

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0109.15
(312 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
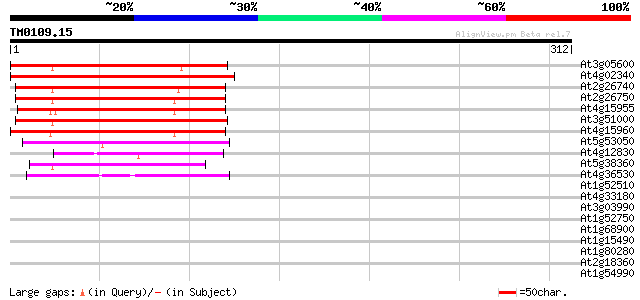
Score E
Sequences producing significant alignments: (bits) Value
At3g05600 putative epoxide hydrolase 186 1e-47
At4g02340 unknown protein 180 8e-46
At2g26740 epoxide hydrolase (ATsEH) 172 2e-43
At2g26750 putative epoxide hydrolase 167 7e-42
At4g15955 unknown protein 166 2e-41
At3g51000 epoxide hydrolase-like protein 164 7e-41
At4g15960 unknown protein 155 2e-38
At5g53050 unknown protein 53 2e-07
At4g12830 hydrolase like protein 49 4e-06
At5g38360 similar to unknown protein (pir||T03994) 47 2e-05
At4g36530 unknown protein 43 3e-04
At1g52510 unknown protein (At1g52510) 41 0.001
At4g33180 putative protein 39 0.004
At3g03990 unknown protein 39 0.005
At1g52750 alpha/beta fold family protein 39 0.005
At1g68900 hypothetical protein 37 0.011
At1g15490 unknown protein 37 0.014
At1g80280 unknown protein 36 0.023
At2g18360 unknown protein 36 0.031
At1g54990 unknown protein 35 0.040
>At3g05600 putative epoxide hydrolase
Length = 331
Score = 186 bits (473), Expect = 1e-47
Identities = 85/125 (68%), Positives = 102/125 (81%), Gaps = 4/125 (3%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPD 58
MEGI+HR V VNGI MHIAEKG +GPVVL LHGFP+LWY+WRHQI LSS GY AVAPD
Sbjct: 1 MEGIDHRMVSVNGITMHIAEKGPKEGPVVLLLHGFPDLWYTWRHQISGLSSLGYRAVAPD 60
Query: 59 LRGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQ--VFLVAHDWGSIIGWYLCMFRPE 116
LRGYGD+DSP S + YTC ++VGD+VAL+D + Q VFLV HDWG+IIGW+LC+FRPE
Sbjct: 61 LRGYGDSDSPESFSEYTCLNVVGDLVALLDSVAGNQEKVFLVGHDWGAIIGWFLCLFRPE 120
Query: 117 RVKAY 121
++ +
Sbjct: 121 KINGF 125
>At4g02340 unknown protein
Length = 324
Score = 180 bits (457), Expect = 8e-46
Identities = 76/125 (60%), Positives = 98/125 (77%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH T+ NGI MH+A G GPV+LF+HGFP+LWYSWRHQ+++ ++ GY A+APDLR
Sbjct: 1 MEKIEHTTISTNGINMHVASIGSGPVILFVHGFPDLWYSWRHQLVSFAALGYRAIAPDLR 60
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGD+D+P S +YT HIVGD+V L+D LG +VFLV HDWG+I+ W+LCM RP+RV A
Sbjct: 61 GYGDSDAPPSRESYTILHIVGDLVGLLDSLGVDRVFLVGHDWGAIVAWWLCMIRPDRVNA 120
Query: 121 YAPSS 125
+S
Sbjct: 121 LVNTS 125
>At2g26740 epoxide hydrolase (ATsEH)
Length = 321
Score = 172 bits (436), Expect = 2e-43
Identities = 78/122 (63%), Positives = 100/122 (81%), Gaps = 5/122 (4%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+EHR V NGI +H+A +G GP+VL LHGFPELWYSWRHQI L++RGY AVAPDLRG
Sbjct: 1 MEHRKVRGNGIDIHVAIQGPSDGPIVLLLHGFPELWYSWRHQIPGLAARGYRAVAPDLRG 60
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHLGA---KQVFLVAHDWGSIIGWYLCMFRPERV 118
YGD+D+P I++YTCF+IVGD++A+I L A ++VF+V HDWG++I WYLC+FRP+RV
Sbjct: 61 YGDSDAPAEISSYTCFNIVGDLIAVISALTASEDEKVFVVGHDWGALIAWYLCLFRPDRV 120
Query: 119 KA 120
KA
Sbjct: 121 KA 122
>At2g26750 putative epoxide hydrolase
Length = 320
Score = 167 bits (423), Expect = 7e-42
Identities = 76/121 (62%), Positives = 98/121 (80%), Gaps = 4/121 (3%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+EHR V NGI +H+A +G G +VL LHGFPELWYSWRHQI L++RGY AVAPDLRG
Sbjct: 1 MEHRNVRGNGIDIHVAIQGPSDGTIVLLLHGFPELWYSWRHQISGLAARGYRAVAPDLRG 60
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
YGD+D+P I+++TCF+IVGD+VA+I L K+VF+V HDWG++I WYLC+FRP++VK
Sbjct: 61 YGDSDAPAEISSFTCFNIVGDLVAVISTLIKEDKKVFVVGHDWGALIAWYLCLFRPDKVK 120
Query: 120 A 120
A
Sbjct: 121 A 121
>At4g15955 unknown protein
Length = 178
Score = 166 bits (419), Expect = 2e-41
Identities = 76/127 (59%), Positives = 97/127 (75%), Gaps = 11/127 (8%)
Query: 5 EHRTVEVNGIKMHIAEK-----GQG----PVVLFLHGFPELWYSWRHQILTLSSRGYHAV 55
+H V+VNGI MH+AEK G G PV+LFLHGFPELWY+WRHQ++ LSS GY +
Sbjct: 6 DHSFVKVNGITMHVAEKSPSVAGNGAIRPPVILFLHGFPELWYTWRHQMVALSSLGYRTI 65
Query: 56 APDLRGYGDTDSPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMF 113
APDLRGYGDTD+P S+ YT H+VGD++ LID + ++VF+V HDWG+II W+LC+F
Sbjct: 66 APDLRGYGDTDAPESVDAYTSLHVVGDLIGLIDAVVGDREKVFVVGHDWGAIIAWHLCLF 125
Query: 114 RPERVKA 120
RP+RVKA
Sbjct: 126 RPDRVKA 132
>At3g51000 epoxide hydrolase-like protein
Length = 323
Score = 164 bits (414), Expect = 7e-41
Identities = 71/120 (59%), Positives = 91/120 (75%), Gaps = 2/120 (1%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+ + ++ NGI +++AEKG +GP+VL LHGFPE WYSWRHQI LSS GYH VAPDLRG
Sbjct: 5 VREKKIKTNGIWLNVAEKGDEEGPLVLLLHGFPETWYSWRHQIDFLSSHGYHVVAPDLRG 64
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKAY 121
YGD+DS S +YT H+V D++ L+DH G Q F+ HDWG+IIGW LC+FRP+RVK +
Sbjct: 65 YGDSDSLPSHESYTVSHLVADVIGLLDHYGTTQAFVAGHDWGAIIGWCLCLFRPDRVKGF 124
>At4g15960 unknown protein
Length = 375
Score = 155 bits (393), Expect = 2e-38
Identities = 70/127 (55%), Positives = 94/127 (73%), Gaps = 7/127 (5%)
Query: 1 MEGIEHRTVEVNGIKMHIAEK-----GQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAV 55
++G+EH+T++VNGI MH+AEK G+ P++LFLHGFPELWY+WRHQ++ LSS GY +
Sbjct: 51 LDGVEHKTLKVNGINMHVAEKPGSGSGEDPIILFLHGFPELWYTWRHQMVALSSLGYRTI 110
Query: 56 APDLRGYGDTDSPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMF 113
APDLRGYGDT++P + YT +VALI + G K V +V HDWG++I W LC +
Sbjct: 111 APDLRGYGDTEAPEKVEDYTLLKRGRSVVALIVAVTGGDKAVSVVGHDWGAMIAWQLCQY 170
Query: 114 RPERVKA 120
RPE+VKA
Sbjct: 171 RPEKVKA 177
>At5g53050 unknown protein
Length = 396
Score = 52.8 bits (125), Expect = 2e-07
Identities = 38/134 (28%), Positives = 56/134 (41%), Gaps = 19/134 (14%)
Query: 8 TVEVNGIKMHIAEKGQGPV-VLFLHGFPELWYSWRHQILTLSSR---------------- 50
T+ IK+ G GP+ L + G SW QI+ L+
Sbjct: 16 TLNNAAIKIFYRTYGHGPIKALLIIGLAGTHESWGPQIMGLTGTDKPNDDDDDDGGIVSD 75
Query: 51 --GYHAVAPDLRGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGW 108
G A D RG G + P + YT + D ++L+DHLG K+ ++ H G++I
Sbjct: 76 DSGIEVCAFDNRGMGRSSVPTHKSEYTTTIMANDSISLLDHLGWKKAHIIGHSMGAMIAC 135
Query: 109 YLCMFRPERVKAYA 122
L PERV + A
Sbjct: 136 KLAAMAPERVLSLA 149
>At4g12830 hydrolase like protein
Length = 393
Score = 48.9 bits (115), Expect = 4e-06
Identities = 32/97 (32%), Positives = 48/97 (48%), Gaps = 3/97 (3%)
Query: 25 PVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYGDTDSPIS--ITTYTCFHIVGD 82
P V+ +HGFP YS+R I L S+ Y A+A D G+G +D P + YT V
Sbjct: 134 PPVILIHGFPSQAYSYRKTIPVL-SKNYRAIAFDWLGFGFSDKPQAGYGFNYTMDEFVSS 192
Query: 83 IVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
+ + ID + +V LV + S RP+++K
Sbjct: 193 LESFIDEVTTSKVSLVVQGYFSAAVVKYARNRPDKIK 229
>At5g38360 similar to unknown protein (pir||T03994)
Length = 242
Score = 46.6 bits (109), Expect = 2e-05
Identities = 31/106 (29%), Positives = 50/106 (46%), Gaps = 8/106 (7%)
Query: 12 NGIKMHIAEKG------QGPVVLFLHGFPELWYSWRHQILTL-SSRGYHAVAPDLRGYGD 64
+G+KM + E+ + P ++F+HG + W L SS G+ + A L G G+
Sbjct: 48 SGLKMEVIEQRKSKSERENPPLVFVHGSYHAAWCWAENWLPFFSSSGFDSYAVSLLGQGE 107
Query: 65 TDSPISITTYTCFHIVGDIVALID-HLGAKQVFLVAHDWGSIIGWY 109
+D P+ T DI I+ +LG+ LV H +G +I Y
Sbjct: 108 SDEPLGTVAGTLQTHASDIADFIESNLGSSPPVLVGHSFGGLIVQY 153
>At4g36530 unknown protein
Length = 378
Score = 42.7 bits (99), Expect = 3e-04
Identities = 27/113 (23%), Positives = 51/113 (44%), Gaps = 3/113 (2%)
Query: 10 EVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYGDTDSPI 69
E G K+H +G+G ++ +HGF + WR+ I L+ + Y A DL G+G +D
Sbjct: 85 EWRGHKIHYVVQGEGSPLVLIHGFGASVFHWRYNIPELAKK-YKVYALDLLGFGWSDK-- 141
Query: 70 SITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKAYA 122
++ Y ++ + + + +V + G + + PE+V A
Sbjct: 142 ALIEYDAMVWTDQVIDFMKEVVKEPAVVVGNSLGGFTALSVAVGLPEQVTGVA 194
>At1g52510 unknown protein (At1g52510)
Length = 380
Score = 40.8 bits (94), Expect = 0.001
Identities = 20/65 (30%), Positives = 34/65 (51%), Gaps = 4/65 (6%)
Query: 8 TVEVNGIKMHIAEKGQGP----VVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYG 63
T++ ++ + E G ++F+HG P +S+R + LS G+H APD G+G
Sbjct: 105 TIKSGKLRWFVRETGSKESRRGTIVFVHGAPTQSFSYRTVMSELSDAGFHCFAPDWIGFG 164
Query: 64 DTDSP 68
+D P
Sbjct: 165 FSDKP 169
>At4g33180 putative protein
Length = 111
Score = 38.9 bits (89), Expect = 0.004
Identities = 18/44 (40%), Positives = 25/44 (55%), Gaps = 1/44 (2%)
Query: 25 PVVLFLHGF-PELWYSWRHQILTLSSRGYHAVAPDLRGYGDTDS 67
PV+L LHGF P + WR Q+ S + +PDL +GD+ S
Sbjct: 56 PVMLLLHGFGPSSMWQWRRQMQAFSPSAFRVYSPDLVFFGDSTS 99
>At3g03990 unknown protein
Length = 267
Score = 38.5 bits (88), Expect = 0.005
Identities = 31/104 (29%), Positives = 47/104 (44%), Gaps = 4/104 (3%)
Query: 16 MHIAEKGQGPVVLFL-HGFPELWYSWRHQILTLSSRGYHAVAPDLRGYGDTDSP-ISITT 73
+++ G G +LFL HGF +W H IL ++ Y V DL G +
Sbjct: 10 LNVRVVGTGDRILFLAHGFGTDQSAW-HLILPYFTQNYRVVLYDLVCAGSVNPDYFDFNR 68
Query: 74 YTCFH-IVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPE 116
YT V D++ ++D LG + V H ++IG + RPE
Sbjct: 69 YTTLDPYVDDLLNIVDSLGIQNCAYVGHSVSAMIGIIASIRRPE 112
>At1g52750 alpha/beta fold family protein
Length = 633
Score = 38.5 bits (88), Expect = 0.005
Identities = 31/107 (28%), Positives = 48/107 (43%), Gaps = 10/107 (9%)
Query: 10 EVNGIKM-HIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSR-GYHAVAPDLRGYGDTDS 67
++N IK+ EK + ++ +HGF +SWRH + LS + G VA D G+G T
Sbjct: 338 QINSIKLGDDMEKDENTGIVLVHGFGGGVFSWRHVMGELSLQLGCRVVAYDRPGWGLTSR 397
Query: 68 PIS--------ITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSII 106
I Y V +++ +G V LV HD G ++
Sbjct: 398 LIRKDWEKRNLANPYKLESQVDLLLSFCSEMGFSSVILVGHDDGGLL 444
>At1g68900 hypothetical protein
Length = 656
Score = 37.4 bits (85), Expect = 0.011
Identities = 33/115 (28%), Positives = 56/115 (48%), Gaps = 12/115 (10%)
Query: 9 VEVNG----IKMH-IAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYG 63
V+V+G I++H + E +G V LFLHGF W I+T S ++ D+ G+G
Sbjct: 360 VDVDGFSHFIRVHDVGENAEGSVALFLHGFLGTGEEW-IPIMTGISGSARCISVDIPGHG 418
Query: 64 DT--DSPISIT----TYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCM 112
+ S S T T++ I + LI+ + +V +V + G+ I Y+ +
Sbjct: 419 RSRVQSHASETQTSPTFSMEMIAEALYKLIEQITPGKVTIVGYSMGARIALYMAL 473
>At1g15490 unknown protein
Length = 648
Score = 37.0 bits (84), Expect = 0.014
Identities = 30/98 (30%), Positives = 46/98 (46%), Gaps = 10/98 (10%)
Query: 18 IAEKGQGPVVLFLHGFPELWYSWRHQILTLSSR-GYHAVAPDLRGYGDTDSPIS------ 70
+ E GQ VVL +HGF +SWRH + +L+ + G A D G+G T P
Sbjct: 361 LGESGQFGVVL-VHGFGGGVFSWRHVMGSLAQQLGCVVTAFDRPGWGLTARPHKNDLEER 419
Query: 71 --ITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSII 106
+ Y+ + V ++A +G V V HD G ++
Sbjct: 420 QLLNPYSLENQVEMLIAFCYEMGFSSVVFVGHDDGGLL 457
>At1g80280 unknown protein
Length = 647
Score = 36.2 bits (82), Expect = 0.023
Identities = 31/98 (31%), Positives = 45/98 (45%), Gaps = 10/98 (10%)
Query: 18 IAEKGQGPVVLFLHGFPELWYSWRHQILTLSSR-GYHAVAPDLRGYGDTDSPIS------ 70
+ GQ VVL +HGF +SWRH + +L+ + G A D G+G T P
Sbjct: 366 VGGNGQFGVVL-VHGFGGGVFSWRHVMSSLAHQLGCVVTAFDRPGWGLTARPHKKDLEER 424
Query: 71 --ITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSII 106
YT + V ++A +G V LV HD G ++
Sbjct: 425 EMPNPYTLDNQVDMLLAFCHEMGFASVVLVGHDDGGLL 462
>At2g18360 unknown protein
Length = 313
Score = 35.8 bits (81), Expect = 0.031
Identities = 30/106 (28%), Positives = 52/106 (48%), Gaps = 4/106 (3%)
Query: 20 EKGQGPVVLFLHGF-PELWYSWRHQILTLSSRGYHAVAPDLRGYGDTDSPISITTYTCFH 78
+K PV+LF+HGF E +W+ Q+ +L+ + Y PDL +G + S + F
Sbjct: 58 QKPTKPVLLFIHGFAAEGIVTWQFQVGSLAKK-YSVYIPDLLFFGGSYSD-NADRSPAFQ 115
Query: 79 IVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKAYAPS 124
+V + LG ++ LV +G ++ + + PE V+A S
Sbjct: 116 -AHCLVKSLRILGIEKFTLVGFSYGGMVAFKIAEEYPEMVQAMVVS 160
>At1g54990 unknown protein
Length = 473
Score = 35.4 bits (80), Expect = 0.040
Identities = 22/69 (31%), Positives = 38/69 (54%), Gaps = 6/69 (8%)
Query: 9 VEVNG----IKMHIAEKGQ--GPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGY 62
V+VN I++ +AE G V+ +HG ++++ I +L S+G H+VA DL G
Sbjct: 96 VQVNSNESPIEVFVAESGSIHTETVVIVHGLGLSSFAFKEMIQSLGSKGIHSVAIDLPGN 155
Query: 63 GDTDSPISI 71
G +D + +
Sbjct: 156 GFSDKSMVV 164
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.340 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,478,565
Number of Sequences: 26719
Number of extensions: 244288
Number of successful extensions: 708
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 673
Number of HSP's gapped (non-prelim): 37
length of query: 312
length of database: 11,318,596
effective HSP length: 99
effective length of query: 213
effective length of database: 8,673,415
effective search space: 1847437395
effective search space used: 1847437395
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0109.15


