

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.1
(915 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
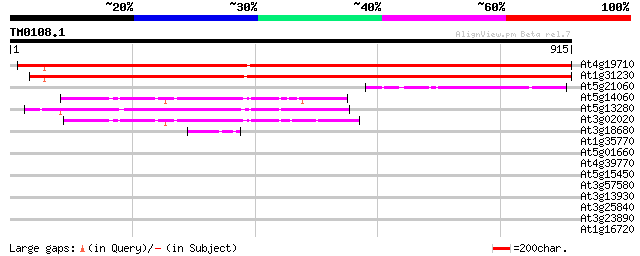
Score E
Sequences producing significant alignments: (bits) Value
At4g19710 aspartate kinase-homoserine dehydrogenase - like protein 1386 0.0
At1g31230 aspartate kinase-homoserine dehydrogenase like protein 1359 0.0
At5g21060 homoserine dehydrogenase-like protein 199 8e-51
At5g14060 lysine-sensitive aspartate kinase (gb|AAB63104.1) 184 3e-46
At5g13280 aspartate kinase 182 6e-46
At3g02020 putative aspartate kinase 179 5e-45
At3g18680 uridylate kinase like protein 44 5e-04
At1g35770 hypothetical protein 37 0.066
At5g01660 putative protein 32 2.1
At4g39770 trehalose-6-phosphate phosphatase like protein 32 2.1
At5g15450 clpB heat shock protein-like 31 3.6
At3g57580 unknown protein 31 3.6
At3g13930 putative acetyltransferase 30 4.7
At3g25840 protein kinase like protein 30 6.2
At3g23890 topoisomerase II 30 8.1
At1g16720 unknown protein (At1g16720) 30 8.1
>At4g19710 aspartate kinase-homoserine dehydrogenase - like protein
Length = 916
Score = 1386 bits (3588), Expect = 0.0
Identities = 702/910 (77%), Positives = 791/910 (86%), Gaps = 10/910 (1%)
Query: 14 TISPSNNTPDITKISQCQCLPFLPSHRSHSLRKALSLLPRGN-----QSPSTKISASLTD 68
T+SP N+ P QC +P R L + G + P +SA+ T
Sbjct: 9 TVSPPNSNPIRFGSFPPQCFLRVPKPRRLILPRFRKTTGGGGGLIRCELPDFHLSATATT 68
Query: 69 VSPSV---LVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMS 125
VS LV++ Q+ KGE WSVHKFGGTC+G+SQRI+NV ++++ D+SERKLVVVSAMS
Sbjct: 69 VSGVSTVNLVDQVQIPKGEMWSVHKFGGTCVGNSQRIRNVAEVIINDNSERKLVVVSAMS 128
Query: 126 KVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKA 185
KVTDMMY+LI KAQSRD+SY+S+L+AVLEKH LTA DLLDGD LA+FLS LH+DISNLKA
Sbjct: 129 KVTDMMYDLIRKAQSRDDSYLSALEAVLEKHRLTARDLLDGDDLASFLSHLHNDISNLKA 188
Query: 186 MLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQ 245
MLRAIYIAGHA+ESF+DFV GHGELWSAQMLS V+RK G +CKWMDTRDVLIVNPTSSNQ
Sbjct: 189 MLRAIYIAGHASESFSDFVAGHGELWSAQMLSYVVRKTGLECKWMDTRDVLIVNPTSSNQ 248
Query: 246 VDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRAR 305
VDPD+ ESEKRL+ WFSLNP K+IIATGFIASTPQ IPTTLKRDGSDFSAAIMGAL RAR
Sbjct: 249 VDPDFGESEKRLDKWFSLNPSKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMGALLRAR 308
Query: 306 QVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPIL 365
QVTIWTDVDGVYSADPRKV+EAVIL+TLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+
Sbjct: 309 QVTIWTDVDGVYSADPRKVNEAVILQTLSYQEAWEMSYFGANVLHPRTIIPVMRYNIPIV 368
Query: 366 IRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASA 425
IRNIFNLSAPGT IC P +D + + + VKGFATIDNLALINVEGTGMAGVPGTAS
Sbjct: 369 IRNIFNLSAPGTIICQPPEDDYD--LKLTTPVKGFATIDNLALINVEGTGMAGVPGTASD 426
Query: 426 IFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIP 485
IFG VKDVGANVIMISQASSEHS+CFAVPEKEV AV+EAL+SRF AL GRLSQ+ +IP
Sbjct: 427 IFGCVKDVGANVIMISQASSEHSVCFAVPEKEVNAVSEALRSRFSEALQAGRLSQIEVIP 486
Query: 486 NCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRA 545
NCSILAAVGQKMASTPGVS TLF+ALAKANINVRAI+QGCSEYN+TVV+KRED VKALRA
Sbjct: 487 NCSILAAVGQKMASTPGVSCTLFSALAKANINVRAISQGCSEYNVTVVIKREDSVKALRA 546
Query: 546 VHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSD 605
VHSRF+LSRTT+AMGI+GPGLIG+TLLDQLRDQA++LK+EFNIDLRV+GI GSK MLLSD
Sbjct: 547 VHSRFFLSRTTLAMGIVGPGLIGATLLDQLRDQAAVLKQEFNIDLRVLGITGSKKMLLSD 606
Query: 606 WGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGI 665
GIDL+RWREL +E G A+L+KF Q VHGNHFIPN+ +VDCTADS IA YYDWLRKGI
Sbjct: 607 IGIDLSRWRELLNEKGTEADLDKFTQQVHGNHFIPNSVVVDCTADSAIASRYYDWLRKGI 666
Query: 666 HVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQI 725
HV+TPNKKANSGPLDQYLKLR LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+I
Sbjct: 667 HVITPNKKANSGPLDQYLKLRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRI 726
Query: 726 EGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGL 785
EGI SGTLSY+FNNF R+FSEVV EAK AG+TEPDPRDDLSGTDVARKVIILARESGL
Sbjct: 727 EGICSGTLSYLFNNFVGDRSFSEVVTEAKNAGFTEPDPRDDLSGTDVARKVIILARESGL 786
Query: 786 KLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVT 845
KL+L+++P+ SLVPEPL+ C S +EFM++LP++D AK+ +AEN+GEVLRYVGVVD
Sbjct: 787 KLDLADLPIRSLVPEPLKGCTSVEEFMEKLPQYDGDLAKERLDAENSGEVLRYVGVVDAV 846
Query: 846 SKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDIL 905
++KG VELRRYKK+HPFAQL+GSDNIIAFTT RYKD PLIVRGPGAGAQVTAGGIFSDIL
Sbjct: 847 NQKGTVELRRYKKEHPFAQLAGSDNIIAFTTTRYKDHPLIVRGPGAGAQVTAGGIFSDIL 906
Query: 906 RLASYLGAPS 915
RLASYLGAPS
Sbjct: 907 RLASYLGAPS 916
>At1g31230 aspartate kinase-homoserine dehydrogenase like protein
Length = 911
Score = 1359 bits (3518), Expect = 0.0
Identities = 683/891 (76%), Positives = 783/891 (87%), Gaps = 10/891 (1%)
Query: 33 LPFLPSHRSHSLRKALSLLPRGN--------QSPSTKISASLTDVSPSVLVEEQQLQKGE 84
+PF+ R S R + L R + + S ++ S+TD++ VE L KG+
Sbjct: 23 VPFIYGKRLVSNRVSFGKLRRRSCIGQCVRSELQSPRVLGSVTDLALDNSVENGHLPKGD 82
Query: 85 TWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDES 144
+W+VHKFGGTC+G+S+RIK+V +V++DDSERKLVVVSAMSKVTDMMY+LI++A+SRD+S
Sbjct: 83 SWAVHKFGGTCVGNSERIKDVAAVVVKDDSERKLVVVSAMSKVTDMMYDLIHRAESRDDS 142
Query: 145 YVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFV 204
Y+S+L VLEKH TA DLLDGD L++FL++L+ DI+NLKAMLRAIYIAGHATESF+DFV
Sbjct: 143 YLSALSGVLEKHRATAVDLLDGDELSSFLARLNDDINNLKAMLRAIYIAGHATESFSDFV 202
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLN 264
VGHGELWSAQML+ V+RK+G DC WMD RDVL+V PTSSNQVDPD++ESEKRLE WF+ N
Sbjct: 203 VGHGELWSAQMLAAVVRKSGLDCTWMDARDVLVVIPTSSNQVDPDFVESEKRLEKWFTQN 262
Query: 265 PCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKV 324
K+IIATGFIASTPQ IPTTLKRDGSDFSAAIM ALFR+ Q+TIWTDVDGVYSADPRKV
Sbjct: 263 SAKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMSALFRSHQLTIWTDVDGVYSADPRKV 322
Query: 325 SEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSV 384
SEAV+LKTLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+IRNIFNLSAPGT IC
Sbjct: 323 SEAVVLKTLSYQEAWEMSYFGANVLHPRTIIPVMKYDIPIVIRNIFNLSAPGTMICRQI- 381
Query: 385 NDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQAS 444
D+ED + VKGFATIDNLAL+NVEGTGMAGVPGTASAIF AVK+VGANVIMISQAS
Sbjct: 382 -DDEDGFKLDAPVKGFATIDNLALVNVEGTGMAGVPGTASAIFSAVKEVGANVIMISQAS 440
Query: 445 SEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVS 504
SEHS+CFAVPEKEVKAV+EAL SRFR AL GRLSQ+ IIPNCSILAAVGQKMASTPGVS
Sbjct: 441 SEHSVCFAVPEKEVKAVSEALNSRFRQALAGGRLSQIEIIPNCSILAAVGQKMASTPGVS 500
Query: 505 ATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGP 564
AT FNALAKANIN+RAIAQGCSE+NITVV+KREDC++ALRAVHSRFYLSRTT+A+GIIGP
Sbjct: 501 ATFFNALAKANINIRAIAQGCSEFNITVVVKREDCIRALRAVHSRFYLSRTTLAVGIIGP 560
Query: 565 GLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVA 624
GLIG TLLDQ+RDQA++LKEEF IDLRV+GI GS ML+S+ GIDL+RWREL E GE A
Sbjct: 561 GLIGGTLLDQIRDQAAVLKEEFKIDLRVIGITGSSKMLMSESGIDLSRWRELMKEEGEKA 620
Query: 625 NLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLK 684
++EKF Q+V GNHFIPN+ +VDCTAD+ IA YYDWL +GIHVVTPNKKANSGPLDQYLK
Sbjct: 621 DMEKFTQYVKGNHFIPNSVMVDCTADADIASCYYDWLLRGIHVVTPNKKANSGPLDQYLK 680
Query: 685 LRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGR 744
+R LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+IEGIFSGTLSY+FNNF R
Sbjct: 681 IRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRIEGIFSGTLSYLFNNFVGTR 740
Query: 745 AFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQA 804
+FSEVV EAK+AG+TEPDPRDDLSGTDVARKV ILARESGLKL+L +PV++LVP+PLQA
Sbjct: 741 SFSEVVAEAKQAGFTEPDPRDDLSGTDVARKVTILARESGLKLDLEGLPVQNLVPKPLQA 800
Query: 805 CASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQ 864
CASA+EFM++LP+FD+ +K+ EEAE AGEVLRYVGVVD KKG VEL+RYKKDHPFAQ
Sbjct: 801 CASAEEFMEKLPQFDEELSKQREEAEAAGEVLRYVGVVDAVEKKGTVELKRYKKDHPFAQ 860
Query: 865 LSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAPS 915
LSG+DNIIAFTT+RYK+QPLIVRGPGAGAQVTAGGIFSDILRLA YLGAPS
Sbjct: 861 LSGADNIIAFTTKRYKEQPLIVRGPGAGAQVTAGGIFSDILRLAFYLGAPS 911
>At5g21060 homoserine dehydrogenase-like protein
Length = 376
Score = 199 bits (505), Expect = 8e-51
Identities = 128/332 (38%), Positives = 192/332 (57%), Gaps = 18/332 (5%)
Query: 581 ILKEEFNIDL-RVMGILGSKSMLLSDWGIDLARWRELRDE--SGEVANLEKFVQHVHGNH 637
+LKEE N +L + ++ S LS G L +R + D S E + K + G
Sbjct: 53 VLKEELNDELLSEVCLIKSTGSALSKLGA-LGGYRVVHDSELSTETEEIAKLLGKSTG-- 109
Query: 638 FIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYF 697
A+VDC+A + G +V NKK + L+ Y KL AL + H
Sbjct: 110 ----LAVVDCSASMETIEILMKAVDLGCCIVLANKKPVTSTLEHYDKL-ALHPRFIRH-- 162
Query: 698 YEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAG 757
E+TVGAGLP++++L ++ +GD + +I G SGTL Y+ + +DG+ S+VVQ AK+ G
Sbjct: 163 -ESTVGAGLPVIASLNRIISSGDPVHRIVGSLSGTLGYVMSELEDGKPLSQVVQAAKKLG 221
Query: 758 YTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPL-QACASAQEFMQR-L 815
YTEPDPRDDL G DVARK +ILAR G ++ + +I +ESL PE + S +F+ +
Sbjct: 222 YTEPDPRDDLGGMDVARKGLILARLLGKRIIMDSIKIESLYPEEMGPGLMSVDDFLHNGI 281
Query: 816 PEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFT 875
+ DQ +++++A + G VLRYV V++ +S + V +R KD P +L GSDNI+
Sbjct: 282 VKLDQNIEERVKKASSKGCVLRYVCVIEGSSVQ--VGIREVSKDSPLGRLRGSDNIVEIY 339
Query: 876 TRRYKDQPLIVRGPGAGAQVTAGGIFSDILRL 907
+R YK+QPL+++G GAG TA G+ +DI+ L
Sbjct: 340 SRCYKEQPLVIQGAGAGNDTTAAGVLADIIDL 371
>At5g14060 lysine-sensitive aspartate kinase (gb|AAB63104.1)
Length = 544
Score = 184 bits (466), Expect = 3e-46
Identities = 139/483 (28%), Positives = 243/483 (49%), Gaps = 38/483 (7%)
Query: 84 ETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDE 143
E V KFGG+ + S++R+K V +++L ER ++V+SAM K T+ + + KA +
Sbjct: 81 ELTCVMKFGGSSVESAERMKEVANLILSFPDERPVIVLSAMGKTTNKLLKAGEKAVTCGV 140
Query: 144 SYVSSLDA---VLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESF 200
+ V S++ + E H TAH+L G+ T + + H + L +L+ I + T
Sbjct: 141 TNVESIEELSFIKELHLRTAHEL----GVETTVIEKH--LEGLHQLLKGISMMKELTLRT 194
Query: 201 TDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLE------SE 254
D++V GE S ++ S + K G + D ++ + T+ + + D LE S+
Sbjct: 195 RDYLVSFGECMSTRLFSAYLNKIGHKARQYDAFEIGFI--TTDDFTNADILEATYPAVSK 252
Query: 255 KRLETWFSLNPCKVIIATGFIASTPQKIP-TTLKRDGSDFSAAIMGALFRARQVTIWTDV 313
+ W N V+ TG++ + TTL R GSD +A +G R++ +W DV
Sbjct: 253 TLVGDWSKENAVPVV--TGYLGKGWRSCAITTLGRGGSDLTATTIGKALGLREIQVWKDV 310
Query: 314 DGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLS 373
DGV + DP A + L++ EA E++YFGA VLHP ++ P IP+ ++N +N +
Sbjct: 311 DGVLTCDPNIYPGAQSVPYLTFDEAAELAYFGAQVLHPLSMRPARDGDIPVRVKNSYNPT 370
Query: 374 APGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDV 433
APGT I + DM K + N+ ++++ T M G G + +F +D+
Sbjct: 371 APGTVITR-----SRDMS--KAVLTSIVLKRNVTMLDIASTRMLGQYGFLAKVFTTFEDL 423
Query: 434 GANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDN-----GRLSQVAIIPNCS 488
G +V ++ A+SE SI + K L R + LDN +++ V ++ S
Sbjct: 424 GISVDVV--ATSEVSISLTL--DPAKLWGRELIQRV-NELDNLVEELEKIAVVKLLQRRS 478
Query: 489 ILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHS 548
I++ +G S+ + +F +NV+ I+QG S+ NI++++ E+ + +RA+HS
Sbjct: 479 IISLIGNVQKSSL-ILEKVFQVFRSNGVNVQMISQGASKVNISLIVNDEEAEQCVRALHS 537
Query: 549 RFY 551
F+
Sbjct: 538 AFF 540
>At5g13280 aspartate kinase
Length = 569
Score = 182 bits (463), Expect = 6e-46
Identities = 146/546 (26%), Positives = 266/546 (47%), Gaps = 37/546 (6%)
Query: 24 ITKISQCQCLPFLPSHRSHSLRKALSLLPRGNQSPSTKISASLTDVSPSVLVEEQQLQ-- 81
+T+ LP + K LSL P G+ S K+S S + ++EE++ +
Sbjct: 20 LTRHRNSPTLPISLNRVDFPTLKKLSL-PIGDGSSIRKVSGSGSRNIVRAVLEEKKTEAI 78
Query: 82 -----KGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELIN 136
KG T V KFGG+ + S++R+K V D++L E ++V+SAM K T+ +
Sbjct: 79 TEVDEKGIT-CVMKFGGSSVASAERMKEVADLILTFPEESPVIVLSAMGKTTNNLLLAGE 137
Query: 137 KAQS---RDESYVSSLDAVLEKHSLTAHDL-LDGDGLATFLSQLHHDISNLKAMLRAIYI 192
KA S + S + L + E H T +L +D + T+L +L +L+ I +
Sbjct: 138 KAVSCGVSNASEIEELSIIKELHIRTVKELNIDPSVILTYLEELEQ-------LLKGIAM 190
Query: 193 AGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNP---TSSNQVDPD 249
T D++V GE S ++ + + G + D ++ + T+ + ++
Sbjct: 191 MKELTLRTRDYLVSFGECLSTRIFAAYLNTIGVKARQYDAFEIGFITTDDFTNGDILEAT 250
Query: 250 YLESEKRLETWFSLNPCKVIIATGFIASTPQK-IPTTLKRDGSDFSAAIMGALFRARQVT 308
Y KRL + +P V I TGF+ + TTL R GSD +A +G +++
Sbjct: 251 YPAVAKRLYDDWMHDPA-VPIVTGFLGKGWKTGAVTTLGRGGSDLTATTIGKALGLKEIQ 309
Query: 309 IWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRN 368
+W DVDGV + DP A + L++ EA E++YFGA VLHP+++ P + IP+ ++N
Sbjct: 310 VWKDVDGVLTCDPTIYKRATPVPYLTFDEAAELAYFGAQVLHPQSMRPAREGEIPVRVKN 369
Query: 369 IFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFG 428
+N APGT + DM K+ + N+ ++++ T M G G + +F
Sbjct: 370 SYNPKAPGT-----IITKTRDM--TKSILTSIVLKRNVTMLDIASTRMLGQVGFLAKVFS 422
Query: 429 AVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDN-GRLSQVAIIPNC 487
+++G +V ++ A+SE SI + ++ + E +Q H ++ +++ V ++
Sbjct: 423 IFEELGISVDVV--ATSEVSISLTLDPSKLWS-RELIQQELDHVVEELEKIAVVNLLKGR 479
Query: 488 SILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVH 547
+I++ +G S+ + F+ L +NV+ I+QG S+ NI+ ++ + ++A+H
Sbjct: 480 AIISLIGNVQHSSL-ILERAFHVLYTKGVNVQMISQGASKVNISFIVNEAEAEGCVQALH 538
Query: 548 SRFYLS 553
F+ S
Sbjct: 539 KSFFES 544
Score = 36.6 bits (83), Expect = 0.066
Identities = 30/93 (32%), Positives = 45/93 (48%), Gaps = 10/93 (10%)
Query: 393 VKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASSEHSICFA 452
V N +KG A I + NV+ + + F + G NV MISQ +S+ +I F
Sbjct: 472 VVNLLKGRAIISLIG--NVQHSSLI-----LERAFHVLYTKGVNVQMISQGASKVNISFI 524
Query: 453 VPEKEVKAVAEALQSRFRHALDNGRLSQVAIIP 485
V E E + +AL F ++G LS++ I P
Sbjct: 525 VNEAEAEGCVQALHKSF---FESGDLSELLIQP 554
>At3g02020 putative aspartate kinase
Length = 559
Score = 179 bits (455), Expect = 5e-45
Identities = 137/494 (27%), Positives = 248/494 (49%), Gaps = 32/494 (6%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESYVS 147
V KFGG+ + S++R+ V ++L E+ +VV+SAM+K T+ + KA + V
Sbjct: 86 VMKFGGSSVASAERMIQVAKLILSFPDEKPVVVLSAMAKTTNKLLMAGEKAVCCGVTNVD 145
Query: 148 SLDA---VLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFV 204
+++ + E H TAH+L G+ T + H + L+ +L+ + + T D++
Sbjct: 146 TIEELSYIKELHIRTAHEL----GVETAVIAEH--LEGLEQLLKGVAMMKELTLRSRDYL 199
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLE------SEKRLE 258
V GE S ++ + + K G + D ++ I+ T+ + + D LE S+K L
Sbjct: 200 VSFGECMSTRLFAAYLNKIGHKARQYDAFEIGII--TTDDFTNADILEATYPAVSKKLLG 257
Query: 259 TWFSLNPCKVIIATGFIASTPQKIP-TTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVY 317
W N V+ TGF+ + TTL R GSD +A +G R++ +W DVDGV
Sbjct: 258 DWSKENALPVV--TGFLGKGWRSCAVTTLGRGGSDLTATTIGKALGLREIQVWKDVDGVL 315
Query: 318 SADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGT 377
+ DP A + L++ EA E++YFGA VLHP ++ P + IP+ ++N +N +APGT
Sbjct: 316 TCDPNIYCGAQPVPHLTFDEAAELAYFGAQVLHPLSMRPAREGNIPVRVKNSYNPTAPGT 375
Query: 378 KICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANV 437
I + DM K + N+ ++++ T M G G + +F + +G +V
Sbjct: 376 VITR-----SRDMS--KAVLTSIVLKRNVTMLDITSTRMLGQYGFLAKVFSTFEKLGISV 428
Query: 438 IMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDN-GRLSQVAIIPNCSILAAVGQK 496
++ A+SE SI + + E +Q ++ +++ V ++ + SI++ +G
Sbjct: 429 DVV--ATSEVSISLTLDPSKF-CSRELIQHELDQVVEELEKIAVVNLLRHRSIISLIGNV 485
Query: 497 MASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTT 556
S+ + F L INV+ I+QG S+ NI++++ ++ ++A+HS F+ + T
Sbjct: 486 QRSS-FILEKGFRVLRTNGINVQMISQGASKVNISLIVNDDEAEHCVKALHSAFFETDTC 544
Query: 557 IAMGIIGPGLIGST 570
A+ G I ++
Sbjct: 545 EAVSECPTGYIAAS 558
>At3g18680 uridylate kinase like protein
Length = 339
Score = 43.5 bits (101), Expect = 5e-04
Identities = 29/86 (33%), Positives = 45/86 (51%), Gaps = 6/86 (6%)
Query: 291 SDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLH 350
+D +AA+ A A V T+VDGV+ DP++ A +L +L+YQE +V+
Sbjct: 233 TDTAAALRCAEINAEVVLKATNVDGVFDDDPKRNPNARLLDSLTYQEVTSKD---LSVMD 289
Query: 351 PRTIIPVMQYGIPILIRNIFNLSAPG 376
I + IP++ +FNLS PG
Sbjct: 290 MTAITLCQENNIPVV---VFNLSEPG 312
>At1g35770 hypothetical protein
Length = 1010
Score = 36.6 bits (83), Expect = 0.066
Identities = 40/147 (27%), Positives = 65/147 (44%), Gaps = 21/147 (14%)
Query: 172 FLSQLHHDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMD 231
F++ H IS L +L+ SF F + HG LW S+ +R+ W
Sbjct: 738 FINVSHFRISQLHPLLKKRCPPSRHLTSFFFFTLAHGALW-----SVPVRQENEIVSWHP 792
Query: 232 TRDVLIVNPTSSNQVDPD--YLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRD 289
+ + VN VDP+ + S K+L+ S++ C G IA + + I + R
Sbjct: 793 SPVINQVN------VDPEERFQNSMKKLKAISSISIC------GGIALSNKDILDLIDRK 840
Query: 290 GSDFSAAIMGALFR-ARQVTIWTDVDG 315
+ ++ +M AL R +R + DVDG
Sbjct: 841 -KNLTSKVMDALIRFSRHLLRTDDVDG 866
>At5g01660 putative protein
Length = 621
Score = 31.6 bits (70), Expect = 2.1
Identities = 22/80 (27%), Positives = 32/80 (39%), Gaps = 5/80 (6%)
Query: 174 SQLHHDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTR 233
S+ H D+ N K + + H E D V H E + V + N CK ++ R
Sbjct: 180 SENHPDVQNPKQIDK-----DHVLEKLKDLVFSHDEHGDNSLTETVEQANIPTCKNLEDR 234
Query: 234 DVLIVNPTSSNQVDPDYLES 253
D L S ++D L S
Sbjct: 235 DTLEEETCSEGKIDGSCLVS 254
>At4g39770 trehalose-6-phosphate phosphatase like protein
Length = 349
Score = 31.6 bits (70), Expect = 2.1
Identities = 27/109 (24%), Positives = 47/109 (42%), Gaps = 14/109 (12%)
Query: 716 LETGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPR---DDLSGTDV 772
L G ++L++ I + G+A +++ A T+ P DD + D
Sbjct: 248 LTQGRKVLEVRPIIK---------WDKGKALEFLLESLGYANCTDVFPLYIGDDRTDEDA 298
Query: 773 ARKVIILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQM 821
+ ++ R GL + +S P E+ LQ EF+QRL E+ Q+
Sbjct: 299 FK--VLRERRQGLGILVSKFPKETSASYSLQEPDEVMEFLQRLVEWKQL 345
>At5g15450 clpB heat shock protein-like
Length = 968
Score = 30.8 bits (68), Expect = 3.6
Identities = 51/233 (21%), Positives = 99/233 (41%), Gaps = 22/233 (9%)
Query: 688 LQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFS 747
L +Q +T +++ V + + ++ET + + +G IF+ K G +
Sbjct: 77 LTQQEFTEMAWQSIVSSPDVAKENKQQIVETEHLMKALLEQKNGLARRIFS--KIGVDNT 134
Query: 748 EVVQEAKEAGYTEPDPRDDLSGTDVARKVIIL---ARESGLKLELSNIPVESLVPEPLQA 804
+V++ ++ +P D +G+ + R + L AR+ L+ S + VE LV A
Sbjct: 135 KVLEATEKFIQRQPKVYGDAAGSMLGRDLEALFQRARQFKKDLKDSYVSVEHLV----LA 190
Query: 805 CASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQ 864
A + F ++L + Q+ + L+ A + + V+D + L +Y KD
Sbjct: 191 FADDKRFGKQLFKDFQISERSLKSAIES--IRGKQSVIDQDPEGKYEALEKYGKDLTAMA 248
Query: 865 LSGS--------DNI---IAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILR 906
G D I I +RR K+ P+++ PG G + G+ I++
Sbjct: 249 REGKLDPVIGRDDEIRRCIQILSRRTKNNPVLIGEPGVGKTAISEGLAQRIVQ 301
>At3g57580 unknown protein
Length = 408
Score = 30.8 bits (68), Expect = 3.6
Identities = 16/35 (45%), Positives = 19/35 (53%)
Query: 820 QMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELR 854
Q F KLEE EN GEV YV V+ S +L+
Sbjct: 348 QGFGAKLEEVENRGEVFAYVDYVEDLSLNDAKQLK 382
>At3g13930 putative acetyltransferase
Length = 539
Score = 30.4 bits (67), Expect = 4.7
Identities = 16/46 (34%), Positives = 24/46 (51%)
Query: 373 SAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAG 418
SAP KI PS +ED + + A +N+ L +++GTG G
Sbjct: 229 SAPEAKISKPSSAPSEDRIFASPLARKLAEDNNVPLSSIKGTGPEG 274
>At3g25840 protein kinase like protein
Length = 935
Score = 30.0 bits (66), Expect = 6.2
Identities = 21/82 (25%), Positives = 36/82 (43%), Gaps = 12/82 (14%)
Query: 39 HRSHSLRKALSLLPRGNQSPSTKISASLTDVSPSVLV------EEQQLQKGETWSVHKFG 92
HR H +K R N+ PS + DV+P+ + EE+ +++GE G
Sbjct: 43 HRHHRDKK------RENEIPSAGDETEILDVTPAAPIVVSNGCEEEDVEEGEILEEDGIG 96
Query: 93 GTCMGSSQRIKNVGDIVLEDDS 114
M ++ G+I ED++
Sbjct: 97 DVLMKTADSDGESGEIKFEDNN 118
>At3g23890 topoisomerase II
Length = 1473
Score = 29.6 bits (65), Expect = 8.1
Identities = 31/135 (22%), Positives = 54/135 (39%), Gaps = 15/135 (11%)
Query: 2 ASLSASSLYHFSTISPSNNTPDITKISQCQCLPFLPSHR-----SHSLRK-----ALSLL 51
AS S ++ +S + NN ++ K Q S SHS A +
Sbjct: 1218 ASESETTEASYSAMDTDNNVAEVVKPKARQGAKKKASESETTEASHSAMDTDNNVAEVVK 1277
Query: 52 PRGNQSPSTKISASLTDVSPSVLVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLE 111
P+G Q K A+ +V E++ L + + + FG SS+ + I ++
Sbjct: 1278 PKGRQGAKKKAPAAAKEVE-----EDEMLDLAQRLAQYNFGSAPADSSKTAETSKAIAVD 1332
Query: 112 DDSERKLVVVSAMSK 126
DD + +V V+ + K
Sbjct: 1333 DDDDDVVVEVAPVKK 1347
>At1g16720 unknown protein (At1g16720)
Length = 598
Score = 29.6 bits (65), Expect = 8.1
Identities = 22/81 (27%), Positives = 34/81 (41%), Gaps = 2/81 (2%)
Query: 13 STISPSNNTPDITKISQCQCLPFLPSHRSHSLRKALSLLPRGNQSPSTKISASLTDVSPS 72
S+ SPS + P K ++ + ++ K L + P QSP + + L DV+P
Sbjct: 48 SSSSPSPSPPSDKKKTKTRPGTITTKESEETVAKKLDVAPPSPQSPPSPPTLKLDDVNPV 107
Query: 73 VLVEEQQLQKGETWSVHKFGG 93
L + E W KF G
Sbjct: 108 GLGRRSRQIFDEVW--RKFSG 126
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,555,514
Number of Sequences: 26719
Number of extensions: 830630
Number of successful extensions: 2213
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 2186
Number of HSP's gapped (non-prelim): 18
length of query: 915
length of database: 11,318,596
effective HSP length: 108
effective length of query: 807
effective length of database: 8,432,944
effective search space: 6805385808
effective search space used: 6805385808
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0108.1


