

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
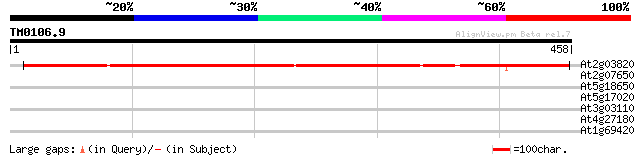
Query= TM0106.9
(458 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g03820 putative nonsense-mediated mRNA decay protein 577 e-165
At2g07650 putative non-LTR retrolelement reverse transcriptase 30 2.8
At5g18650 unknown protein 29 4.7
At5g17020 Exportin1 (XPO1) protein 29 4.7
At3g03110 exportin1 (XPO1) like protein 29 4.7
At4g27180 heavy chain polypeptide of kinesin-like protein (katB) 28 8.1
At1g69420 hypothetical protein 28 8.1
>At2g03820 putative nonsense-mediated mRNA decay protein
Length = 516
Score = 577 bits (1487), Expect = e-165
Identities = 284/450 (63%), Positives = 348/450 (77%), Gaps = 11/450 (2%)
Query: 12 EAGMFSVPQTICSVLCCMCGIPMQPNAANMCVKCLSSQEDITEGLQKHINIFHCPQCESY 71
E+GMF+V QTI SVLCC CG+PM PNAANMCV CL S+ DITEGLQK I IF+CP+C Y
Sbjct: 6 ESGMFNVQQTIGSVLCCKCGVPMAPNAANMCVNCLRSEVDITEGLQKSIQIFYCPECTCY 65
Query: 72 FQHPKTWIKHLKPESDELLKFCLKMMKKTNKVRMVDARFIWTEPHSKRIKINVKVQKEVL 131
Q PKTWIK + ES ELL FC+K +K NKV++ +A F+WTEPHSKRIK+ + VQ EVL
Sbjct: 66 LQPPKTWIK-CQWESKELLTFCIKRLKNLNKVKLKNAEFVWTEPHSKRIKVKLTVQAEVL 124
Query: 132 NGVILEQSYLVEYVQLEHLCDSCTRVAANPDQWVAVVQLRQHVSHRRTFFYLEQLILKHG 191
NG +LEQSY VEY ++LC+SC+R ANPDQWVA +QLRQHVSHRRTFFYLEQLIL+H
Sbjct: 125 NGAVLEQSYPVEYTVRDNLCESCSRFQANPDQWVASIQLRQHVSHRRTFFYLEQLILRHD 184
Query: 192 AAARAIRIKQMHEGIDFYFSNEDHGEKFVQFIGKVAPIRSKGYNKQLVSHDTKSNNYNYR 251
AA+RAIRI+Q+ +GIDF+F N+ H FV+F+ KV PI + ++QLVSHD KS+ YNY+
Sbjct: 185 AASRAIRIQQVDQGIDFFFGNKSHANSFVEFLRKVVPIEYR-QDQQLVSHDVKSSLYNYK 243
Query: 252 YTFSVEISPICREDLICLPHHVALSLGNLGPLVICTKVTNSIVLLDPFTLRHCFLDASQY 311
YT+SV+I P+CREDL+CLP VA LGNLGPLV+CTKV+++I LLDP TLR FLDA QY
Sbjct: 244 YTYSVKICPVCREDLVCLPSKVASGLGNLGPLVVCTKVSDNITLLDPRTLRCAFLDARQY 303
Query: 312 WRASFKSLLTSRQLVEYVVLDVEEVVPSEVVTVGGTKYVLAVAQVARVSDFGKNDTIFSI 371
WR+ F+S LTSRQLV+Y V DVE P TVGG KY L+ Q+AR SD GK +F +
Sbjct: 304 WRSGFRSALTSRQLVKYFVFDVEP--PVGEATVGGQKYALSYVQIARESDIGK---MFYV 358
Query: 372 KTHLGHLLNPGDYALGYDLYGANCNDIELDKYK---GGDLPHAILVKKSYVEKRLKKRGK 428
+THLGH+L PGD ALGYD+YGAN ND E++KY+ LP AIL+KK Y E+R +K+ K
Sbjct: 359 QTHLGHILKPGDQALGYDIYGANVNDNEMEKYRLSVKNGLPEAILIKKCYEEQRERKQKK 418
Query: 429 PRSWKLKSLNMEIDD-KGRTDPDKMSSEYE 457
R+WKLKSL ME+DD +GR DP+K EYE
Sbjct: 419 SRNWKLKSLPMEMDDSRGRVDPEKTDKEYE 448
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 30.0 bits (66), Expect = 2.8
Identities = 18/87 (20%), Positives = 38/87 (42%), Gaps = 12/87 (13%)
Query: 37 NAANMCVKCLSSQEDITEGLQKHINIFHCPQCESYFQHPKTWIKHLKPESDELLKFCLKM 96
+ +C C ++E I L+ CP E W++ + ++E+ LK+
Sbjct: 601 STTTICQVCKGAKESIIHVLRD------CPAMEGI------WLRLISDLANEVSAAHLKL 648
Query: 97 MKKTNKVRMVDARFIWTEPHSKRIKIN 123
T ++ V+ + W +P+ +K+N
Sbjct: 649 SGNTKEIVQVERQVSWRKPNEGWVKLN 675
>At5g18650 unknown protein
Length = 267
Score = 29.3 bits (64), Expect = 4.7
Identities = 22/76 (28%), Positives = 30/76 (38%), Gaps = 25/76 (32%)
Query: 18 VPQTICSV---------LCCMCGIPMQPNAANMCVKCLSSQEDITEGLQKHI-------- 60
V Q ICSV +C CG+ M ++C+ +D TE Q H
Sbjct: 63 VKQVICSVCDTEQPAAQVCSNCGVNMGEYFCSICI----FYDDDTEKQQFHCDDCGICRV 118
Query: 61 ----NIFHCPQCESYF 72
N FHC +C S +
Sbjct: 119 GGRENFFHCKKCGSCY 134
>At5g17020 Exportin1 (XPO1) protein
Length = 1075
Score = 29.3 bits (64), Expect = 4.7
Identities = 28/95 (29%), Positives = 43/95 (44%), Gaps = 19/95 (20%)
Query: 159 ANPDQWVAVVQLRQHVSHRRTFFYLEQL---ILKHGAAARAIRIKQMHEGIDFYFSNEDH 215
ANPD W+ VV + Q+ + T F+ Q+ ++K+ A + + +G+ Y S
Sbjct: 46 ANPDMWLQVVHILQNTNSLDTKFFALQVLEGVIKYRWNALPV---EQRDGMKNYIS---- 98
Query: 216 GEKFVQFIGKVAPIRSKG--YNK------QLVSHD 242
E VQ A RS+ NK Q+V HD
Sbjct: 99 -EVIVQLSSNEASFRSERLYVNKLNVILVQIVKHD 132
>At3g03110 exportin1 (XPO1) like protein
Length = 1022
Score = 29.3 bits (64), Expect = 4.7
Identities = 12/29 (41%), Positives = 18/29 (61%)
Query: 159 ANPDQWVAVVQLRQHVSHRRTFFYLEQLI 187
ANPD W+ VV + Q+ S T F+ Q++
Sbjct: 46 ANPDTWLQVVHILQNTSSTHTKFFALQVL 74
>At4g27180 heavy chain polypeptide of kinesin-like protein (katB)
Length = 745
Score = 28.5 bits (62), Expect = 8.1
Identities = 23/94 (24%), Positives = 43/94 (45%), Gaps = 7/94 (7%)
Query: 67 QCESYFQHPKTWIKHLKPESDELLKFCLKMMKKTNKVRMVDARFIWTEPHSKRIKINVKV 126
+CE+ + K ++ + L + + K N + M E H +++N+KV
Sbjct: 58 RCENTMDYVKRLRLCIRWFQELELDYAFEQEKLKNAMEM-------NEKHCADLEVNLKV 110
Query: 127 QKEVLNGVILEQSYLVEYVQLEHLCDSCTRVAAN 160
++E LN VI E VQ++ + ++AAN
Sbjct: 111 KEEELNMVIDELRKNFASVQVQLAKEQTEKLAAN 144
>At1g69420 hypothetical protein
Length = 596
Score = 28.5 bits (62), Expect = 8.1
Identities = 17/42 (40%), Positives = 22/42 (51%), Gaps = 3/42 (7%)
Query: 415 KKSYVEKRLKKRGKP---RSWKLKSLNMEIDDKGRTDPDKMS 453
KKS VE+R+KK+ +P W L LN E K + K S
Sbjct: 367 KKSVVEERVKKKPQPVKISPWTLARLNAEEVSKAAAEARKKS 408
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,663,524
Number of Sequences: 26719
Number of extensions: 459508
Number of successful extensions: 1132
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1124
Number of HSP's gapped (non-prelim): 7
length of query: 458
length of database: 11,318,596
effective HSP length: 103
effective length of query: 355
effective length of database: 8,566,539
effective search space: 3041121345
effective search space used: 3041121345
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0106.9


