
Length = 587
Score = 424 bits (1091), Expect = e-119
Identities = 221/487 (45%), Positives = 304/487 (62%), Gaps = 20/487 (4%)
Query: 72 VLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRRI 131
++ + TL + A+ + + + R + A C EL+D SV L +L + +
Sbjct: 106 LIHISFNATLQKFSKALYTSSTITYTQMPPRVRSAYDSCLELLDDSVDALTRALSSVVVV 165
Query: 132 RAGDTSVQYEGNLEAWLSAALSNQDTCLEGF---EGTDRRLESYISGSLTQVTQLISNVL 188
+GD S ++ WLS+A++N DTC +GF EG ++ + G++ +++++SN L
Sbjct: 166 -SGDES---HSDVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQVIGAVKDLSEMVSNCL 221
Query: 189 SLYT-QLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALD 247
+++ ++ L P NN + + E P W+ D+ELL + +AD V+ D
Sbjct: 222 AIFAGKVKDLSGVPVVNN--RKLLGTEETEELPNWLKREDRELLGTPTSAIQADITVSKD 279
Query: 248 GSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRE-NVDMKRKMTNIMLVGDGIGQTIITS 306
GSG ++TI EA+ AP HS+RR+VIYVK G Y E N+ + RK TN+M +GDG G+T+IT
Sbjct: 280 GSGTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEENLKVGRKKTNLMFIGDGKGKTVITG 339
Query: 307 NRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEG 366
++ TTF TAT A +G GFI RDM+F N AGP HQAVALRV D + YRC+I G
Sbjct: 340 GKSIADDLTTFHTATFAATGAGFIVRDMTFENYAGPAKHQAVALRVGGDHAVVYRCNIIG 399
Query: 367 FQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSP 426
+QD LY HS RQF+RECEIYGT+DFIFGN A +LQ+C IY R P+ QK+T+TAQ RK P
Sbjct: 400 YQDALYVHSNRQFFRECEIYGTVDFIFGNAAVILQSCNIYARKPMAQQKITITAQNRKDP 459
Query: 427 HQSTGFTIQDSFILAT---------QPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWF 477
+Q+TG +I +LAT PTYLGRPWK YSR VY+ + M + PRGWLEW
Sbjct: 460 NQNTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRVVYMMSDMGDHIDPRGWLEWN 519
Query: 478 GDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGV 537
G FAL +L+YGEY N G G+ + RVKWPGYH++ AS FTV +FI G SWLP TGV
Sbjct: 520 GPFALDSLYYGEYMNKGLGSGIGQRVKWPGYHVITSTVEASKFTVAQFISGSSWLPSTGV 579
Query: 538 KFTAGLT 544
F +GL+
Sbjct: 580 SFFSGLS 586
>At4g33220 pectinesterase like protein
Length = 404
Score = 419 bits (1077), Expect = e-117
Identities = 209/406 (51%), Positives = 278/406 (67%), Gaps = 25/406 (6%)
Query: 159 LEGFEGTDRRLESYISGSLTQVTQLISNVLSLYTQ------------LHALPFRPPRNNT 206
+EGF+GT ++S ++GSL Q+ ++ +L L + P PP
Sbjct: 1 MEGFDGTSGLVKSLVAGSLDQLYSMLRELLPLVQPEQKPKAVSKPGPIAKGPKAPPGRKL 60
Query: 207 HKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHS 266
T E L +FP+W+ D++LL S D VALDG+G++ I +A+ AP +S
Sbjct: 61 RDTDEDESL--QFPDWVRPDDRKLLESNGRTY--DVSVALDGTGNFTKIMDAIKKAPDYS 116
Query: 267 NRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSG 326
+ R+VIY+KKGLY ENV++K+K NI+++GDGI T+I+ NR+F+ GWTTFR+AT AVSG
Sbjct: 117 STRFVIYIKKGLYLENVEIKKKKWNIVMLGDGIDVTVISGNRSFIDGWTTFRSATFAVSG 176
Query: 327 KGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIY 386
+GF+ARD++F+NTAGP HQAVALR DSD S F+RC++ G+QDTLY H++RQFYREC I
Sbjct: 177 RGFLARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYTHTMRQFYRECTIT 236
Query: 387 GTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFI------- 439
GT+DFIFG+G V QNC+I + LP QK T+TAQGRK +Q +GF+IQ S I
Sbjct: 237 GTVDFIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQPSGFSIQFSNISADADLV 296
Query: 440 --LATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGA 497
L T TYLGRPWK YSRTV+I MS +V+P GWLEW DFAL TL+YGE+ NYGPG+
Sbjct: 297 PYLNTTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNADFALDTLFYGEFMNYGPGS 356
Query: 498 SLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
L+ RVKWPGYH+ ++ A+ FTV +FI G+ WLP TGV F+ GL
Sbjct: 357 GLSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTGVTFSDGL 402
>At1g23200 putative pectinesterase
Length = 554
Score = 407 bits (1047), Expect = e-114
Identities = 220/461 (47%), Positives = 284/461 (60%), Gaps = 32/461 (6%)
Query: 106 AIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGT 165
A+ DC EL + ++ +L S R G S ++ LSAA++NQDTC GF
Sbjct: 103 ALFDCLELYEDTIDQLNHS-----RRSYGQYSSPHDRQTS--LSAAIANQDTCRNGFR-- 153
Query: 166 DRRLESYISGSLT-----QVTQLISNVLSLY---TQLHALPFRPPRNNTHKTSES----- 212
D +L S S +T+ ISN L++ + A+ + P K S+
Sbjct: 154 DFKLTSSYSKYFPVQFHRNLTKSISNSLAVTKAAAEAEAVAEKYPSTGFTKFSKQRSSAG 213
Query: 213 --------LDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPS 264
L D +FP W D++LL ++AD VVA DGSGHY +I +AVNAA
Sbjct: 214 GGSHRRLLLFSDEKFPSWFPLSDRKLLEDSKTTAKADLVVAKDGSGHYTSIQQAVNAAAK 273
Query: 265 --HSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATV 322
N+R VIYVK G+YRENV +K+ + N+M++GDGI TI+T NRN G TTFR+AT
Sbjct: 274 LPRRNQRLVIYVKAGVYRENVVIKKSIKNVMVIGDGIDSTIVTGNRNVQDGTTTFRSATF 333
Query: 323 AVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRE 382
AVSG GFIA+ ++F NTAGP HQAVALR SD S FY CS +G+QDTLY HS RQF R
Sbjct: 334 AVSGNGFIAQGITFENTAGPEKHQAVALRSSSDFSVFYACSFKGYQDTLYLHSSRQFLRN 393
Query: 383 CEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILAT 442
C IYGT+DFIFG+ A+LQNC IY R P+ QK T+TAQ RK P ++TGF IQ S +
Sbjct: 394 CNIYGTVDFIFGDATAILQNCNIYARKPMSGQKNTITAQSRKEPDETTGFVIQSSTVATA 453
Query: 443 QPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGR 502
TYLGRPW+ +SRTV++ + +V P GWL W G FAL+TL+YGEY N G GAS++GR
Sbjct: 454 SETYLGRPWRSHSRTVFMKCNLGALVSPAGWLPWSGSFALSTLYYGEYGNTGAGASVSGR 513
Query: 503 VKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
VKWPGYH++K A FTV+ F+ G+ W+ TGV GL
Sbjct: 514 VKWPGYHVIKTVTEAEKFTVENFLDGNYWITATGVPVNDGL 554
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 400 bits (1029), Expect = e-112
Identities = 209/554 (37%), Positives = 313/554 (55%), Gaps = 21/554 (3%)
Query: 1 MALVNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNE 60
+++ + L I M++ + S + SD E T ++A I C + + +C + +
Sbjct: 19 ISISSVLLISMVVAVTIGVSVNKSDNAGDEEITTSVKA-IKDVCAPTDYKETCEDTLRKD 77
Query: 61 LTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSE 120
P ++ A T+ + R ++A+ CKEL+D+++ E
Sbjct: 78 AKDTS--DPLELVKTAFNATMKQISDVAKKSQTMIELQKDPRAKMALDQCKELMDYAIGE 135
Query: 121 LAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQV 180
L+ S E+ + L WLSA +S++ TCL+GF+GT I +L
Sbjct: 136 LSKSFEELGKFEFHKVDEALV-KLRIWLSATISHEQTCLDGFQGTQGNAGETIKKALKTA 194
Query: 181 TQLISNVLSLYTQL--HALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRS 238
QL N L++ T++ + + P N+ + L EFP WM + LL +
Sbjct: 195 VQLTHNGLAMVTEMSNYLGQMQIPEMNSRRL-----LSQEFPSWMDARARRLLNAPMSEV 249
Query: 239 RADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDG 298
+ D VVA DGSG Y+TI EA+N P N +V+++K+G+Y+E V + R MT+++ +GDG
Sbjct: 250 KPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIKEGIYKEYVQVNRSMTHLVFIGDG 309
Query: 299 IGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSA 358
+T+I+ ++++ G TT++TATVA+ G FIA++++F NTAG + HQAVA+RV +D+S
Sbjct: 310 PDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNIAFENTAGAIKHQAVAIRVLADESI 369
Query: 359 FYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTV 418
FY C +G+QDTLYAHS RQFYR+C I GTIDF+FG+ AAV QNC + R PL Q +
Sbjct: 370 FYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGDAAAVFQNCTLLVRKPLLNQACPI 429
Query: 419 TAQGRKSPHQSTGFTIQDSFILA---------TQPTYLGRPWKQYSRTVYINTYMSGMVQ 469
TA GRK P +STGF +Q I+ TYLGRPWK+YSRT+ +NT++ V
Sbjct: 430 TAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKTYLGRPWKEYSRTIIMNTFIPDFVP 489
Query: 470 PRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGD 529
P GW W G+F L TL+Y E +N GPGA++ RV WPG + D FT ++I GD
Sbjct: 490 PEGWQPWLGEFGLNTLFYSEVQNTGPGAAITKRVTWPGIKKLSDEEILK-FTPAQYIQGD 548
Query: 530 SWLPGTGVKFTAGL 543
+W+PG GV + GL
Sbjct: 549 AWIPGKGVPYILGL 562
>At4g02300 putative pectinesterase
Length = 532
Score = 395 bits (1014), Expect = e-110
Identities = 210/487 (43%), Positives = 304/487 (62%), Gaps = 27/487 (5%)
Query: 72 VLSAALRTTLNEAQGAIDNLTKFSTF---SVSNREQIAIGDCKELVDFSVSELAWSLGEM 128
++ A L T+ + A N + T ++++ E+ A DC L+D ++S+L ++ ++
Sbjct: 56 LIIADLNLTILKVNLASSNFSDLQTRLFPNLTHYERCAFEDCLGLLDDTISDLETAVSDL 115
Query: 129 RRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRR----LESYISGSLTQVTQLI 184
R +S+++ ++ L+ ++ QDTCL+GF +D + + +L ++ I
Sbjct: 116 R-----SSSLEFN-DISMLLTNVMTYQDTCLDGFSTSDNENNNDMTYELPENLKEIILDI 169
Query: 185 SNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVV 244
SN LS LH L + + K+SE +D E+P W++E DQ LL + + + V
Sbjct: 170 SNNLS--NSLHMLQVISRKKPSPKSSE---VDVEYPSWLSENDQRLLEAPVQETNYNLSV 224
Query: 245 ALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTII 304
A+DG+G++ TI +AV AAP+ S R++IY+K G Y ENV++ +K T IM +GDGIG+T+I
Sbjct: 225 AIDGTGNFTTINDAVFAAPNMSETRFIIYIKGGEYFENVELPKKKTMIMFIGDGIGKTVI 284
Query: 305 TSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSI 364
+NR+ + GW+TF+T TV V GKG+IA+D+SF N+AGP QAVA R SD SAFYRC
Sbjct: 285 KANRSRIDGWSTFQTPTVGVKGKGYIAKDISFVNSAGPAKAQAVAFRSGSDHSAFYRCEF 344
Query: 365 EGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRK 424
+G+QDTLY HS +QFYREC+IYGTIDFIFGN A V QN +Y R P P K+ TAQ R
Sbjct: 345 DGYQDTLYVHSAKQFYRECDIYGTIDFIFGNAAVVFQNSSLYARKPNPGHKIAFTAQSRN 404
Query: 425 SPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSGMVQPRGWLE 475
Q TG +I + ILA YLGRPW++YSRTV I +++ ++ P GWLE
Sbjct: 405 QSDQPTGISILNCRILAAPDLIPVKENFKAYLGRPWRKYSRTVIIKSFIDDLIHPAGWLE 464
Query: 476 WFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGT 535
DFAL TL+YGEY N GPGA++A RV WPG+ +++ A+ FTV FI G +WL T
Sbjct: 465 GKKDFALETLYYGEYMNEGPGANMAKRVTWPGFRRIENQTEATQFTVGPFIDGSTWLNST 524
Query: 536 GVKFTAG 542
G+ F+ G
Sbjct: 525 GIPFSLG 531
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 393 bits (1009), Expect = e-109
Identities = 227/580 (39%), Positives = 330/580 (56%), Gaps = 44/580 (7%)
Query: 1 MALVNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHN- 59
+ L++A L+ + + S S + A++ +C C+ +
Sbjct: 20 LVLLSAAVALLFVAAVAGISAGASKANEKRTLSPSSHAVLRSSCSSTRYPELCISAVVTA 79
Query: 60 ---ELTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKF--STFSVSNREQIAIGDCKELV 114
ELT S V+ A++ T+ + + K ++ RE+ A+ DC E +
Sbjct: 80 GGVELT-----SQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREKTALHDCLETI 134
Query: 115 DFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGF--EGTDRRLESY 172
D ++ EL ++ ++ T ++ G+L+ +S+A++NQ+TCL+GF + D+++
Sbjct: 135 DETLDELHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSHDDADKQVRKA 194
Query: 173 ISGSLTQVTQLISNVLSLYTQLHALPF-------------RPPRNNTHKTSESLD----- 214
+ V + SN L++ + R + +T+ ++D
Sbjct: 195 LLKGQIHVEHMCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAG 254
Query: 215 -LDSE-FPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVI 272
LDSE +P W++ GD+ LL+ +ADA VA DGSG ++T+ AV AAP +SN+RYVI
Sbjct: 255 ELDSEGWPTWLSAGDRRLLQGSG--VKADATVAADGSGTFKTVAAAVAAAPENSNKRYVI 312
Query: 273 YVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIAR 332
++K G+YRENV++ +K NIM +GDG +TIIT +RN + G TTF +ATVA G+ F+AR
Sbjct: 313 HIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAAVGERFLAR 372
Query: 333 DMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFI 392
D++F+NTAGP HQAVALRV SD SAFY C + +QDTLY HS RQF+ +C I GT+DFI
Sbjct: 373 DITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAGTVDFI 432
Query: 393 FGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ--------- 443
FGN A VLQ+C I+ R P QK VTAQGR P+Q+TG IQ I AT
Sbjct: 433 FGNAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQSVKGSF 492
Query: 444 PTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRV 503
PTYLGRPWK+YS+TV + + +S +++P GW EW G FAL TL Y EY N G GA A RV
Sbjct: 493 PTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGAGAGTANRV 552
Query: 504 KWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
KW G+ ++ AA A +T +FI G WL TG F+ GL
Sbjct: 553 KWRGFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLGL 592
>At1g53830 unknown protein
Length = 587
Score = 390 bits (1003), Expect = e-109
Identities = 216/465 (46%), Positives = 293/465 (62%), Gaps = 25/465 (5%)
Query: 102 REQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEG 161
RE A+ DC E +D ++ EL ++ ++ + + ++ +L+ +S+A++NQ TCL+G
Sbjct: 125 REVTALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDG 184
Query: 162 F--EGTDRRLESYISGSLTQVTQLISNVLSLY-----TQLHALPFRPP-----RNNTHKT 209
F + DR++ + V + SN L++ T + R NN K
Sbjct: 185 FSYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKL 244
Query: 210 SESL-DLDSE-FPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSN 267
E DLDS+ +P+W++ GD+ LL+ +ADA VA DGSG + T+ AV AAP SN
Sbjct: 245 KEVTGDLDSDGWPKWLSVGDRRLLQGST--IKADATVADDGSGDFTTVAAAVAAAPEKSN 302
Query: 268 RRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGK 327
+R+VI++K G+YRENV++ +K TNIM +GDG G+TIIT +RN + G TTF +ATVA G+
Sbjct: 303 KRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGE 362
Query: 328 GFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYG 387
F+ARD++F+NTAGP HQAVALRV SD SAFY+C + +QDTLY HS RQF+ +C I G
Sbjct: 363 RFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITG 422
Query: 388 TIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQD------SFILA 441
T+DFIFGN AAVLQ+C I R P QK VTAQGR P+Q+TG IQ+ S +LA
Sbjct: 423 TVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLA 482
Query: 442 ---TQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGAS 498
T PTYLGRPWK+YSRTV + + +S +++P GW EW G FAL TL Y EY N G GA
Sbjct: 483 VKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAG 542
Query: 499 LAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
A RVKW GY ++ A FT +FI G WL TG F+ L
Sbjct: 543 TANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>At3g05610 putative pectinesterase
Length = 669
Score = 390 bits (1002), Expect = e-108
Identities = 201/490 (41%), Positives = 293/490 (59%), Gaps = 24/490 (4%)
Query: 69 PTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEM 128
P ++ A T+ + A +R ++A+ CKEL+D+++ EL+ S E+
Sbjct: 85 PMELVKTAFNVTMKQITDAAKKSQTIMELQKDSRTRMALDQCKELMDYALDELSNSFEEL 144
Query: 129 RRIRAG--DTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISN 186
+ D ++ NL WLSAA+S+++TCLEGF+GT + +L +L N
Sbjct: 145 GKFEFHLLDEALI---NLRIWLSAAISHEETCLEGFQGTQGNAGETMKKALKTAIELTHN 201
Query: 187 VLSLYTQLHAL--PFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRS--RADA 242
L++ +++ + P N+ + L FP W+ + ++LL++ S + D
Sbjct: 202 GLAIISEMSNFVGQMQIPGLNSRRL-----LAEGFPSWVDQRGRKLLQAAAAYSDVKPDI 256
Query: 243 VVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQT 302
VVA DGSG Y+TI EA+ P N +V+++K GLY+E V + + M++++ +GDG +T
Sbjct: 257 VVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKAGLYKEYVQVNKTMSHLVFIGDGPDKT 316
Query: 303 IITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRC 362
II+ N+N+ G TT+RTATVA+ G FIA+++ F NTAG + HQAVA+RV SD+S F+ C
Sbjct: 317 IISGNKNYKDGITTYRTATVAIVGNYFIAKNIGFENTAGAIKHQAVAVRVQSDESIFFNC 376
Query: 363 SIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQG 422
+G+QDTLY HS RQF+R+C I GTIDF+FG+ AAV QNC + R PLP Q +TA G
Sbjct: 377 RFDGYQDTLYTHSHRQFFRDCTISGTIDFLFGDAAAVFQNCTLLVRKPLPNQACPITAHG 436
Query: 423 RKSPHQSTGFTIQDSFILA---------TQPTYLGRPWKQYSRTVYINTYMSGMVQPRGW 473
RK P +STGF Q I T YLGRPWK+YSRT+ +NT++ VQP+GW
Sbjct: 437 RKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGRPWKEYSRTIIMNTFIPDFVQPQGW 496
Query: 474 LEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLP 533
W GDF L TL+Y E +N GPG++LA RV W G + + FT ++I GD W+P
Sbjct: 497 QPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIKTLSEEDILK-FTPAQYIQGDDWIP 555
Query: 534 GTGVKFTAGL 543
G GV +T GL
Sbjct: 556 GKGVPYTTGL 565
>At4g02320 pectinesterase - like protein
Length = 518
Score = 389 bits (1000), Expect = e-108
Identities = 207/491 (42%), Positives = 300/491 (60%), Gaps = 27/491 (5%)
Query: 70 TSVLSAALRTTL---NEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLG 126
T ++ AL T+ N + +L + ++S+R+ A DC EL+D +V +L ++
Sbjct: 38 TELVVTALNQTISNVNLSSSNFSDLLQRLGSNLSHRDLCAFDDCLELLDDTVFDLTTAIS 97
Query: 127 EMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDR-----RLESYISGSLTQVT 181
++R + N++ LSAA++N TCL+GF +D ++ G +
Sbjct: 98 KLR------SHSPELHNVKMLLSAAMTNTRTCLDGFASSDNDENLNNNDNKTYGVAESLK 151
Query: 182 QLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRAD 241
+ + N+ S + A+ P + K E D FP W++ D+ LL+ ++ +
Sbjct: 152 ESLFNISSHVSDSLAMLENIPGHIPGKVKE----DVGFPMWVSGSDRNLLQDPVDETKVN 207
Query: 242 AVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQ 301
VVA +G+G+Y TI EA++AAP+ S R+VIY+K G Y EN+++ R+ T IM +GDGIG+
Sbjct: 208 LVVAQNGTGNYTTIGEAISAAPNSSETRFVIYIKCGEYFENIEIPREKTMIMFIGDGIGR 267
Query: 302 TIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYR 361
T+I +NR++ GWT F +ATV V G GFIA+D+SF N AGP HQAVALR SD SA+YR
Sbjct: 268 TVIKANRSYADGWTAFHSATVGVRGSGFIAKDLSFVNYAGPEKHQAVALRSSSDLSAYYR 327
Query: 362 CSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQ 421
CS E +QDT+Y HS +QFYREC+IYGT+DFIFG+ + V QNC +Y R P P QK+ TAQ
Sbjct: 328 CSFESYQDTIYVHSHKQFYRECDIYGTVDFIFGDASVVFQNCSLYARRPNPNQKIIYTAQ 387
Query: 422 GRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSGMVQPRG 472
GR++ + TG +I S ILA YLGRPW+ YSRTV + +++ +V P G
Sbjct: 388 GRENSREPTGISIISSRILAAPDLIPVQANFKAYLGRPWQLYSRTVIMKSFIDDLVDPAG 447
Query: 473 WLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWL 532
WL+W DFAL TL+YGEY N GPG+++ RV+WPG+ ++ AS F+V FI G+ WL
Sbjct: 448 WLKWKDDFALETLYYGEYMNEGPGSNMTNRVQWPGFKRIETVEEASQFSVGPFIDGNKWL 507
Query: 533 PGTGVKFTAGL 543
T + FT L
Sbjct: 508 NSTRIPFTLDL 518
>At1g11580 unknown protein
Length = 557
Score = 389 bits (1000), Expect = e-108
Identities = 216/549 (39%), Positives = 315/549 (57%), Gaps = 28/549 (5%)
Query: 5 NALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALI-AQACMDIENQNSCLQNIHNELT- 62
N +L + IL S + + I D L +Q C +Q+SC Q + +E T
Sbjct: 19 NLCLVLSFVAILGSVAFFTAQLISVNTNNNDDSLLTTSQICHGAHDQDSC-QALLSEFTT 77
Query: 63 -KIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSEL 121
+ + +L L+ ++ + + +++ S R++ DC+E++D S +
Sbjct: 78 LSLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNGVRDKAGFADCEEMMDVSKDRM 137
Query: 122 AWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVT 181
+ M +R G+ +++ N+ WLS+ L+N TCLE + + L +
Sbjct: 138 ---MSSMEELRGGNYNLESYSNVHTWLSSVLTNYMTCLESISDVSVNSKQIVKPQLEDLV 194
Query: 182 QLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSR-- 239
L+++ + P R++ + + + FP W+T D++LL S P +
Sbjct: 195 SRARVALAIFVSV-----LPARDDL-----KMIISNRFPSWLTALDRKLLESSPKTLKVT 244
Query: 240 ADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGI 299
A+ VVA DG+G ++T+ EAV AAP +SN RYVIYVKKG+Y+E +D+ +K N+MLVGDG
Sbjct: 245 ANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKGVYKETIDIGKKKKNLMLVGDGK 304
Query: 300 GQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAF 359
TIIT + N + G TTFR+ATVA +G GF+A+D+ F+NTAGP HQAVALRV +DQ+
Sbjct: 305 DATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQNTAGPAKHQAVALRVSADQTVI 364
Query: 360 YRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVT 419
RC I+ +QDTLY H+LRQFYR+ I GT+DFIFGN A V QNC I R P QK +T
Sbjct: 365 NRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSAVVFQNCDIVARNPGAGQKNMLT 424
Query: 420 AQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSGMVQP 470
AQGR+ +Q+T +IQ I A+ T+LGRPWK YSRTV + +++ + P
Sbjct: 425 AQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLGRPWKLYSRTVIMQSFIDNHIDP 484
Query: 471 RGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDS 530
GW W G+FAL+TL+YGEY N GPGA + RV W G+ ++KD+ A FTV + I G
Sbjct: 485 AGWFPWDGEFALSTLYYGEYANTGPGADTSKRVNWKGFKVIKDSKEAEQFTVAKLIQGGL 544
Query: 531 WLPGTGVKF 539
WL TGV F
Sbjct: 545 WLKPTGVTF 553
>At2g45220 pectinesterase like protein
Length = 511
Score = 387 bits (995), Expect = e-108
Identities = 225/516 (43%), Positives = 312/516 (59%), Gaps = 54/516 (10%)
Query: 44 CMDIENQNSC----LQNIHNELTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSV 99
C N C N +NE K S + L +++ L+ A A +
Sbjct: 34 CSQTPNPKPCEYFLTHNSNNEPIK----SESEFLKISMKLVLDRAILAKTHAFTLGPKCR 89
Query: 100 SNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCL 159
RE+ A DC +L D +VS++ ++ + D + WLS AL+N DTC
Sbjct: 90 DTREKAAWEDCIKLYDLTVSKINETMDPNVKCSKLDA--------QTWLSTALTNLDTCR 141
Query: 160 EGFEGTDRRLESYISGS-LTQVTQLISNVLSLYTQLHALPFR--PPRNNTHKTSESLDLD 216
GF LE ++ L ++ +SN+L ++ +PF PP +
Sbjct: 142 AGF------LELGVTDIVLPLMSNNVSNLLCNTLAINKVPFNYTPPEKDG---------- 185
Query: 217 SEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKK 276
FP W+ GD++LL+S + +AVVA DGSG+++TI EA++AA R+VIYVK+
Sbjct: 186 --FPSWVKPGDRKLLQSSTPKD--NAVVAKDGSGNFKTIKEAIDAASGSG--RFVIYVKQ 239
Query: 277 GLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSF 336
G+Y EN+++++K N+ML GDGIG+TIIT +++ G TTF +ATVA G GFIAR ++F
Sbjct: 240 GVYSENLEIRKK--NVMLRGDGIGKTIITGSKSVGGGTTTFNSATVAAVGDGFIARGITF 297
Query: 337 RNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNG 396
RNTAG N QAVALR SD S FY+CS E +QDTLY HS RQFYR+C++YGT+DFIFGN
Sbjct: 298 RNTAGASNEQAVALRSGSDLSVFYQCSFEAYQDTLYVHSNRQFYRDCDVYGTVDFIFGNA 357
Query: 397 AAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSF---------ILATQPTYL 447
AAVLQNC I+ R P + T+TAQGR P+Q+TG I +S +L + TYL
Sbjct: 358 AAVLQNCNIFARRPRS-KTNTITAQGRSDPNQNTGIIIHNSRVTAASDLRPVLGSTKTYL 416
Query: 448 GRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPG 507
GRPW+QYSRTV++ T + ++ PRGWLEW G+FAL TL+Y E++N GPGAS +GRV WPG
Sbjct: 417 GRPWRQYSRTVFMKTSLDSLIDPRGWLEWDGNFALKTLFYAEFQNTGPGASTSGRVTWPG 476
Query: 508 YHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
+ ++ A+ AS FTV F+ G SW+P + V FT+GL
Sbjct: 477 FRVLGSASEASKFTVGTFLAGGSWIP-SSVPFTSGL 511
>At2g43050 putative pectinesterase
Length = 518
Score = 377 bits (968), Expect = e-105
Identities = 218/492 (44%), Positives = 289/492 (58%), Gaps = 40/492 (8%)
Query: 68 SPTSVLSAALRTTLNE-------AQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSE 120
SP+S T+ E A+ NLT S +V I DC EL+D ++
Sbjct: 53 SPSSKTKQGFLATVQESMNHALLARSLAFNLT-LSHRTVQTHTFDPIHDCLELLDDTLDM 111
Query: 121 LAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQ- 179
L+ RI A + E ++ WLSAAL+NQDTC + + + ESY G
Sbjct: 112 LS-------RIHADND----EEDVHTWLSAALTNQDTCEQSLQ---EKSESYKHGLAMDF 157
Query: 180 ----VTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKP 235
+T L+++ L L+ + + K + L FP ++ +Q L P
Sbjct: 158 VARNLTGLLTSSLDLFVSVKS-----------KHRKLLSKQEYFPTFVPSSEQRRLLEAP 206
Query: 236 -HRSRADAVVALDGSGHYRTITEAV-NAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIM 293
DAVVA DGSG ++TI EA+ + + + S R IY+K G Y EN+++ K N+M
Sbjct: 207 VEELNVDAVVAPDGSGTHKTIGEALLSTSLASSGGRTKIYLKAGTYHENINIPTKQKNVM 266
Query: 294 LVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVD 353
LVGDG G+T+I +R+ GWTT++TATVA G+GFIARDM+F N AGP + QAVALRV
Sbjct: 267 LVGDGKGKTVIVGSRSNRGGWTTYKTATVAAMGEGFIARDMTFVNNAGPKSEQAVALRVG 326
Query: 354 SDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPL 413
+D+S +RCS+EG+QD+LY HS RQFYRE +I GT+DFIFGN A V Q+C I R PLP
Sbjct: 327 ADKSVVHRCSVEGYQDSLYTHSKRQFYRETDITGTVDFIFGNSAVVFQSCNIAARKPLPG 386
Query: 414 QKVTVTAQGRKSPHQSTGFTIQDSFILATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGW 473
Q+ VTAQGR +P Q+TG IQ+ I A TYLGRPWK+YSRTV + +++ G + P GW
Sbjct: 387 QRNFVTAQGRSNPGQNTGIAIQNCRITAESMTYLGRPWKEYSRTVVMQSFIGGSIHPSGW 446
Query: 474 LEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLP 533
W G F L +L+YGEY N GPG+S++GRVKW G H A FTV FI G+ WLP
Sbjct: 447 SPWSGGFGLKSLFYGEYGNSGPGSSVSGRVKWSGCHPSLTVTEAEKFTVASFIDGNIWLP 506
Query: 534 GTGVKFTAGLTN 545
TGV F GL N
Sbjct: 507 STGVSFDPGLVN 518
>At2g26440 putative pectinesterase
Length = 547
Score = 377 bits (968), Expect = e-105
Identities = 226/559 (40%), Positives = 315/559 (55%), Gaps = 38/559 (6%)
Query: 5 NALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNELTKI 64
N +L +L PS FS + + A C + ++C ++ ++
Sbjct: 7 NLSSLLFLLFFTPSV---FSYSYQPSLNPHETSA--TSFCKNTPYPDACFTSLKLSISIN 61
Query: 65 GPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNR----EQIAIGDCKELVDFSVSE 120
P+ S L L+T L+EA D L S VSN ++ ++ DCK+L + S
Sbjct: 62 ISPNILSFLLQTLQTALSEAGKLTDLL---SGAGVSNNLVEGQRGSLQDCKDLHHITSSF 118
Query: 121 LAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQV 180
L S+ +I+ G + + A+LSAAL+N+ TCLEG E L+ + S T
Sbjct: 119 LKRSIS---KIQDGVNDSRKLADARAYLSAALTNKITCLEGLESASGPLKPKLVTSFTTT 175
Query: 181 TQLISNVLSLYTQLHALPFRPPRNN--THKTSESLDLDSEFPEWMTEGDQELLRSKP--- 235
+ ISN LS ALP + N T +++ L FP+W+ + D L
Sbjct: 176 YKHISNSLS------ALPKQRRTTNPKTGGNTKNRRLLGLFPDWVYKKDHRFLEDSSDGY 229
Query: 236 --HRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIM 293
+ VVA DG+G++ TI EA++ AP+ SN R +IYVK+G+Y EN+D+ TNI+
Sbjct: 230 DEYDPSESLVVAADGTGNFSTINEAISFAPNMSNDRVLIYVKEGVYDENIDIPIYKTNIV 289
Query: 294 LVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVD 353
L+GDG T IT NR+ GWTTFR+AT+AVSG+GF+ARD+ NTAGP HQAVALRV+
Sbjct: 290 LIGDGSDVTFITGNRSVGDGWTTFRSATLAVSGEGFLARDIMITNTAGPEKHQAVALRVN 349
Query: 354 SDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPL 413
+D A YRC I+G+QDTLY HS RQFYREC+IYGTID+IFGN A V Q C I ++ P+P
Sbjct: 350 ADFVALYRCVIDGYQDTLYTHSFRQFYRECDIYGTIDYIFGNAAVVFQGCNIVSKLPMPG 409
Query: 414 QKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYM 464
Q +TAQ R + + TG ++Q+ ILA++ +YLGRPW+++SRTV + +Y+
Sbjct: 410 QFTVITAQSRDTQDEDTGISMQNCSILASEDLFNSSNKVKSYLGRPWREFSRTVVMESYI 469
Query: 465 SGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQR 524
+ GW +W G AL TL+YGEY N GPG+ RV WPG+HIM A FT
Sbjct: 470 DEFIDGSGWSKWNGGEALDTLYYGEYNNNGPGSETVKRVNWPGFHIM-GYEDAFNFTATE 528
Query: 525 FIHGDSWLPGTGVKFTAGL 543
FI GD WL T + G+
Sbjct: 529 FITGDGWLGSTSFPYDNGI 547
>At3g60730 pectinesterase - like protein
Length = 496
Score = 372 bits (955), Expect = e-103
Identities = 203/475 (42%), Positives = 283/475 (58%), Gaps = 25/475 (5%)
Query: 78 RTTLNEAQGAIDNL--TKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGD 135
RT + EA+ + ++ T+ ++ + ++ + +C++L D S + L+ + + D
Sbjct: 38 RTAVVEAKTSFGSMAVTEATSEVAGSYYKLGLSECEKLYDESEARLSKLVVDHENFTVED 97
Query: 136 TSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISNVLSLYTQLH 195
+ WLS L+N TCL+G + + + ++T V + L+ Y +
Sbjct: 98 --------VRTWLSGVLANHHTCLDGLIQQRQGHKPLVHSNVTFV---LHEALAFYKKSR 146
Query: 196 AL----PFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGH 251
A P RP T + + P + L+ P SRAD VVA DGS
Sbjct: 147 ARQGHGPTRPKHRPT-RPNHGPGRSHHGPSRPNQNGGMLVSWNPTSSRADFVVARDGSAT 205
Query: 252 YRTITEAVNAAPSHSN---RRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNR 308
+RTI +A+ A R +IY+K G+Y E +++ R M NIMLVGDG+ +TI+T+NR
Sbjct: 206 HRTINQALAAVSRMGKSRLNRVIIYIKAGVYNEKIEIDRHMKNIMLVGDGMDRTIVTNNR 265
Query: 309 NFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQ 368
N G TT+ +AT VSG GF ARD++F NTAGP HQAVALRV SD S FYRCS +G+Q
Sbjct: 266 NVPDGSTTYGSATFGVSGDGFWARDITFENTAGPHKHQAVALRVSSDLSLFYRCSFKGYQ 325
Query: 369 DTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQ 428
DTL+ HSLRQFYR+C IYGTIDFIFG+ AAV QNC I+ R P+ Q +TAQGR PH
Sbjct: 326 DTLFTHSLRQFYRDCHIYGTIDFIFGDAAAVFQNCDIFVRRPMDHQGNMITAQGRDDPHT 385
Query: 429 STGFTIQDSFILATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYG 488
++ F + +YLGRPWK+YSRTV++ T + ++ PRGW EW G +AL+TL+YG
Sbjct: 386 NSEF----EAVKGRFKSYLGRPWKKYSRTVFLKTDIDELIDPRGWREWSGSYALSTLYYG 441
Query: 489 EYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
E+ N G GA RV WPG+H+++ AS FTV RFI GDSW+P TGV F+AG+
Sbjct: 442 EFMNTGAGAGTGRRVNWPGFHVLRGEEEASPFTVSRFIQGDSWIPITGVPFSAGV 496
>At5g51490 pectinesterase
Length = 536
Score = 369 bits (946), Expect = e-102
Identities = 207/482 (42%), Positives = 290/482 (59%), Gaps = 31/482 (6%)
Query: 77 LRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDT 136
+ ++ A A LT +++Q + DC +L ++ +L +L + +AG
Sbjct: 71 VEAAMDRAISARAELTNSGKNCTDSKKQAVLADCIDLYGDTIMQLNRTLHGVSP-KAGAA 129
Query: 137 SVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISG--SLTQVTQLISNVLSLYTQL 194
+ + + WLS AL+N +TC G +D + +I+ S T+++ LISN L++
Sbjct: 130 KSCTDFDAQTWLSTALTNTETCRRG--SSDLNVTDFITPIVSNTKISHLISNCLAVNG-- 185
Query: 195 HALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRT 254
AL + NT + FP W++ D+ LLR+ RA+ VVA DGSGH+ T
Sbjct: 186 -ALLTAGNKGNTTANQKG------FPTWLSRKDKRLLRAV----RANLVVAKDGSGHFNT 234
Query: 255 ITEAVNAAPSH--SNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQ 312
+ A++ A ++ R+VIYVK+G+Y+EN++++ +IMLVGDG+ TIIT R+
Sbjct: 235 VQAAIDVAGRRKVTSGRFVIYVKRGIYQENINVRLNNDDIMLVGDGMRSTIITGGRSVQG 294
Query: 313 GWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLY 372
G+TT+ +AT + G FIA+ ++FRNTAGP QAVALR SD S FY+CSIEG+QDTL
Sbjct: 295 GYTTYNSATAGIEGLHFIAKGITFRNTAGPAKGQAVALRSSSDLSIFYKCSIEGYQDTLM 354
Query: 373 AHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGF 432
HS RQFYREC IYGT+DFIFGN AAV QNC I R PL Q +TAQGR P Q+TG
Sbjct: 355 VHSQRQFYRECYIYGTVDFIFGNAAAVFQNCLILPRRPLKGQANVITAQGRADPFQNTGI 414
Query: 433 TIQDSFIL---------ATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWF--GDFA 481
+I +S IL T TY+GRPW ++SRTV + TY+ +V P GW W F
Sbjct: 415 SIHNSRILPAPDLKPVVGTVKTYMGRPWMKFSRTVVLQTYLDNVVSPVGWSPWIEGSVFG 474
Query: 482 LTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTA 541
L TL+Y EY+N GP +S RV W G+H++ A+ AS FTV +FI G +WLP TG+ FT+
Sbjct: 475 LDTLFYAEYKNTGPASSTRWRVSWKGFHVLGRASDASAFTVGKFIAGTAWLPRTGIPFTS 534
Query: 542 GL 543
GL
Sbjct: 535 GL 536
>At3g10710 putative pectinesterase
Length = 561
Score = 365 bits (938), Expect = e-101
Identities = 208/483 (43%), Positives = 287/483 (59%), Gaps = 25/483 (5%)
Query: 68 SPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGE 127
+P + A++ T+ E AI+ FS+ + I + C EL+D ++ L +L
Sbjct: 97 NPEELFRYAVKITIAEVSKAIN---AFSSSLGDEKNNITMNACAELLDLTIDNLNNTLTS 153
Query: 128 MRRIRAGDTSV-QYEGNLEAWLSAALSNQDTCLEGFEGTDRRL-ESYISGSLTQVTQLIS 185
GD +V + +L WLS+A + Q TC+E R ES++ S T+L S
Sbjct: 154 SSN---GDVTVPELVDDLRTWLSSAGTYQRTCVETLAPDMRPFGESHLKNS----TELTS 206
Query: 186 NVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVA 245
N L++ T L + T+ +++D + LL+S R AD VVA
Sbjct: 207 NALAIITWLGKIADSFKLRRRLLTTADVEVDFH-------AGRRLLQSTDLRKVADIVVA 259
Query: 246 LDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIIT 305
DGSG YRTI A+ P S +R +IYVKKG+Y ENV +++KM N+++VGDG ++I++
Sbjct: 260 KDGSGKYRTIKRALQDVPEKSEKRTIIYVKKGVYFENVKVEKKMWNVIVVGDGESKSIVS 319
Query: 306 SNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIE 365
N + G TF+TAT AV GKGF+ARDM F NTAGP HQAVAL V +D +AFYRC++
Sbjct: 320 GRLNVIDGTPTFKTATFAVFGKGFMARDMGFINTAGPSKHQAVALMVSADLTAFYRCTMN 379
Query: 366 GFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKS 425
+QDTLY H+ RQFYREC I GT+DFIFGN A+VLQ+C+I R P+ Q+ T+TAQGR
Sbjct: 380 AYQDTLYVHAQRQFYRECTIIGTVDFIFGNSASVLQSCRILPRRPMKGQQNTITAQGRTD 439
Query: 426 PHQSTGFTIQDSFI-----LATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDF 480
P+ +TG +I I L T+LGRPWK +S TV +++Y+ G + +GWL W GD
Sbjct: 440 PNMNTGISIHRCNISPLGDLTDVMTFLGRPWKNFSTTVIMDSYLHGFIDRKGWLPWTGDS 499
Query: 481 ALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFT 540
A T++YGEY+N GPGAS RVKW G + A+ FTV+ FI G WLP T V F
Sbjct: 500 APDTIFYGEYKNTGPGASTKNRVKWKGLRFL-STKEANRFTVKPFIDGGRWLPATKVPFR 558
Query: 541 AGL 543
+GL
Sbjct: 559 SGL 561
>At5g04960 pectinesterase
Length = 564
Score = 363 bits (933), Expect = e-100
Identities = 206/545 (37%), Positives = 309/545 (55%), Gaps = 27/545 (4%)
Query: 7 LFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNELTKIGP 66
L +++ ++ ++++ S + P+E + + + C ++ C + +
Sbjct: 35 LVCIVVGAVVGTTARDNSKKPPTENNGEPISVSVKALCDVTLHKEKCFETL-GSAPNASR 93
Query: 67 PSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLG 126
SP + A++ T+ E +D + + N A+G C EL+ +V +L ++
Sbjct: 94 SSPEELFKYAVKVTITELSKVLDGFSNGE--HMDNATSAAMGACVELIGLAVDQLNETM- 150
Query: 127 EMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGF-EGTDRRLESYISGSLTQVTQLIS 185
+S++ +L WLS+ + Q+TC++ E L ++ L T++ S
Sbjct: 151 --------TSSLKNFDDLRTWLSSVGTYQETCMDALVEANKPSLTTFGENHLKNSTEMTS 202
Query: 186 NVLSLYTQL----HALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRAD 241
N L++ T L + FR R +T + + ++ P M EG + LL S + +A
Sbjct: 203 NALAIITWLGKIADTVKFR--RRRLLETGNAKVVVADLP--MMEG-RRLLESGDLKKKAT 257
Query: 242 AVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQ 301
VVA DGSG YRTI EA+ + + +IYVKKG+Y ENV +++ N+++VGDG +
Sbjct: 258 IVVAKDGSGKYRTIGEALAEVEEKNEKPTIIYVKKGVYLENVRVEKTKWNVVMVGDGQSK 317
Query: 302 TIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYR 361
TI+++ NF+ G TF TAT AV GKGF+ARDM F NTAGP HQAVAL V +D S FY+
Sbjct: 318 TIVSAGLNFIDGTPTFETATFAVFGKGFMARDMGFINTAGPAKHQAVALMVSADLSVFYK 377
Query: 362 CSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQ 421
C+++ FQDT+YAH+ RQFYR+C I GT+DFIFGN A V Q C+I R P+ Q+ T+TAQ
Sbjct: 378 CTMDAFQDTMYAHAQRQFYRDCVILGTVDFIFGNAAVVFQKCEILPRRPMKGQQNTITAQ 437
Query: 422 GRKSPHQSTGFTIQDSFI-----LATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEW 476
GRK P+Q+TG +I + I L T+LGRPWK +S TV + ++M + P+GWL W
Sbjct: 438 GRKDPNQNTGISIHNCTIKPLDNLTDIQTFLGRPWKDFSTTVIMKSFMDKFINPKGWLPW 497
Query: 477 FGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTG 536
GD A T++Y EY N GPGAS RVKW G A+ FTV+ FI G++WLP T
Sbjct: 498 TGDTAPDTIFYAEYLNSGPGASTKNRVKWQGLKTSLTKKEANKFTVKPFIDGNNWLPATK 557
Query: 537 VKFTA 541
V F +
Sbjct: 558 VPFNS 562
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,776,317
Number of Sequences: 26719
Number of extensions: 485520
Number of successful extensions: 1499
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1174
Number of HSP's gapped (non-prelim): 116
length of query: 545
length of database: 11,318,596
effective HSP length: 104
effective length of query: 441
effective length of database: 8,539,820
effective search space: 3766060620
effective search space used: 3766060620
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0109.6


