

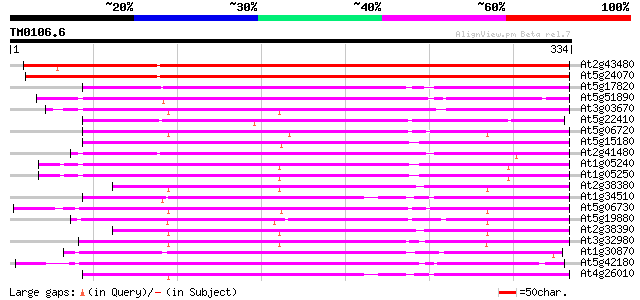
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.6
(334 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g43480 putative peroxidase 439 e-123
At5g24070 peroxidase-like protein 425 e-119
At5g17820 peroxidase ATP13a 227 6e-60
At5g51890 peroxidase 216 1e-56
At3g03670 putative peroxidase 216 1e-56
At5g22410 peroxidase ATP14a homolog 215 3e-56
At5g06720 peroxidase (emb|CAA68212.1) 214 5e-56
At5g15180 prx10 peroxidase - like protein 214 7e-56
At2g41480 peroxidase like protein 210 1e-54
At1g05240 putative peroxidase ATP12a 209 2e-54
At1g05250 putative peroxidase ATP12a 209 2e-54
At2g38380 peroxidase like protein 208 3e-54
At1g34510 peroxidase ATP13a, putative 208 3e-54
At5g06730 peroxidase 208 4e-54
At5g19880 peroxidase 207 8e-54
At2g38390 peroxidase 205 2e-53
At3g32980 peroxidase, putative 204 7e-53
At1g30870 unknown protein 203 1e-52
At5g42180 peroxidase (emb|CAA66960.1) 202 2e-52
At4g26010 peroxidase like protein 202 2e-52
>At2g43480 putative peroxidase
Length = 335
Score = 439 bits (1129), Expect = e-123
Identities = 211/330 (63%), Positives = 262/330 (78%), Gaps = 6/330 (1%)
Query: 9 FLLVVAFAALSSSPVAAAT----PPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKS 64
FL V+ +S P A P KL WHYYK +N C +AE +VR+QV++F+K DKS
Sbjct: 7 FLTVMVVGGVSLFPETAEAIVMGPSMQKLTWHYYKVYNTCENAENFVRHQVEIFYKNDKS 66
Query: 65 ITAKLLRLVYSDCFITGCDASILLDEGPNTEKKAPQNRGLGAFVLIDNIKTFVERQCPGV 124
I KLLRL+YSDCF++GCDAS+LL EGPN+EK APQNRGLG FVLID IK +E++CPGV
Sbjct: 67 IAPKLLRLLYSDCFVSGCDASVLL-EGPNSEKMAPQNRGLGGFVLIDKIKIVLEQRCPGV 125
Query: 125 VSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAASVDIPSPSVSWQEALAYFKSRG 184
VSCADIL LATRDAV LAG P YPVFTGR+DG+ SD +VD+PSPS+SW +A++YFKSRG
Sbjct: 126 VSCADILNLATRDAVHLAGAPSYPVFTGRRDGLTSDKQTVDLPSPSISWDQAMSYFKSRG 185
Query: 185 LNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPR-KKGQ 243
LNVLDM TLLG+H++GRTHCSY+ DRLYNYN TG P+MN FL M K CPPR +KGQ
Sbjct: 186 LNVLDMATLLGSHSMGRTHCSYVVDRLYNYNKTGKPSPTMNKYFLSEMAKQCPPRTRKGQ 245
Query: 244 SDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFKK 303
+DPLVYLNPDSGS++ FT S+Y RIL++++VL +DQQLL DDTK+I++EF+ G +DF+K
Sbjct: 246 TDPLVYLNPDSGSNHSFTSSFYSRILSNKSVLEVDQQLLYNDDTKQISKEFSEGFEDFRK 305
Query: 304 SFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
SFA+SM MG I VLT +G+IR++CR IN
Sbjct: 306 SFALSMSKMGAINVLTKTEGEIRKDCRHIN 335
>At5g24070 peroxidase-like protein
Length = 340
Score = 425 bits (1093), Expect = e-119
Identities = 203/325 (62%), Positives = 254/325 (77%), Gaps = 2/325 (0%)
Query: 10 LLVVAFAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKL 69
L+V++ A ++ A P KL WHYYK N C DAE Y+R QV+ F+K D SI KL
Sbjct: 12 LVVISLAGKATVEAATGLNPPVKLVWHYYKLTNTCDDAETYIRYQVEKFYKNDSSIAPKL 71
Query: 70 LRLVYSDCFITGCDASILLDEGPNTEKKAPQNRGLGAFVLIDNIKTFVERQCPGVVSCAD 129
LRL+YSDC + GCD SILL +GPN+E+ APQNRGLG FV+ID IK +E +CPGVVSCAD
Sbjct: 72 LRLLYSDCMVNGCDGSILL-QGPNSERTAPQNRGLGGFVIIDKIKQVLESRCPGVVSCAD 130
Query: 130 ILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAASVDIPSPSVSWQEALAYFKSRGLNVLD 189
IL LATRDAV +AG P YPVFTGR+DG +A +VD+PSPS+S E+LAYFKS+GL+VLD
Sbjct: 131 ILNLATRDAVHMAGAPSYPVFTGRRDGGTLNADAVDLPSPSISVDESLAYFKSKGLDVLD 190
Query: 190 MGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPR-KKGQSDPLV 248
M TLLGAH++G+THCSY+ DRLYN+ TG DP+MN + +R LCPPR +KGQ+DPLV
Sbjct: 191 MTTLLGAHSMGKTHCSYVVDRLYNFKNTGKPDPTMNTTLVSQLRYLCPPRTQKGQTDPLV 250
Query: 249 YLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFKKSFAVS 308
YLNPDSGSS +FT SYY R+L+H AVL +DQ+LLN DD+KEIT+EFA+G +DF+KSFA++
Sbjct: 251 YLNPDSGSSNRFTSSYYSRVLSHNAVLRVDQELLNNDDSKEITQEFASGFEDFRKSFALA 310
Query: 309 MYNMGNIKVLTGNQGQIRQNCRFIN 333
M MG+I VLTG G+IR++CR N
Sbjct: 311 MSRMGSINVLTGTAGEIRRDCRVTN 335
>At5g17820 peroxidase ATP13a
Length = 313
Score = 227 bits (579), Expect = 6e-60
Identities = 120/290 (41%), Positives = 172/290 (58%), Gaps = 9/290 (3%)
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPNTEKKAPQNRG 103
C AE VRN V+ + ++TA LLR+ + DCF+ GCDAS+L+D N+EK A N
Sbjct: 33 CPQAETIVRNLVRQRFGVTPTVTAALLRMHFHDCFVKGCDASLLIDS-TNSEKTAGPNGS 91
Query: 104 LGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAAS 163
+ F LID IK +E CP VSCADI+ LATRD+V LAGGP Y + TGR+DG S+
Sbjct: 92 VREFDLIDRIKAQLEAACPSTVSCADIVTLATRDSVALAGGPSYSIPTGRRDGRVSNNLD 151
Query: 164 VDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPS 223
V +P P++S A++ F ++G+N D LLGAHT+G+ +C +DR+ ++ GTG DPS
Sbjct: 152 VTLPGPTISVSGAVSLFTNKGMNTFDAVALLGAHTVGQGNCGLFSDRITSFQGTGRPDPS 211
Query: 224 MNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLN 283
M+ A + ++R C + + L D S +F ++K+I VL +DQ+L +
Sbjct: 212 MDPALVTSLRNTC---RNSATAAL-----DQSSPLRFDNQFFKQIRKRRGVLQVDQRLAS 263
Query: 284 GDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
T+ I +A FK+ F +M MG + VLTG G+IR+NCR N
Sbjct: 264 DPQTRGIVARYANNNAFFKRQFVRAMVKMGAVDVLTGRNGEIRRNCRRFN 313
>At5g51890 peroxidase
Length = 322
Score = 216 bits (550), Expect = 1e-56
Identities = 132/321 (41%), Positives = 179/321 (55%), Gaps = 12/321 (3%)
Query: 17 ALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSD 76
A+ + +A P L+ HYY C AE + V+ +D + A+LLR+ + D
Sbjct: 10 AMIFAVLAIVKPSEAALDAHYYDQS--CPAAEKIILETVRNATLYDPKVPARLLRMFFHD 67
Query: 77 CFITGCDASILLDE--GPNTEKKAPQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLA 134
CFI GCDASILLD EK P N + +F +I++ K +E+ CP VSCAD++ +A
Sbjct: 68 CFIRGCDASILLDSTRSNQAEKDGPPNISVRSFYVIEDAKRKLEKACPRTVSCADVIAIA 127
Query: 135 TRDAVQLAGGPGYPVFTGRKDGMRSDAASV-DIPSPSVSWQEALAYFKSRGLNVLDMGTL 193
RD V L+GGP + V GRKDG S A ++P P+ + + + F +RGL+V DM TL
Sbjct: 128 ARDVVTLSGGPYWSVLKGRKDGTISRANETRNLPPPTFNVSQLIQSFAARGLSVKDMVTL 187
Query: 194 LGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCP-PRKKGQSDPLVYLNP 252
G HTIG +HCS RL N++ DPSMN AF T++K CP +G++ V
Sbjct: 188 SGGHTIGFSHCSSFESRLQNFSKFHDIDPSMNYAFAQTLKKKCPRTSNRGKNAGTVL--- 244
Query: 253 DSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFKKSFAVSMYNM 312
DS SS F YYK+IL+ + V G DQ LL TK I E FA + F + FA SM +
Sbjct: 245 DSTSSV-FDNVYYKQILSGKGVFGSDQALLGDSRTKWIVETFAQDQKAFFREFAASMVKL 303
Query: 313 GNIKVLTGNQGQIRQNCRFIN 333
GN V GQ+R N RF+N
Sbjct: 304 GNFGV--KETGQVRVNTRFVN 322
>At3g03670 putative peroxidase
Length = 321
Score = 216 bits (550), Expect = 1e-56
Identities = 128/317 (40%), Positives = 182/317 (57%), Gaps = 17/317 (5%)
Query: 22 PVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITG 81
PVA A +L++ +Y C +AE V N V+ + D SITA L R+ + DCF+ G
Sbjct: 17 PVALA-----QLKFKFYSES--CPNAETIVENLVRQQFARDPSITAALTRMHFHDCFVQG 69
Query: 82 CDASILLDEGPN--TEKKAPQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAV 139
CDAS+L+D + +EK A N + F LID IKT +E QCP VSC+DI+ LATRDAV
Sbjct: 70 CDASLLIDPTTSQLSEKNAGPNFSVRGFELIDEIKTALEAQCPSTVSCSDIVTLATRDAV 129
Query: 140 QLAGGPGYPVFTGRKDGMRS--DAASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAH 197
L GGP Y V TGR+DG S + A+ +P P +S + L++F ++G+NV D LLGAH
Sbjct: 130 FLGGGPSYVVPTGRRDGFVSNPEDANEILPPPFISVEGMLSFFGNKGMNVFDSVALLGAH 189
Query: 198 TIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCP-PRKKGQSDPLVYLNPDSGS 256
T+G C DR+ N+ GTG DPSM+ +R C P D + + P S
Sbjct: 190 TVGIASCGNFVDRVTNFQGTGLPDPSMDPTLAGRLRNTCAVPGGFAALDQSMPVTPVS-- 247
Query: 257 SYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIK 316
F ++ +I + +L IDQ + + T + ++A+ + FK+ FA++M MG +
Sbjct: 248 ---FDNLFFGQIRERKGILLIDQLIASDPATSGVVLQYASNNELFKRQFAIAMVKMGAVD 304
Query: 317 VLTGNQGQIRQNCRFIN 333
VLTG+ G+IR NCR N
Sbjct: 305 VLTGSAGEIRTNCRAFN 321
>At5g22410 peroxidase ATP14a homolog
Length = 331
Score = 215 bits (547), Expect = 3e-56
Identities = 125/290 (43%), Positives = 174/290 (59%), Gaps = 7/290 (2%)
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPNTEKKAPQNRG 103
C++ E V V + D SI ++RL + DCF GCDAS+LLD G N+EKKA N
Sbjct: 37 CQNVENIVSKVVGEAFIKDSSIAPAMIRLYFHDCFSNGCDASLLLD-GSNSEKKASPNLS 95
Query: 104 LGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGG--PGYPVFTGRKDGMRSDA 161
+ + +ID+IK+ VE++C VVSCADI+ LATRD V LA G Y + TGR DG S A
Sbjct: 96 VRGYEVIDDIKSAVEKECDRVVSCADIIALATRDLVTLASGGKTRYEIPTGRLDGKISSA 155
Query: 162 ASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSD 221
VD+PSP ++ E A F R L++ DM LLG HTIG THCS+I DRLYN+ T D
Sbjct: 156 LLVDLPSPKMTVAETAAKFDQRKLSLNDMVLLLGGHTIGVTHCSFIMDRLYNFQNTQKPD 215
Query: 222 PSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQL 281
PSM+ ++ + CP K +D ++ L+ ++ SS S+YK I VL IDQ+L
Sbjct: 216 PSMDPKLVEELSAKCP--KSSSTDGIISLDQNATSSNTMDVSFYKEIKVSRGVLHIDQKL 273
Query: 282 LNGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLT-GNQGQIRQNCR 330
D T ++ + A G DF F +M N+G+++V++ G+IR++CR
Sbjct: 274 AIDDLTSKMVTDIANG-NDFLVRFGQAMVNLGSVRVISKPKDGEIRRSCR 322
>At5g06720 peroxidase (emb|CAA68212.1)
Length = 335
Score = 214 bits (545), Expect = 5e-56
Identities = 119/297 (40%), Positives = 177/297 (59%), Gaps = 11/297 (3%)
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPN--TEKKA-PQ 100
C +A VR+ ++ + D I A L+RL + DCF+ GCDASILLD+ + +EK A P
Sbjct: 41 CPNASAIVRSTIQQALQSDTRIGASLIRLHFHDCFVNGCDASILLDDTGSIQSEKNAGPN 100
Query: 101 NRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSD 160
F ++DNIKT +E CPGVVSC+D+L LA+ +V LAGGP + V GR+D + ++
Sbjct: 101 VNSARGFNVVDNIKTALENACPGVVSCSDVLALASEASVSLAGGPSWTVLLGRRDSLTAN 160
Query: 161 AASVD--IPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTG 218
A + IPSP S F + GLN D+ L GAHT GR C +RL+N++GTG
Sbjct: 161 LAGANSSIPSPIESLSNITFKFSAVGLNTNDLVALSGAHTFGRARCGVFNNRLFNFSGTG 220
Query: 219 SSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGID 278
+ DP++N+ L T+++LCP + G + + N D + F +Y+ + +++ +L D
Sbjct: 221 NPDPTLNSTLLSTLQQLCP--QNGSASTIT--NLDLSTPDAFDNNYFANLQSNDGLLQSD 276
Query: 279 QQLLN--GDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
Q+L + G T I FA+ F ++FA SM NMGNI LTG+ G+IR +C+ +N
Sbjct: 277 QELFSTTGSSTIAIVTSFASNQTLFFQAFAQSMINMGNISPLTGSNGEIRLDCKKVN 333
>At5g15180 prx10 peroxidase - like protein
Length = 329
Score = 214 bits (544), Expect = 7e-56
Identities = 115/294 (39%), Positives = 169/294 (57%), Gaps = 9/294 (3%)
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLD-EGPNTEKKAPQNR 102
C AEL V+ V K D++I A LLR+ + DCF+ GC+ S+LL+ + EK + N
Sbjct: 41 CPKAELIVKKSVFEAVKNDRTIAAPLLRMFFHDCFVRGCEGSVLLELKNKKDEKNSIPNL 100
Query: 103 GLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSD-- 160
L F +IDN+K +E++CPG+VSC+D+L L RDA+ GP + V TGR+DG+ ++
Sbjct: 101 TLRGFEIIDNVKAALEKECPGIVSCSDVLALVARDAMVALNGPSWEVETGRRDGLVTNIT 160
Query: 161 AASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSS 220
A +++PSP + + F+S+GL+ D+ L G HTIG HC IT+RLYN+ G G S
Sbjct: 161 EALLNLPSPFNNISSLITQFQSKGLDKKDLVVLSGGHTIGNGHCPQITNRLYNFTGKGDS 220
Query: 221 DPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQ 280
DP+++ + +R C P +D L D GS F ESY+K + + D
Sbjct: 221 DPNLDTEYAVKLRGKCKP-----TDTTTALEMDPGSFKTFDESYFKLVSQRRGLFQSDAA 275
Query: 281 LLNGDDTKE-ITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
LL+ +TK + + + F K F VSM MG I VLTG G++R+ CR +N
Sbjct: 276 LLDNQETKSYVLKSLNSDGSTFFKDFGVSMVKMGRIGVLTGQVGEVRKKCRMVN 329
>At2g41480 peroxidase like protein
Length = 328
Score = 210 bits (534), Expect = 1e-54
Identities = 123/303 (40%), Positives = 178/303 (58%), Gaps = 13/303 (4%)
Query: 37 YYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPNTEK 96
YY T C AE VR+ V+ + D +I+ LLRL + DCF+ GCD S+L+ +G + E+
Sbjct: 33 YYSTS--CPKAESIVRSTVESHFDSDPTISPGLLRLHFHDCFVQGCDGSVLI-KGKSAEQ 89
Query: 97 KAPQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDG 156
A N GL +ID+ K +E CPGVVSCADIL LA RD+V L+ GP + V TGRKDG
Sbjct: 90 AALPNLGLRGLEVIDDAKARLEAVCPGVVSCADILALAARDSVDLSDGPSWRVPTGRKDG 149
Query: 157 MRSDAASV-DIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYN 215
S A ++PSP S F+ +GL+ D+ TLLGAHTIG+T C + RLYN+
Sbjct: 150 RISLATEASNLPSPLDSVAVQKQKFQDKGLDTHDLVTLLGAHTIGQTDCLFFRYRLYNFT 209
Query: 216 GTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVL 275
TG+SDP+++ +FL ++ LCPP G + D GS KF ES++K + + A+L
Sbjct: 210 VTGNSDPTISPSFLTQLKTLCPPNGDGSKRVAL----DIGSPSKFDESFFKNLRDGNAIL 265
Query: 276 GIDQQLLNGDDTKEITEEFAAGLQD-----FKKSFAVSMYNMGNIKVLTGNQGQIRQNCR 330
DQ+L + +T + +++A+ L+ F F +M M +I V T G++R+ C
Sbjct: 266 ESDQRLWSDAETNAVVKKYASRLRGLLGFRFDYEFGKAMIKMSSIDVKTDVDGEVRKVCS 325
Query: 331 FIN 333
+N
Sbjct: 326 KVN 328
>At1g05240 putative peroxidase ATP12a
Length = 325
Score = 209 bits (531), Expect = 2e-54
Identities = 121/322 (37%), Positives = 178/322 (54%), Gaps = 15/322 (4%)
Query: 18 LSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDC 77
LS V+ A P L+ YY++ C AE VR + K++ AKLLR+ + DC
Sbjct: 13 LSVVGVSVAIPQL--LDLDYYRSK--CPKAEEIVRGVTVQYVSRQKTLAAKLLRMHFHDC 68
Query: 78 FITGCDASILLDEGPN-TEKKAPQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATR 136
F+ GCD S+LL N E+ A N L + ++D KT +ER+CP ++SCAD+L L R
Sbjct: 69 FVRGCDGSVLLKSAKNDAERDAVPNLTLKGYEVVDAAKTALERKCPNLISCADVLALVAR 128
Query: 137 DAVQLAGGPGYPVFTGRKDGMRS--DAASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLL 194
DAV + GGP +PV GR+DG S + A +++PSP + F ++GLN D+ L
Sbjct: 129 DAVAVIGGPWWPVPLGRRDGRISKLNDALLNLPSPFADIKTLKKNFANKGLNAKDLVVLS 188
Query: 195 GAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDS 254
G HTIG + C+ + RLYN+ G G SDPSMN +++ +++ CPP +D LN D
Sbjct: 189 GGHTIGISSCALVNSRLYNFTGKGDSDPSMNPSYVRELKRKCPP-----TDFRTSLNMDP 243
Query: 255 GSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFA---AGLQDFKKSFAVSMYN 311
GS+ F Y+K + + + D LL+ +TK + A F K F+ SM
Sbjct: 244 GSALTFDTHYFKVVAQKKGLFTSDSTLLDDIETKNYVQTQAILPPVFSSFNKDFSDSMVK 303
Query: 312 MGNIKVLTGNQGQIRQNCRFIN 333
+G +++LTG G+IR+ C F N
Sbjct: 304 LGFVQILTGKNGEIRKRCAFPN 325
>At1g05250 putative peroxidase ATP12a
Length = 325
Score = 209 bits (531), Expect = 2e-54
Identities = 121/322 (37%), Positives = 178/322 (54%), Gaps = 15/322 (4%)
Query: 18 LSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDC 77
LS V+ A P L+ YY++ C AE VR + K++ AKLLR+ + DC
Sbjct: 13 LSVVGVSVAIPQL--LDLDYYRSK--CPKAEEIVRGVTVQYVSRQKTLAAKLLRMHFHDC 68
Query: 78 FITGCDASILLDEGPN-TEKKAPQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATR 136
F+ GCD S+LL N E+ A N L + ++D KT +ER+CP ++SCAD+L L R
Sbjct: 69 FVRGCDGSVLLKSAKNDAERDAVPNLTLKGYEVVDAAKTALERKCPNLISCADVLALVAR 128
Query: 137 DAVQLAGGPGYPVFTGRKDGMRS--DAASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLL 194
DAV + GGP +PV GR+DG S + A +++PSP + F ++GLN D+ L
Sbjct: 129 DAVAVIGGPWWPVPLGRRDGRISKLNDALLNLPSPFADIKTLKKNFANKGLNAKDLVVLS 188
Query: 195 GAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDS 254
G HTIG + C+ + RLYN+ G G SDPSMN +++ +++ CPP +D LN D
Sbjct: 189 GGHTIGISSCALVNSRLYNFTGKGDSDPSMNPSYVRELKRKCPP-----TDFRTSLNMDP 243
Query: 255 GSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFA---AGLQDFKKSFAVSMYN 311
GS+ F Y+K + + + D LL+ +TK + A F K F+ SM
Sbjct: 244 GSALTFDTHYFKVVAQKKGLFTSDSTLLDDIETKNYVQTQAILPPVFSSFNKDFSDSMVK 303
Query: 312 MGNIKVLTGNQGQIRQNCRFIN 333
+G +++LTG G+IR+ C F N
Sbjct: 304 LGFVQILTGKNGEIRKRCAFPN 325
>At2g38380 peroxidase like protein
Length = 349
Score = 208 bits (530), Expect = 3e-54
Identities = 117/280 (41%), Positives = 165/280 (58%), Gaps = 12/280 (4%)
Query: 62 DKSITAKLLRLVYSDCFITGCDASILLDEGPN--TEKKA-PQNRGLGAFVLIDNIKTFVE 118
D I A LLRL + DCF+ GCDASILLD + TEK A P F +ID +K +E
Sbjct: 58 DPRIAASLLRLHFHDCFVRGCDASILLDNSTSFRTEKDAAPNANSARGFNVIDRMKVALE 117
Query: 119 RQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRS--DAASVDIPSPSVSWQEA 176
R CPG VSCADIL +A++ +V L+GGP +PV GR+D + + A+ +PSP + +
Sbjct: 118 RACPGRVSCADILTIASQISVLLSGGPWWPVPLGRRDSVEAFFALANTALPSPFFNLTQL 177
Query: 177 LAYFKSRGLN-VLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKL 235
F GLN D+ L G HT GR C ++T RLYN+NGT S DPS+N +L +R+L
Sbjct: 178 KTAFADVGLNRTSDLVALSGGHTFGRAQCQFVTPRLYNFNGTNSPDPSLNPTYLVELRRL 237
Query: 236 CPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLN--GDDTKEITEE 293
CP G V +N D + F YY + N + ++ DQ+L + G DT + +
Sbjct: 238 CPQNGNG----TVLVNFDVVTPDAFDSQYYTNLRNGKGLIQSDQELFSTPGADTIPLVNQ 293
Query: 294 FAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
+++ + F ++F +M MGN++ LTG QG+IRQNCR +N
Sbjct: 294 YSSDMSVFFRAFIDAMIRMGNLRPLTGTQGEIRQNCRVVN 333
>At1g34510 peroxidase ATP13a, putative
Length = 310
Score = 208 bits (530), Expect = 3e-54
Identities = 117/294 (39%), Positives = 171/294 (57%), Gaps = 19/294 (6%)
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLD---EGPNTEKKAPQ 100
C AE V V W ++++TA LLR+ + DC + GCDAS+L+D E P+ EK +
Sbjct: 31 CPPAESIVGRVVFNHWDRNRTVTAALLRMQFHDCVVKGCDASLLIDPTTERPS-EKSVGR 89
Query: 101 NRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSD 160
N G+ F +ID K +E CP VSCADI+ +ATRD++ LAGGP + V TGR+DG+RS+
Sbjct: 90 NAGVRGFEIIDEAKKELELVCPKTVSCADIVTIATRDSIALAGGPKFKVRTGRRDGLRSN 149
Query: 161 AASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLL-GAHTIGRTHCSYITDRLYNYNGTGS 219
+ V + P+VS ++ FKS G NV M L+ G HT+G HCS DR+
Sbjct: 150 PSDVKLLGPTVSVATSIKAFKSIGFNVSTMVALIGGGHTVGVAHCSLFQDRI-------- 201
Query: 220 SDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQ 279
DP M++ ++K C +G +DP V++ D + ++ Y++++ A+L ID
Sbjct: 202 KDPKMDSKLRAKLKKSC----RGPNDPSVFM--DQNTPFRVDNEIYRQMIQQRAILRIDD 255
Query: 280 QLLNGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
L+ T+ I +FA + FK+SFA +M MG I VLTG+ G+IR NCR N
Sbjct: 256 NLIRDGSTRSIVSDFAYNNKLFKESFAEAMQKMGEIGVLTGDSGEIRTNCRAFN 309
>At5g06730 peroxidase
Length = 358
Score = 208 bits (529), Expect = 4e-54
Identities = 121/338 (35%), Positives = 190/338 (55%), Gaps = 17/338 (5%)
Query: 3 SSMCVAFLLVVAFAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFD 62
SS C F ++ +SS ++ +L +Y C +A VR+ ++ + D
Sbjct: 7 SSTCDGFFIISLIVIVSSLFGTSSA----QLNATFYS--GTCPNASAIVRSTIQQALQSD 60
Query: 63 KSITAKLLRLVYSDCFITGCDASILLDEGPN--TEKKAPQN-RGLGAFVLIDNIKTFVER 119
I L+RL + DCF+ GCD S+LLD+ + +EK AP N F ++D+IKT +E
Sbjct: 61 ARIGGSLIRLHFHDCFVNGCDGSLLLDDTSSIQSEKNAPANANSTRGFNVVDSIKTALEN 120
Query: 120 QCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSD--AASVDIPSPSVSWQEAL 177
CPG+VSC+DIL LA+ +V LAGGP + V GR+DG+ ++ A+ +PSP
Sbjct: 121 ACPGIVSCSDILALASEASVSLAGGPSWTVLLGRRDGLTANLSGANSSLPSPFEGLNNIT 180
Query: 178 AYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCP 237
+ F + GL D+ +L GAHT GR C +RL+N+NGTG+ DP++N+ L ++++LCP
Sbjct: 181 SKFVAVGLKTTDVVSLSGAHTFGRGQCVTFNNRLFNFNGTGNPDPTLNSTLLSSLQQLCP 240
Query: 238 PRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLN--GDDTKEITEEFA 295
+ G + + N D + F +Y+ + ++ +L DQ+L + G T I FA
Sbjct: 241 --QNGSNTGIT--NLDLSTPDAFDNNYFTNLQSNNGLLQSDQELFSNTGSATVPIVNSFA 296
Query: 296 AGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
+ F ++F SM MGNI LTG+ G+IRQ+C+ +N
Sbjct: 297 SNQTLFFEAFVQSMIKMGNISPLTGSSGEIRQDCKVVN 334
>At5g19880 peroxidase
Length = 329
Score = 207 bits (526), Expect = 8e-54
Identities = 124/308 (40%), Positives = 176/308 (56%), Gaps = 18/308 (5%)
Query: 37 YYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGP---- 92
+Y T C + R ++ + D +TAK++RL + DCF+ GCD S+LLD P
Sbjct: 29 FYST--TCPNVTAIARGLIERASRNDVRLTAKVMRLHFHDCFVNGCDGSVLLDAAPADGV 86
Query: 93 NTEKKAPQNRG-LGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFT 151
EK+A QN G L F +ID+IKT +E CPGVVSCADIL +A +V LAGGP V
Sbjct: 87 EGEKEAFQNAGSLDGFEVIDDIKTALENVCPGVVSCADILAIAAEISVALAGGPSLDVLL 146
Query: 152 GRKDG---MRSDAASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYIT 208
GR+DG +R+DA + +P S + + F L+ D+ L GAHT GR C I
Sbjct: 147 GRRDGRTAIRADAVAA-LPLGPDSLEILTSKFSVHNLDTTDLVALSGAHTFGRVQCGVIN 205
Query: 209 DRLYNYNG-TGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKR 267
+RL+N++G +G SDPS+ FL T+R+ CP + G L+P S S F Y+K
Sbjct: 206 NRLHNFSGNSGQSDPSIEPEFLQTLRRQCP--QGGDLTARANLDPTSPDS--FDNDYFKN 261
Query: 268 ILNHEAVLGIDQQLLN--GDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQI 325
+ N+ V+ DQ L + G T + FA +F +FA SM MGN+++LTG +G+I
Sbjct: 262 LQNNRGVIESDQILFSSTGAPTVSLVNRFAENQNEFFTNFARSMIKMGNVRILTGREGEI 321
Query: 326 RQNCRFIN 333
R++CR +N
Sbjct: 322 RRDCRRVN 329
>At2g38390 peroxidase
Length = 349
Score = 205 bits (522), Expect = 2e-53
Identities = 116/280 (41%), Positives = 163/280 (57%), Gaps = 12/280 (4%)
Query: 62 DKSITAKLLRLVYSDCFITGCDASILLDEGPN--TEKKA-PQNRGLGAFVLIDNIKTFVE 118
D I A LLRL + DCF+ GCDASILLD + TEK A P + F +ID +K +E
Sbjct: 58 DPRIAASLLRLHFHDCFVRGCDASILLDNSTSFRTEKDAAPNKNSVRGFDVIDRMKAAIE 117
Query: 119 RQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRS--DAASVDIPSPSVSWQEA 176
R CP VSCADI+ +A++ +V L+GGP +PV GR+D + + A+ +PSP + +
Sbjct: 118 RACPRTVSCADIITIASQISVLLSGGPWWPVPLGRRDSVEAFFALANTALPSPFSTLTQL 177
Query: 177 LAYFKSRGLN-VLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKL 235
F GLN D+ L G HT G+ C ++T RLYN+NGT DPS+N +L +R+L
Sbjct: 178 KTAFADVGLNRPSDLVALSGGHTFGKAQCQFVTPRLYNFNGTNRPDPSLNPTYLVELRRL 237
Query: 236 CPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLN--GDDTKEITEE 293
CP G V +N DS + F YY +LN + ++ DQ L + G DT + +
Sbjct: 238 CPQNGNG----TVLVNFDSVTPTTFDRQYYTNLLNGKGLIQSDQVLFSTPGADTIPLVNQ 293
Query: 294 FAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
+++ F +F +M MGN+K LTG QG+IRQNCR +N
Sbjct: 294 YSSNTFVFFGAFVDAMIRMGNLKPLTGTQGEIRQNCRVVN 333
>At3g32980 peroxidase, putative
Length = 352
Score = 204 bits (518), Expect = 7e-53
Identities = 121/301 (40%), Positives = 169/301 (55%), Gaps = 13/301 (4%)
Query: 42 NICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPN--TEKKA- 98
N C VR+ + + D I A +LRL + DCF+ GCDASILLD + TEK A
Sbjct: 38 NTCPSVFTIVRDTIVNELRSDPRIAASILRLHFHDCFVNGCDASILLDNTTSFRTEKDAA 97
Query: 99 PQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMR 158
P F +ID +K VE CP VSCADIL +A + AV LAGGP + V GR+D ++
Sbjct: 98 PNANSARGFPVIDRMKAAVETACPRTVSCADILTIAAQQAVNLAGGPSWRVPLGRRDSLQ 157
Query: 159 S--DAASVDIPSPSVSWQEALAYFKSRGLN-VLDMGTLLGAHTIGRTHCSYITDRLYNYN 215
+ A+ ++P+P + + A F++ GL+ D+ L G HT G+ C +I DRLYN++
Sbjct: 158 AFFALANTNLPAPFFTLPQLKASFQNVGLDRPSDLVALSGGHTFGKNQCQFIMDRLYNFS 217
Query: 216 GTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVL 275
TG DP++N +L T+R C PR Q+ V ++ D + F YY + + ++
Sbjct: 218 NTGLPDPTLNTTYLQTLRGQC-PRNGNQT---VLVDFDLRTPTVFDNKYYVNLKELKGLI 273
Query: 276 GIDQQLL---NGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFI 332
DQ+L N DT + E+A G Q F +F +M MGNI LTG QGQIRQNCR +
Sbjct: 274 QTDQELFSSPNATDTIPLVREYADGTQKFFNAFVEAMNRMGNITPLTGTQGQIRQNCRVV 333
Query: 333 N 333
N
Sbjct: 334 N 334
>At1g30870 unknown protein
Length = 349
Score = 203 bits (516), Expect = 1e-52
Identities = 128/302 (42%), Positives = 175/302 (57%), Gaps = 16/302 (5%)
Query: 33 LEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLD-EG 91
L +YY IC D E V +V+ + K D S+ LLRL++ DC +TGCDAS+LLD EG
Sbjct: 51 LSLNYYD--RICPDFEKIVVTKVREWTKSDSSLGPALLRLIFHDCGVTGCDASVLLDYEG 108
Query: 92 PNTEKKAPQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFT 151
TE+++P ++ L F LID+IK+ +E+ CPG VSCADIL A+R A GGP +P
Sbjct: 109 --TERRSPASKTLRGFELIDDIKSEMEKSCPGKVSCADILTSASRAATVQLGGPYWPNVY 166
Query: 152 GRKDGMRSDAASVD-IPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDR 210
GR+D S A V+ +PS L F+S GLNVLD+ L GAHTIG+ C I R
Sbjct: 167 GRRDSKHSYARDVEKVPSGRRDVTALLETFQSYGLNVLDLVVLSGAHTIGKASCGTIQSR 226
Query: 211 LYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILN 270
LYNYN T SDPS++A + D +++ C + V L+P + + F YY +
Sbjct: 227 LYNYNATSGSDPSIDAKYADYLQRRC-----RWASETVDLDPVTPA--VFDNQYYINLQK 279
Query: 271 HEAVLGIDQQLLNGDDTKEITEEFA-AGLQDFKKSFAVSMYNMGNIKVLTGNQ--GQIRQ 327
H VL DQ+L+ T + + FA Q F++ FAVSM + N+ VLTG G+IR+
Sbjct: 280 HMGVLSTDQELVKDPRTAPLVKTFAEQSPQIFRQQFAVSMAKLVNVGVLTGEDRVGEIRK 339
Query: 328 NC 329
C
Sbjct: 340 VC 341
>At5g42180 peroxidase (emb|CAA66960.1)
Length = 317
Score = 202 bits (515), Expect = 2e-52
Identities = 122/330 (36%), Positives = 181/330 (53%), Gaps = 25/330 (7%)
Query: 4 SMCVAFLLVVAFAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDK 63
++ V + VV+F + SP HYY + C A+ V N VK D+
Sbjct: 7 NLLVIVIFVVSFDVQALSP-------------HYYD--HTCPQADHIVTNAVKKAMSNDQ 51
Query: 64 SITAKLLRLVYSDCFITGCDASILLD-EGPN-TEKKAPQNRGLGAFVLIDNIKTFVERQC 121
++ A LLR+ + DCF+ GCD S+LLD +G N EK P N L AF +IDN K +E QC
Sbjct: 52 TVPAALLRMHFHDCFVRGCDGSVLLDSKGKNKAEKDGPPNISLHAFYVIDNAKKALEEQC 111
Query: 122 PGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAASV-DIPSPSVSWQEALAYF 180
PG+VSCADIL LA RDAV L+GGP + V GRKDG S A +P+P+ + + F
Sbjct: 112 PGIVSCADILSLAARDAVALSGGPTWAVPKGRKDGRISKAIETRQLPAPTFNISQLRQNF 171
Query: 181 KSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRK 240
RGL++ D+ L G HT+G HCS +RL+ +N DP++N +F + +CP
Sbjct: 172 GQRGLSMHDLVALSGGHTLGFAHCSSFQNRLHKFNTQKEVDPTLNPSFAARLEGVCPAHN 231
Query: 241 KGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQD 300
++ N D G+ F YYK ++ +++ D+ LL TK++ ++A ++
Sbjct: 232 TVKN---AGSNMD-GTVTSFDNIYYKMLIQGKSLFSSDESLLAVPSTKKLVAKYANSNEE 287
Query: 301 FKKSFAVSMYNMGNIKVLTGNQGQIRQNCR 330
F+++F SM M +I +GN ++R NCR
Sbjct: 288 FERAFVKSMIKMSSI---SGNGNEVRLNCR 314
>At4g26010 peroxidase like protein
Length = 310
Score = 202 bits (515), Expect = 2e-52
Identities = 116/293 (39%), Positives = 165/293 (55%), Gaps = 17/293 (5%)
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPN--TEKKAPQN 101
C AE V + V ++ DKSITA LR+ + DCF+ GCDAS+L+D P +EK N
Sbjct: 31 CPRAESIVASVVANRFRSDKSITAAFLRMQFHDCFVRGCDASLLIDPRPGRPSEKSTGPN 90
Query: 102 RGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDA 161
+ + +ID K +E CP VSCADI+ LATRD+V LAGGP + V TGR+DG+RS+
Sbjct: 91 ASVRGYEIIDEAKRQLEAACPRTVSCADIVTLATRDSVALAGGPRFSVPTGRRDGLRSNP 150
Query: 162 ASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLL-GAHTIGRTHCSYITDRLYNYNGTGSS 220
V++P P++ ++ F ++G+N DM TL+ G H++G HCS DRL S
Sbjct: 151 NDVNLPGPTIPVSASIQLFAAQGMNTNDMVTLIGGGHSVGVAHCSLFQDRL--------S 202
Query: 221 DPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQ 280
D +M + ++R+ C +DP +L D +S+ + Y I +L IDQ
Sbjct: 203 DRAMEPSLKSSLRRKC----SSPNDPTTFL--DQKTSFTVDNAIYGEIRRQRGILRIDQN 256
Query: 281 LLNGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
L T I +A+ F+K FA ++ MG IKVLTG G+IR+NCR N
Sbjct: 257 LGLDRSTSGIVSGYASSNTLFRKRFAEALVKMGTIKVLTGRSGEIRRNCRVFN 309
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,802,537
Number of Sequences: 26719
Number of extensions: 341070
Number of successful extensions: 1025
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 84
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 685
Number of HSP's gapped (non-prelim): 90
length of query: 334
length of database: 11,318,596
effective HSP length: 100
effective length of query: 234
effective length of database: 8,646,696
effective search space: 2023326864
effective search space used: 2023326864
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0106.6


