

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
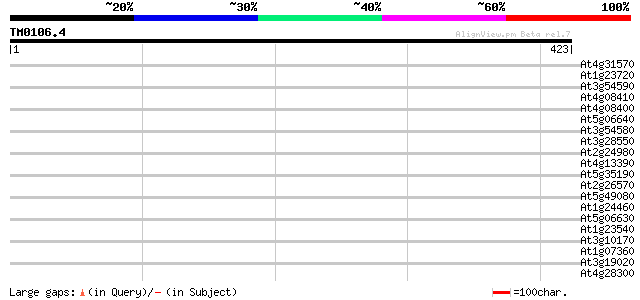
Query= TM0106.4
(423 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g31570 putative protein 41 0.001
At1g23720 unknown protein 40 0.002
At3g54590 extensin precursor -like protein 40 0.003
At4g08410 extensin-like protein 39 0.004
At4g08400 extensin-like protein 39 0.004
At5g06640 putative protein 39 0.007
At3g54580 extensin precursor -like protein 38 0.009
At3g28550 unknown protein 38 0.009
At2g24980 unknown protein 38 0.009
At4g13390 extensin-like protein 38 0.012
At5g35190 extensin -like protein 37 0.016
At2g26570 unknown protein 37 0.016
At5g49080 putative protein 37 0.021
At1g24460 unknown protein 37 0.021
At5g06630 putative protein 37 0.027
At1g23540 putative serine/threonine protein kinase 36 0.035
At3g10170 hypothetical protein 36 0.046
At1g07360 unknown protein 35 0.060
At3g19020 hypothetical protein 35 0.078
At4g28300 predicted proline-rich protein 34 0.13
>At4g31570 putative protein
Length = 2712
Score = 40.8 bits (94), Expect = 0.001
Identities = 49/193 (25%), Positives = 86/193 (44%), Gaps = 22/193 (11%)
Query: 219 AQSGGVPDDSRIEAALVKLKGLVFDAGFIGSVRVNPQLGQEVKELLAFLLSQQLDQIQAD 278
AQ+G +++A +++ + G + VR + + VKELLA Q
Sbjct: 2312 AQAGAKIFAEDLQSASARMRDMQDQLGIL--VRERDSMKERVKELLA-------GQASHS 2362
Query: 279 ALVELQTLVVDIVATLDKAI-AVEQSIETKKAQVASNTNGLLPAKEEVL-------KLTA 330
L E T + D++A D I A+ Q+++ +++Q+ + ++EV K A
Sbjct: 2363 ELQEKVTSLSDLLAAKDLEIEALMQALDEEESQMEDLKLRVTELEQEVQQKNLDLQKAEA 2422
Query: 331 RRTLIAAELSSVDARLDELH----REVARLEKQKAMLIDEGPAVNSRLNTLAAECTGVML 386
R I+ +LS + DELH +A +EK + + D V S L CT L
Sbjct: 2423 SRGKISKKLSITVDKFDELHHLSENLLAEIEKLQQQVQDRDTEV-SFLRQEVTRCTNEAL 2481
Query: 387 STSQLSKEVAAEQ 399
+ SQ+ + +E+
Sbjct: 2482 AASQMGTKRDSEE 2494
Score = 30.8 bits (68), Expect = 1.5
Identities = 24/74 (32%), Positives = 35/74 (46%), Gaps = 5/74 (6%)
Query: 315 TNGLLPAKEEVLKLTARRTLIAAELSSVDARLDELHREVARLEK----QKAMLIDEGPA- 369
T LL +E + + RT + EL ARLD+L E L +KA ++D G A
Sbjct: 732 TQELLTLQEHMSTVEEERTHLEVELREAIARLDKLAEENTSLTSSIMVEKARMVDNGSAD 791
Query: 370 VNSRLNTLAAECTG 383
V+ +N +E G
Sbjct: 792 VSGLINQEISEKLG 805
>At1g23720 unknown protein
Length = 895
Score = 40.4 bits (93), Expect = 0.002
Identities = 37/141 (26%), Positives = 51/141 (35%), Gaps = 25/141 (17%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + +NS P + P +P Y
Sbjct: 154 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYNSPPPPYYSPSPKPTYKSPP 208
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQ---PDQVIGVMPPPKADVLAS- 127
+ + PP + PSP P+ + P V PPP
Sbjct: 209 PPYIYSSPPPPYYS-PSPK---------------PVYKSPPPPYVYSSPPPPYYSPSPKP 252
Query: 128 AHSEPPPRQLQTSPQQPRLSP 148
A+ PPP + +SP P SP
Sbjct: 253 AYKSPPPPYVYSSPPPPYYSP 273
Score = 38.1 bits (87), Expect = 0.009
Identities = 37/141 (26%), Positives = 50/141 (35%), Gaps = 25/141 (17%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S P + +NS P + P +P Y
Sbjct: 254 YKSPPPPYVYSSPPPPYYSPSPKPIYKSPPP-----PYVYNSPPPPYYSPSPKPAYKSPP 308
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQ---PDQVIGVMPPPKADVLAS- 127
+ +FPP + PSP P+ + P V PPP
Sbjct: 309 PPYVYSFPPPPYYS-PSPK---------------PVYKSPPPPYVYNSPPPPYYSPSPKP 352
Query: 128 AHSEPPPRQLQTSPQQPRLSP 148
A+ PPP + +SP P SP
Sbjct: 353 AYKSPPPPYVYSSPPPPYYSP 373
Score = 37.0 bits (84), Expect = 0.021
Identities = 34/139 (24%), Positives = 50/139 (35%), Gaps = 18/139 (12%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P P + P PYY+
Sbjct: 757 YKSPPPPYVYSSPPPPYYSPSPKVEYKSPPP------------PYVYSSPPPPPYYSPSP 804
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPK--ADVLASAH 129
+ + PP + + P P + +V+ P P V PPP + +
Sbjct: 805 KVEYKSPPPPYVYSSPPPPTYYSP-SPKVEYKSPP---PPYVYNSPPPPAYYSPSPKIEY 860
Query: 130 SEPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 861 KSPPPPYVYSSPPPPSYSP 879
Score = 35.0 bits (79), Expect = 0.078
Identities = 34/140 (24%), Positives = 49/140 (34%), Gaps = 20/140 (14%)
Query: 7 PRATCFSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPY 66
P+ T + S PPP SS P S S K T+ P + P PY
Sbjct: 702 PKPT-YKSPPPPYVYSSPPPPYYSPSP------------KPTYKSPPPPYVYSSPPPPPY 748
Query: 67 YTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPII--QPDQVIGVMPPPKADV 124
Y+ + + PP + + P P E + P + P P PK +
Sbjct: 749 YSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPPYYSPSPKVE- 807
Query: 125 LASAHSEPPPRQLQTSPQQP 144
+ PPP + +SP P
Sbjct: 808 ----YKSPPPPYVYSSPPPP 823
Score = 32.3 bits (72), Expect = 0.51
Identities = 37/156 (23%), Positives = 53/156 (33%), Gaps = 21/156 (13%)
Query: 7 PRATCFSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPY 66
P+ T + S PPP SS P S S P + +NS P + P +P
Sbjct: 375 PKPT-YKSPPPPYVYSSPPPPYYSPSPKPVYKSPPP-----PYIYNSPPPPYYSPSPKPS 428
Query: 67 YTKECRDNLSNFPPQDMQNLPSP--AREENILHDEVQGGEDPIIQPDQVIGVMPPPKADV 124
Y + + PP + PSP + + P P + PP V
Sbjct: 429 YKSPPPPYVYSSPPPPYYS-PSPKLTYKSSPPPYVYSSPPPPYYSPSPKVVYKSPPPPYV 487
Query: 125 LAS------------AHSEPPPRQLQTSPQQPRLSP 148
+S ++ PPP + SP P SP
Sbjct: 488 YSSPPPPYYSPSPKPSYKSPPPPYVYNSPPPPYYSP 523
Score = 32.0 bits (71), Expect = 0.66
Identities = 34/154 (22%), Positives = 50/154 (32%), Gaps = 49/154 (31%)
Query: 7 PRATCFSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPY 66
P +SS PPP++ S SP +D P + ++S P PY
Sbjct: 32 PPPYVYSSPPPPLSYSPSP--------KVDYKSPPP-----PYVYSSPP--------PPY 70
Query: 67 YTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLA 126
Y+ + + PP + + P P P P + PP V +
Sbjct: 71 YSPSPKVEYKSPPPPYVYSSPPP----------------PYYSPSPKVDYKSPPPPYVYS 114
Query: 127 S------------AHSEPPPRQLQTSPQQPRLSP 148
S + PPP + SP P SP
Sbjct: 115 SPPPPYYSPSPKPTYKSPPPPYVYNSPPPPYYSP 148
Score = 31.2 bits (69), Expect = 1.1
Identities = 30/137 (21%), Positives = 45/137 (31%), Gaps = 17/137 (12%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP +S P S S K T+ P + P PYY+
Sbjct: 606 YKSPPPPYVYNSPPPPYYSPSP------------KPTYKSPPPPYVYSSPP-PPYYSPTP 652
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSE 131
+ + PP + + P P + P + PP + +
Sbjct: 653 KPTYKSPPPPYVYSSPPPPYYSPSPKPTYKSPPPPYVYSSPP----PPYYSPAPKPTYKS 708
Query: 132 PPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 709 PPPPYVYSSPPPPYYSP 725
Score = 30.8 bits (68), Expect = 1.5
Identities = 32/141 (22%), Positives = 49/141 (34%), Gaps = 25/141 (17%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP +S P S S P + ++S P + P +P Y
Sbjct: 329 YKSPPPPYVYNSPPPPYYSPSPKPAYKSPPP-----PYVYSSPPPPYYSPSPKPTYKSPP 383
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQ---PDQVIGVMPPPKADVLAS- 127
+ + PP + PSP P+ + P + PPP
Sbjct: 384 PPYVYSSPPPPYYS-PSPK---------------PVYKSPPPPYIYNSPPPPYYSPSPKP 427
Query: 128 AHSEPPPRQLQTSPQQPRLSP 148
++ PPP + +SP P SP
Sbjct: 428 SYKSPPPPYVYSSPPPPYYSP 448
Score = 29.6 bits (65), Expect = 3.3
Identities = 32/145 (22%), Positives = 48/145 (33%), Gaps = 18/145 (12%)
Query: 14 SKPPPIALSSSPSGLESSSSSLDSTESDPV------WDKLTFYFNSKPIAWQHPGCEPYY 67
+ PPP S SP + S P K+ + P + P PYY
Sbjct: 514 NSPPPPYYSPSPKVIYKSPPHPHVCVCPPPPPCYSHSPKIEYKSPPTPYVYHSPPPPPYY 573
Query: 68 TKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQ---PDQVIGVMPPP-KAD 123
+ + + PP + + P P P+ + P V PPP +
Sbjct: 574 SPSPKPAYKSSPPPYVYSSPPP--------PYYSPAPKPVYKSPPPPYVYNSPPPPYYSP 625
Query: 124 VLASAHSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 626 SPKPTYKSPPPPYVYSSPPPPYYSP 650
>At3g54590 extensin precursor -like protein
Length = 743
Score = 39.7 bits (91), Expect = 0.003
Identities = 35/138 (25%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + +NS P PYY+
Sbjct: 93 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYNSPP--------PPYYSPSP 139
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P E + P + P PK D +
Sbjct: 140 KVDYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVD-----YK 194
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 195 SPPPPYVYSSPPPPYYSP 212
Score = 37.0 bits (84), Expect = 0.021
Identities = 33/138 (23%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 268 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 314
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 315 KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPTYSPSPKVD-----YK 369
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 370 SPPPPYVYSSPPPPYYSP 387
Score = 37.0 bits (84), Expect = 0.021
Identities = 35/138 (25%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 293 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 339
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + + PP + + P P +V P P V PPP +
Sbjct: 340 KVDYKSPPPPYVYSSPPPPTYSP--SPKVDYKSPP---PPYVYSSPPPPYYSPSPKVEYK 394
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 395 SPPPPYVYSSPPPPTYSP 412
Score = 36.2 bits (82), Expect = 0.035
Identities = 33/138 (23%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P PYY+
Sbjct: 143 YKSPPPPYVYSSPPPPYYSPSPKVEYKSPPP-----PYVYSSPP--------PPYYSPSP 189
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P E + P + P PK D +
Sbjct: 190 KVDYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVD-----YK 244
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 245 SPPPPYVYSSPPPPYYSP 262
Score = 36.2 bits (82), Expect = 0.035
Identities = 36/138 (26%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 443 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 497
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP H
Sbjct: 498 PPYVYSSPPPPYYS-PSP---------KVYYKSPP---PPYVYSSPPPPYYSPSPKVHYK 544
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 545 SPPPPYVYSSPPPPYYSP 562
Score = 35.0 bits (79), Expect = 0.078
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P PYY+
Sbjct: 518 YKSPPPPYVYSSPPPPYYSPSPKVHYKSPPP-----PYVYSSPP--------PPYYSPSP 564
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + + PP + N P P +V P P V PPP +
Sbjct: 565 KVHYKSPPPPYVYNSPPPPYYSP--SPKVYYKSPP---PPYVYSSPPPPYYSPSPKVYYK 619
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 620 SPPPPYVYSSPPPPYYSP 637
Score = 33.9 bits (76), Expect = 0.17
Identities = 34/138 (24%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P P Y+
Sbjct: 318 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPTYSPSP 364
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + + PP + + P P +V+ P P V PPP +
Sbjct: 365 KVDYKSPPPPYVYSSPPPPYYSP--SPKVEYKSPP---PPYVYSSPPPPTYSPSPKVYYK 419
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 420 SPPPPYVYSSPPPPYYSP 437
Score = 33.9 bits (76), Expect = 0.17
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 418 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 472
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP +
Sbjct: 473 PPYVYSSPPPPYYS-PSP---------KVYYKSPP---PPYVYSSPPPPYYSPSPKVYYK 519
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 520 SPPPPYVYSSPPPPYYSP 537
Score = 32.7 bits (73), Expect = 0.39
Identities = 34/138 (24%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P PYY+
Sbjct: 493 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPP--------PPYYSPSP 539
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + + PP + + P P +V P P V PPP +
Sbjct: 540 KVHYKSPPPPYVYSSPPPPYYSP--SPKVHYKSPP---PPYVYNSPPPPYYSPSPKVYYK 594
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 595 SPPPPYVYSSPPPPYYSP 612
Score = 30.8 bits (68), Expect = 1.5
Identities = 33/143 (23%), Positives = 48/143 (33%), Gaps = 12/143 (8%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP +S P S S + P + ++S P + P + YY
Sbjct: 568 YKSPPPPYVYNSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 622
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQ---PDQVIGVMPPPKADVLAS- 127
+ + PP + PSP + P + V PPP S
Sbjct: 623 PPYVYSSPPPPYYS-PSPKVYYKSPPPPYYSPSPKVYYKSPPHPHVCVCPPPPPCYSPSP 681
Query: 128 --AHSEPPPRQLQTSPQQPRLSP 148
+ PPP + SP P SP
Sbjct: 682 KVVYKSPPPPYVYNSPPPPYYSP 704
>At4g08410 extensin-like protein
Length = 707
Score = 39.3 bits (90), Expect = 0.004
Identities = 38/148 (25%), Positives = 57/148 (37%), Gaps = 12/148 (8%)
Query: 7 PRATCFSSKPPPIALSSSPSGLESS--SSSLDSTESDPVWD---KLTFYFNSKPIAWQHP 61
P+ + +SS PPP S SP S S + S+ P + K+ + P + P
Sbjct: 76 PKPSIYSSSPPPSYYSPSPKVDYKSPPPSYVYSSPPPPYYSPSPKVDYKSLPPPYVYSSP 135
Query: 62 GCEPYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPK 121
PYY+ + N + PP + + P P + G+ P V PPP
Sbjct: 136 P-PPYYSPSPKVNYKSPPPPYVYSSPPPP-----YYSPSPKGDYKSPPPPYVYSSPPPPY 189
Query: 122 ADVLASA-HSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 190 YSPSPKVDYKSPPPPYVYSSPPPPYYSP 217
Score = 37.7 bits (86), Expect = 0.012
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP S P S S +D P + ++S P PYY+
Sbjct: 498 YKSPPPPYVYSFPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 544
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ N + PP + + P P E + P I P PK D +
Sbjct: 545 KVNYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYIYSSPPPPYYAPSPKVD-----YK 599
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 600 SPPPPYVYSSPPPPYYSP 617
Score = 36.6 bits (83), Expect = 0.027
Identities = 35/154 (22%), Positives = 52/154 (33%), Gaps = 42/154 (27%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCE------- 64
+ S PPP SS+P S S +D P + ++S P + P +
Sbjct: 373 YKSPPPPYVYSSTPPPYYSPSPKVDYKSPPP-----PYVYSSPPPPYYSPSPKVDYKSPP 427
Query: 65 ----------PYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVI 114
PYY+ + + + PP + + P P P P +
Sbjct: 428 PPYIYSSTPLPYYSPSPKVDYKSPPPPYVYSSPPP----------------PYYSPSPKV 471
Query: 115 GVMPPPKADVLASAHSEPPPRQLQTSPQQPRLSP 148
PPP V +S PPP SP+ SP
Sbjct: 472 DYKPPPPPYV----YSSPPPPYYSPSPKVDYKSP 501
Score = 36.2 bits (82), Expect = 0.035
Identities = 35/138 (25%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + + S P PYY+
Sbjct: 248 YKSPPPPYVYSSPPPPYYSPSPKVNYKSPPP-----PYVYGSPP--------PPYYSPSP 294
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + + PP + + P P +V P P V G PPP +
Sbjct: 295 KVDYKSPPPPYVYSSPPPPYYSP--SPKVNYKSPP---PPYVYGSPPPPYYSPSPKVDYK 349
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 350 SPPPPYVYSSPPPPYYSP 367
Score = 35.0 bits (79), Expect = 0.078
Identities = 32/138 (23%), Positives = 50/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P PYY+
Sbjct: 123 YKSLPPPYVYSSPPPPYYSPSPKVNYKSPPP-----PYVYSSPP--------PPYYSPSP 169
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 170 KGDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPTPKVD-----YK 224
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 225 SPPPPYVYSSPPPPYYSP 242
Score = 34.3 bits (77), Expect = 0.13
Identities = 34/138 (24%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P PYY
Sbjct: 548 YKSPPPPYVYSSPPPPYYSPSPKVEYKSPPP-----PYIYSSPP--------PPYYAPSP 594
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + + PP + + P P +V P P V PPP +
Sbjct: 595 KVDYKSPPPPYVYSSPPPPYYSP--SPKVDYKSPP---PPYVYSSPPPPYYSPSPKVNYK 649
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 650 SPPPPYVYSSPPPPYYSP 667
>At4g08400 extensin-like protein
Length = 513
Score = 39.3 bits (90), Expect = 0.004
Identities = 38/148 (25%), Positives = 57/148 (37%), Gaps = 12/148 (8%)
Query: 7 PRATCFSSKPPPIALSSSPSGLESS--SSSLDSTESDPVWD---KLTFYFNSKPIAWQHP 61
P+ + +SS PPP S SP S S + S+ P + K+ + P + P
Sbjct: 58 PKPSIYSSSPPPSYYSPSPKVDYKSPPPSYVYSSPPPPYYSPSPKVDYKSLPPPYVYSSP 117
Query: 62 GCEPYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPK 121
PYY+ + N + PP + + P P + G+ P V PPP
Sbjct: 118 P-PPYYSPSPKVNYKSPPPPYVYSSPPPP-----YYSPSPKGDYKSPPPPYVYSSPPPPY 171
Query: 122 ADVLASA-HSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 172 YSPSPKVDYKSPPPPYVYSSPPPPYYSP 199
Score = 38.9 bits (89), Expect = 0.005
Identities = 36/138 (26%), Positives = 50/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP +S P S S +D P + +NS P PYY+
Sbjct: 379 YKSPPPPYVYNSPPPPYYSPSPKVDYKSPPP-----PYIYNSPP--------PPYYSPSP 425
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ N PP + + P P +V P P V PPP + +
Sbjct: 426 KVNYKTPPPPYVYSSPPPPYYSP--SPKVNYKSPP---PPYVYSSPPPPYYSPSPNVDYK 480
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 481 SPPPPYVYSSPPTPYYSP 498
Score = 35.8 bits (81), Expect = 0.046
Identities = 36/157 (22%), Positives = 52/157 (32%), Gaps = 33/157 (21%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 205 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 251
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDE--------VQGGEDPIIQPDQVIGVMPPPKAD 123
+ N + PP ++ P P + D P P + PP
Sbjct: 252 KVNYKSPPPPYYRSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPY 311
Query: 124 VLASA------------HSEPPPRQLQTSPQQPRLSP 148
V +S + PPP + +SP P SP
Sbjct: 312 VYSSPPPPYYSPTPKVDYKSPPPPYVYSSPPPPYYSP 348
Score = 35.8 bits (81), Expect = 0.046
Identities = 33/137 (24%), Positives = 45/137 (32%), Gaps = 42/137 (30%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 329 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 375
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSE 131
+ + + PP + N P P P PK D +
Sbjct: 376 KVDYKSPPPPYVYNSPPPPYYS------------------------PSPKVD-----YKS 406
Query: 132 PPPRQLQTSPQQPRLSP 148
PPP + SP P SP
Sbjct: 407 PPPPYIYNSPPPPYYSP 423
Score = 35.0 bits (79), Expect = 0.078
Identities = 34/137 (24%), Positives = 48/137 (34%), Gaps = 18/137 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 155 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPTP 201
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSE 131
+ + + PP + + P P +V P P V PPP + +
Sbjct: 202 KVDYKSPPPPYVYSSPPPPYYSP--SPKVDYKSPP---PPYVYSSPPPPYYSPSPKVNYK 256
Query: 132 PPPRQLQTSPQQPRLSP 148
PP SP P SP
Sbjct: 257 SPPPPYYRSPPPPYYSP 273
>At5g06640 putative protein
Length = 689
Score = 38.5 bits (88), Expect = 0.007
Identities = 36/138 (26%), Positives = 51/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + +NS P PYY+
Sbjct: 200 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYNSPP--------PPYYSPSP 246
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + + PP + + P P +V+ P P V PPP +
Sbjct: 247 KVDYKSPPPPYVYSSPPPPYFSP--SPKVEYKSPP---PPYVYNSPPPPYYSPSPKVEYK 301
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 302 SPPPPYVYSSPPPPYYSP 319
Score = 37.4 bits (85), Expect = 0.016
Identities = 36/138 (26%), Positives = 51/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 325 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 371
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLAS-AHS 130
+ + + PP + + P P +V P P V PPP A+
Sbjct: 372 KVDYKSPPPPYVYSSPPPQYYSP--SPKVAYKSPP---PPYVYSSPPPPYYSPSPKVAYK 426
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 427 SPPPPYVYSSPPPPYYSP 444
Score = 36.6 bits (83), Expect = 0.027
Identities = 33/138 (23%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + +NS P PYY+
Sbjct: 150 YKSPPPPYVYSSPPPPYYSPSPKVEYKSPPP-----PYVYNSPP--------PPYYSPSP 196
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + PP + + P P + + P + P PK D +
Sbjct: 197 KIEYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYNSPPPPYYSPSPKVD-----YK 251
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 252 SPPPPYVYSSPPPPYFSP 269
Score = 34.7 bits (78), Expect = 0.10
Identities = 37/150 (24%), Positives = 54/150 (35%), Gaps = 18/150 (12%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLT----FYFNSKPIAWQHPGCE--- 64
+ S PPP SS P S S +D P + + +Y S +A++ P
Sbjct: 350 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPQYYSPSPKVAYKSPPPPYVY 409
Query: 65 -----PYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP 119
PYY+ + + PP + + P P +V P P V PP
Sbjct: 410 SSPPPPYYSPSPKVAYKSPPPPYVYSSPPPPYYSP--SPKVDYKSPP---PPYVYSSPPP 464
Query: 120 PKADVLASA-HSEPPPRQLQTSPQQPRLSP 148
P + PPP + +SP P SP
Sbjct: 465 PYYSPSPKVEYKSPPPPYVYSSPPPPYYSP 494
Score = 32.7 bits (73), Expect = 0.39
Identities = 31/137 (22%), Positives = 46/137 (32%), Gaps = 42/137 (30%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P PYY+
Sbjct: 500 YKSPPPPYVYSSPPPPYHSPSPKVNYKSPPP-----PYVYSSHP--------PPYYSPSP 546
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSE 131
+ N + PP + + P P P PK + +
Sbjct: 547 KVNYKSPPPPYVYSSPPPPYYS------------------------PSPKVN-----YKS 577
Query: 132 PPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 578 PPPPYVYSSPPPPYYSP 594
Score = 32.3 bits (72), Expect = 0.51
Identities = 24/96 (25%), Positives = 36/96 (37%), Gaps = 6/96 (6%)
Query: 54 KPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQV 113
KP + P YY+ + N + PP ++ N P P + + P +
Sbjct: 79 KPYVYISPPPPSYYSPSPKVNYKSPPPPNVYNSPPPPYYSPSPKVDYKSPPPPYVYSSPP 138
Query: 114 IG-VMPPPKADVLASAHSEPPPRQLQTSPQQPRLSP 148
P PK D + PPP + +SP P SP
Sbjct: 139 PPYYSPSPKVD-----YKSPPPPYVYSSPPPPYYSP 169
Score = 32.3 bits (72), Expect = 0.51
Identities = 34/158 (21%), Positives = 52/158 (32%), Gaps = 34/158 (21%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P PYY+
Sbjct: 525 YKSPPPPYVYSSHPPPYYSPSPKVNYKSPPP-----PYVYSSPP--------PPYYSPSP 571
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPII---------QPDQVIGVMPPPKA 122
+ N + PP + + P P + + P + P + PP
Sbjct: 572 KVNYKSPPPPYVYSSPPPPYYSPSPMVDYKSTPPPYVYSFPPLPYYSPSPKVDYKSPPLP 631
Query: 123 DVLASA------------HSEPPPRQLQTSPQQPRLSP 148
V +S + PPP + SP P SP
Sbjct: 632 YVYSSPPPLYYSPSPKVHYKSPPPPYVYNSPPPPYYSP 669
>At3g54580 extensin precursor -like protein
Length = 951
Score = 38.1 bits (87), Expect = 0.009
Identities = 35/149 (23%), Positives = 50/149 (33%), Gaps = 41/149 (27%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 826 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 872
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA--- 128
+ + + PP + + P P P P V+ PP V +S
Sbjct: 873 KVDYKSPPPPYVYSSPPP----------------PYYSPSPVVDYKSPPPPYVYSSPPPP 916
Query: 129 ---------HSEPPPRQLQTSPQQPRLSP 148
+ PPP + SP P SP
Sbjct: 917 YYSPSPKVEYKSPPPPYVYKSPPPPSYSP 945
Score = 38.1 bits (87), Expect = 0.009
Identities = 35/138 (25%), Positives = 51/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 268 YKSPPPPYVYSSPPPPTYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 314
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + + PP + + P P +V+ P P V PPP +
Sbjct: 315 KVDYKSPPPPYVYSSPPPPTYSP--SPKVEYKSPP---PPYVYSSPPPPTYSPSPKVEYK 369
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 370 SPPPPYVYSSPPPPTYSP 387
Score = 36.2 bits (82), Expect = 0.035
Identities = 35/138 (25%), Positives = 48/138 (34%), Gaps = 18/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 634 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 688
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
++ PP PSP +V P P V PPP +
Sbjct: 689 HPHVCVCPPPPPCYSPSP---------KVVYKSPP---PPYVYSSPPPPHYSPSPKVYYK 736
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 737 SPPPPYVYSSPPPPYYSP 754
Score = 34.7 bits (78), Expect = 0.10
Identities = 33/149 (22%), Positives = 50/149 (33%), Gaps = 41/149 (27%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P PYY+
Sbjct: 801 YKSPPPPYVYSSPPPPYYSPSPKVEYKSPPP-----PYVYSSPP--------PPYYSPSP 847
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA--- 128
+ + + PP + + P P P P + PP V +S
Sbjct: 848 KVDYKSPPPPYVYSSPPP----------------PYYSPSPKVDYKSPPPPYVYSSPPPP 891
Query: 129 ---------HSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 892 YYSPSPVVDYKSPPPPYVYSSPPPPYYSP 920
Score = 34.3 bits (77), Expect = 0.13
Identities = 34/140 (24%), Positives = 50/140 (35%), Gaps = 22/140 (15%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 609 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 663
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLAS---A 128
+ + PP + PSP + + P + V PPP S
Sbjct: 664 PPYVYSSPPPPYYS-PSP----KVYYKS---------PPHPHVCVCPPPPPCYSPSPKVV 709
Query: 129 HSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 710 YKSPPPPYVYSSPPPPHYSP 729
Score = 33.9 bits (76), Expect = 0.17
Identities = 32/141 (22%), Positives = 49/141 (34%), Gaps = 19/141 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P PYY+
Sbjct: 776 YKSPPPPYVYSSPPPPYYSPSPKVHYKSPPP-----PYVYSSPP--------PPYYSPSP 822
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + PP + + P P + + P + P PK D +
Sbjct: 823 KVEYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVD-----YK 877
Query: 131 EPPPRQLQTSPQQPRLSPQRI 151
PPP + +SP P SP +
Sbjct: 878 SPPPPYVYSSPPPPYYSPSPV 898
Score = 33.1 bits (74), Expect = 0.30
Identities = 34/141 (24%), Positives = 49/141 (34%), Gaps = 9/141 (6%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVW---DKLTFYFNSKPIAWQHPGCEPYYT 68
+ S PPP SS P S S + P + Y++ P + PYY+
Sbjct: 493 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYYS 552
Query: 69 KECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA 128
+ + PP + + P P +V P P V PPP
Sbjct: 553 PSPKVYYKSPPPPYVYSSPPPPYYSP--SPKVYYKSPP---PPYVYSSPPPPYHSPSPKV 607
Query: 129 -HSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 608 QYKSPPPPYVYSSPPPPYYSP 628
Score = 32.7 bits (73), Expect = 0.39
Identities = 34/138 (24%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P P + Y
Sbjct: 68 YKSPPPPYVYSSPPPPTYSPSPKVEYKSPPP-----PYVYSSPPPPTYSPSPKVEYKSPP 122
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + PP + PSP +V+ P P V PPP +
Sbjct: 123 PPYVYSSPPPPTYS-PSP---------KVEYKSPP---PPYVYSSPPPPTYSPSPKVEYK 169
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 170 SPPPPYVYSSPPPPTYSP 187
Score = 32.7 bits (73), Expect = 0.39
Identities = 32/138 (23%), Positives = 48/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P P Y+
Sbjct: 143 YKSPPPPYVYSSPPPPTYSPSPKVEYKSPPP-----PYVYSSPP--------PPTYSPSP 189
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + PP + + P P E + P + P PK D +
Sbjct: 190 KVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVD-----YK 244
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 245 SPPPPYVYSSPPPPYYSP 262
Score = 32.7 bits (73), Expect = 0.39
Identities = 32/138 (23%), Positives = 48/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P P Y+
Sbjct: 368 YKSPPPPYVYSSPPPPTYSPSPKVEYKSPPP-----PYVYSSPP--------PPTYSPSP 414
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + PP + + P P E + P + P PK D +
Sbjct: 415 KVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVD-----YK 469
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 470 SPPPPYVYSSPPPPYYSP 487
Score = 32.3 bits (72), Expect = 0.51
Identities = 34/138 (24%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P P + Y
Sbjct: 343 YKSPPPPYVYSSPPPPTYSPSPKVEYKSPPP-----PYVYSSPPPPTYSPSPKVEYKSPP 397
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + PP + PSP +V+ P P V PPP +
Sbjct: 398 PPYVYSSPPPPTYS-PSP---------KVEYKSPP---PPYVYSSPPPPYYSPSPKVEYK 444
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 445 SPPPPYVYSSPPPPTYSP 462
Score = 32.3 bits (72), Expect = 0.51
Identities = 34/138 (24%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P P + Y
Sbjct: 93 YKSPPPPYVYSSPPPPTYSPSPKVEYKSPPP-----PYVYSSPPPPTYSPSPKVEYKSPP 147
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + PP + PSP +V+ P P V PPP +
Sbjct: 148 PPYVYSSPPPPTYS-PSP---------KVEYKSPP---PPYVYSSPPPPTYSPSPKVEYK 194
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 195 SPPPPYVYSSPPPPYYSP 212
Score = 32.3 bits (72), Expect = 0.51
Identities = 34/138 (24%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S ++ P + ++S P P + Y
Sbjct: 318 YKSPPPPYVYSSPPPPTYSPSPKVEYKSPPP-----PYVYSSPPPPTYSPSPKVEYKSPP 372
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + PP + PSP +V+ P P V PPP +
Sbjct: 373 PPYVYSSPPPPTYS-PSP---------KVEYKSPP---PPYVYSSPPPPTYSPSPKVEYK 419
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 420 SPPPPYVYSSPPPPYYSP 437
>At3g28550 unknown protein
Length = 1018
Score = 38.1 bits (87), Expect = 0.009
Identities = 36/138 (26%), Positives = 50/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P + S S +D P + ++S P + P + Y
Sbjct: 110 YKSPPPPYVYSSPPPPIYSPSPKVDYKSPPP-----PYVYSSPPPPYYSPSPKVEYKSPP 164
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ N PP + PSP +V P P V PPP +
Sbjct: 165 SPYVYNSPPPSYYS-PSP---------KVDYKSPP---PPYVYSSPPPPYYSPSPKVVYK 211
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 212 SPPPPYVYSSPPPPYYSP 229
Score = 37.4 bits (85), Expect = 0.016
Identities = 33/137 (24%), Positives = 49/137 (35%), Gaps = 26/137 (18%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 902 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPAP 948
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSE 131
+ + + PP + + P P + + P P PK D +
Sbjct: 949 KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYYSPS--------PKVD-----YKS 995
Query: 132 PPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 996 PPPPYVYSSPPPPSYSP 1012
Score = 35.4 bits (80), Expect = 0.060
Identities = 33/138 (23%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 260 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 306
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + PP + + P P + + P + P PK D +
Sbjct: 307 KVVYKSPPPPYVYSSPPPPYYSPTPKVDYKSPPPPYVYSSPPPPYYSPSPKVD-----YK 361
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 362 SPPPPYVYSSPPPPYYSP 379
Score = 35.0 bits (79), Expect = 0.078
Identities = 34/149 (22%), Positives = 49/149 (32%), Gaps = 41/149 (27%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 210 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 256
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLAS---- 127
+ + PP + + P P P P + PP V +S
Sbjct: 257 KIVYKSPPPPYVYSSPPP----------------PYYSPSPKVDYKSPPPPYVYSSPPPP 300
Query: 128 --------AHSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 301 YYSPSPKVVYKSPPPPYVYSSPPPPYYSP 329
Score = 34.7 bits (78), Expect = 0.10
Identities = 35/154 (22%), Positives = 53/154 (33%), Gaps = 26/154 (16%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVW-------------DKLTFYFNSKPIAW 58
+ S PPP SS P S S +D P + K+ + P +
Sbjct: 410 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVY 469
Query: 59 QHPGCEPYYTKECRDNLSNFPPQDMQNLPSP---AREENILHDEVQGGEDPIIQPDQVIG 115
P PYY+ + + + PP + + P P + +L+ P V
Sbjct: 470 SSPP-PPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVLYKSPP--------PPYVYS 520
Query: 116 VMPPPKADVLASA-HSEPPPRQLQTSPQQPRLSP 148
PPP + PPP + +SP P SP
Sbjct: 521 SPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSP 554
Score = 34.3 bits (77), Expect = 0.13
Identities = 33/149 (22%), Positives = 49/149 (32%), Gaps = 41/149 (27%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P PYY+
Sbjct: 652 YKSSPPPYVYSSPPPPYHSPSPKVHYKSPPP-----PYVYSSPP--------PPYYSPSP 698
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLAS---- 127
+ + + PP + + P P P P + PP V +S
Sbjct: 699 KVHYKSPPPPYVYSSPPP----------------PYYSPSPKVVYKSPPPPYVYSSPPPP 742
Query: 128 --------AHSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 743 YYSPSPKVVYKSPPPPYVYSSPPPPYYSP 771
Score = 33.9 bits (76), Expect = 0.17
Identities = 34/142 (23%), Positives = 51/142 (34%), Gaps = 25/142 (17%)
Query: 7 PRATCFSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPY 66
P +SS PPP+ S +P ++ S P K+ + P + P PY
Sbjct: 47 PLPDVYSSPPPPLEYSPAPK-VDYKSPPPPYYSPSP---KVEYKSPPPPYVYNSPP-PPY 101
Query: 67 YTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLA 126
Y+ + + + PP + + P P PI P + PP V
Sbjct: 102 YSPSPKVDYKSPPPPYVYSSPPP----------------PIYSPSPKVDYKSPPPPYV-- 143
Query: 127 SAHSEPPPRQLQTSPQQPRLSP 148
+S PPP SP+ SP
Sbjct: 144 --YSSPPPPYYSPSPKVEYKSP 163
Score = 32.3 bits (72), Expect = 0.51
Identities = 35/162 (21%), Positives = 51/162 (30%), Gaps = 42/162 (25%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVW-------------DKLTFYFNSKPIAW 58
+ S PPP SS P S S +D P + K+ + P +
Sbjct: 460 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVLYKSPPPPYVY 519
Query: 59 QHPGCEPYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMP 118
P PYY+ + + PP + + P P P P +
Sbjct: 520 SSPP-PPYYSPSPKVVYKSPPPPYVYSSPPP----------------PYYSPSPKVVYKS 562
Query: 119 PPKADVLAS------------AHSEPPPRQLQTSPQQPRLSP 148
PP V +S + PPP + +SP P SP
Sbjct: 563 PPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPYYSP 604
Score = 30.8 bits (68), Expect = 1.5
Identities = 33/149 (22%), Positives = 48/149 (32%), Gaps = 41/149 (27%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P PYY+
Sbjct: 752 YKSPPPPYVYSSPPPPYYSPSPKVVYKSPPP-----PYVYSSPP--------PPYYSPSP 798
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLAS---- 127
+ + PP + + P P P P + PP V +S
Sbjct: 799 KVVYKSPPPPYVYSSPPP----------------PYYSPSPKVVYKSPPPPYVYSSPPPP 842
Query: 128 --------AHSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 843 YYSPSPKVVYKSPPPPYVYSSPPPPYYSP 871
Score = 30.8 bits (68), Expect = 1.5
Identities = 33/149 (22%), Positives = 48/149 (32%), Gaps = 41/149 (27%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P PYY+
Sbjct: 702 YKSPPPPYVYSSPPPPYYSPSPKVVYKSPPP-----PYVYSSPP--------PPYYSPSP 748
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLAS---- 127
+ + PP + + P P P P + PP V +S
Sbjct: 749 KVVYKSPPPPYVYSSPPP----------------PYYSPSPKVVYKSPPPPYVYSSPPPP 792
Query: 128 --------AHSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 793 YYSPSPKVVYKSPPPPYVYSSPPPPYYSP 821
Score = 30.4 bits (67), Expect = 1.9
Identities = 34/138 (24%), Positives = 48/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + Y
Sbjct: 777 YKSPPPPYVYSSPPPPYYSPSPKVVYKSPPP-----PYVYSSPPPPYYSPSPKVVYKSPP 831
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA-HS 130
+ + PP + PSP +V P P V PPP +
Sbjct: 832 PPYVYSSPPPPYYS-PSP---------KVVYKSPP---PPYVYSSPPPPYYSPSPKVDYK 878
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 879 SPPPPYVYSSPPPPYYSP 896
>At2g24980 unknown protein
Length = 559
Score = 38.1 bits (87), Expect = 0.009
Identities = 36/138 (26%), Positives = 50/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 95 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 141
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + + PP + N P P +V P P V PPP +
Sbjct: 142 KVDYKSPPPPYVYNSPPPPYYSP--SPKVDYKSPP---PPYVYSSPPPPYYSPSPKVYYK 196
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 197 SPPPPYVYSSPPPPYYSP 214
Score = 36.2 bits (82), Expect = 0.035
Identities = 36/138 (26%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + +NS P + P + YY
Sbjct: 370 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYNSPPPPYYSPSPKVYYKSPP 424
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP +
Sbjct: 425 PPYVYSSPPPPYYS-PSP---------KVYYKSPP---PPYVYSSPPPPYYSPSPKVYYK 471
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 472 SPPPSYVYSSPPPPYYSP 489
Score = 36.2 bits (82), Expect = 0.035
Identities = 33/138 (23%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 70 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 116
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 117 KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYNSPPPPYYSPSPKVD-----YK 171
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 172 SPPPPYVYSSPPPPYYSP 189
Score = 34.7 bits (78), Expect = 0.10
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 420 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 474
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP +
Sbjct: 475 PSYVYSSPPPPYYS-PSP---------KVYYKSPP---PSYVYSSPPPPYYSPSPKVYYK 521
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 522 SPPPPYVYSSPPPPYYSP 539
Score = 33.9 bits (76), Expect = 0.17
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 245 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 299
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP +
Sbjct: 300 PPYVYSSPPPPYYS-PSP---------KVYYKSPP---PPYVYSSPPPPYYSPSPKVYYK 346
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 347 SPPPPYVYSSPPPPYYSP 364
Score = 33.9 bits (76), Expect = 0.17
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 220 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 274
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP +
Sbjct: 275 PPYVYSSPPPPYYS-PSP---------KVYYKSPP---PPYVYSSPPPPYYSPSPKVYYK 321
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 322 SPPPPYVYSSPPPPYYSP 339
Score = 33.5 bits (75), Expect = 0.23
Identities = 34/138 (24%), Positives = 49/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP +S P S S + P + ++S P + P + YY
Sbjct: 395 YKSPPPPYVYNSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 449
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP +
Sbjct: 450 PPYVYSSPPPPYYS-PSP---------KVYYKSPP---PSYVYSSPPPPYYSPSPKVYYK 496
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 497 SPPPSYVYSSPPPPYYSP 514
>At4g13390 extensin-like protein
Length = 429
Score = 37.7 bits (86), Expect = 0.012
Identities = 35/150 (23%), Positives = 49/150 (32%), Gaps = 39/150 (26%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
++S PPP SS P S S +S P P + P PYY+
Sbjct: 147 YNSPPPPYIYSSPPPPPYYSPSPKVDYKSPP-----------PPYVYSSPPPPPYYSPSP 195
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASA--- 128
+ + PP + + P P P P +G PP V +S
Sbjct: 196 KVEYKSPPPPYVYSFPPP---------------PPYYSPSPKVGYKSPPAPYVYSSPPPP 240
Query: 129 ----------HSEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 241 PYYSPSPKVNYKSPPPPYVYSSPPPPPYSP 270
Score = 37.0 bits (84), Expect = 0.021
Identities = 36/139 (25%), Positives = 48/139 (33%), Gaps = 24/139 (17%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNS--KPIAWQHPGCEPYYTK 69
+ S PPP SS P S S +S P + +NS P + P PYY+
Sbjct: 302 YKSPPPPYVYSSPPPPTYYSPSPRVDYKSPPP----PYVYNSLPPPYVYNSPPPPPYYSP 357
Query: 70 ECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH 129
N + PP + N P P P P + PP + +
Sbjct: 358 SPTVNYKSPPPPYVYNSPPP---------------PPYYSPFPKVEYKSPPPPYIY---N 399
Query: 130 SEPPPRQLQTSPQQPRLSP 148
S PPP SP+ SP
Sbjct: 400 SPPPPPYYSPSPKITYKSP 418
Score = 36.2 bits (82), Expect = 0.035
Identities = 34/135 (25%), Positives = 46/135 (33%), Gaps = 18/135 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP S P S S +S P P + P PYY+
Sbjct: 199 YKSPPPPYVYSFPPPPPYYSPSPKVGYKSPPA-----------PYVYSSPPPPPYYSPSP 247
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPII--QPDQVIGVMPPPKADVLASAH 129
+ N + PP + + P P E + P I P P PK D +
Sbjct: 248 KVNYKSPPPPYVYSSPPPPPYSPSPKVEFKSPPPPYIYNSPPPPSYYSPSPKID-----Y 302
Query: 130 SEPPPRQLQTSPQQP 144
PPP + +SP P
Sbjct: 303 KSPPPPYVYSSPPPP 317
>At5g35190 extensin -like protein
Length = 328
Score = 37.4 bits (85), Expect = 0.016
Identities = 34/137 (24%), Positives = 45/137 (32%), Gaps = 42/137 (30%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP +S P S S +D P + +NS P PYY+
Sbjct: 145 YKSPPPPYVYNSPPPPYYSLSPKVDYKSPPP-----PYVYNSPP--------PPYYSPSP 191
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSE 131
+ + PP + N PSP P PK D +
Sbjct: 192 KVDYKFSPPPYVYNSPSPPYYS------------------------PSPKVD-----YKS 222
Query: 132 PPPRQLQTSPQQPRLSP 148
PPP + SP P SP
Sbjct: 223 PPPPYVYNSPPPPYFSP 239
Score = 35.0 bits (79), Expect = 0.078
Identities = 21/83 (25%), Positives = 35/83 (41%), Gaps = 5/83 (6%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLT-----FYFNSKPIAWQHPGCEPY 66
+ S PPP +S P S S +D P + + +Y S ++++ P PY
Sbjct: 220 YKSPPPPYVYNSPPPPYFSPSPKVDYKSPPPPYVYSSPPPPPYYSPSPEVSYKSPPPPPY 279
Query: 67 YTKECRDNLSNFPPQDMQNLPSP 89
Y+ + + PP + N P P
Sbjct: 280 YSPSLEVSYKSPPPLFVYNFPPP 302
Score = 29.3 bits (64), Expect = 4.3
Identities = 24/100 (24%), Positives = 35/100 (35%), Gaps = 28/100 (28%)
Query: 49 FYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPII 108
+ FNS P PYY+ +++ + PP + N P P P
Sbjct: 77 YLFNSPP--------PPYYSPSPKEDYKSPPPPYVYNSPPP----------------PYY 112
Query: 109 QPDQVIGVMPPPKADVLASAHSEPPPRQLQTSPQQPRLSP 148
P + PP V ++ PPP SP+ SP
Sbjct: 113 SPSPKVDYKSPPPPYV----YNSPPPPYYSPSPKVEYKSP 148
>At2g26570 unknown protein
Length = 807
Score = 37.4 bits (85), Expect = 0.016
Identities = 54/242 (22%), Positives = 98/242 (40%), Gaps = 31/242 (12%)
Query: 164 AKGGSASSKTAASSHTSSERLSALLVEDPLSIFQSFFDGTLDLESPPRQAEIAETAQSGG 223
A+ G S +T +S S +S ++ S G +D +P + A ++ GG
Sbjct: 130 ARNGGGSPRTVSSPRFSGSPVSTGTPKNVDS-----HRGLIDTAAPFESVKEA-VSKFGG 183
Query: 224 VPDDSRIEAALVKLKGLVFDAGFIGSVRVNPQLGQEVKELLAFLLSQQLDQIQADALVEL 283
+ D V+ + L+ + ++ +E+ E S+ + + L EL
Sbjct: 184 ITDWKSHRMQAVERRKLIEEE--------LKKIHEEIPEYKTH--SETAEAAKLQVLKEL 233
Query: 284 QT---LVVDIVATLDKAIAVEQSI----ETKKAQVASNTNGL-----LPAKEEVLKLTAR 331
++ L+ + LDKA EQ E K +V G+ + AK ++ AR
Sbjct: 234 ESTKRLIEQLKLNLDKAQTEEQQAKQDSELAKLRVEEMEQGIAEDVSVAAKAQLEVAKAR 293
Query: 332 RTLIAAELSSVDARLDELHREVARLEKQKAML---IDEGPAVNSRLNTLAAECTGVMLST 388
T ELSSV L+ LH+E L + K + ++E + + E T +++T
Sbjct: 294 HTTAITELSSVKEELETLHKEYDALVQDKDVAVKKVEEAMLASKEVEKTVEELTIELIAT 353
Query: 389 SQ 390
+
Sbjct: 354 KE 355
>At5g49080 putative protein
Length = 609
Score = 37.0 bits (84), Expect = 0.021
Identities = 33/138 (23%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 295 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 341
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 342 KVDYKSPPPPYVYSSPPPPYYSPTPKVDYKSPPPPYVYSSPPPPYYSPSPKVD-----YK 396
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 397 SPPPPYVYSSPPPPYYSP 414
Score = 36.2 bits (82), Expect = 0.035
Identities = 33/138 (23%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 320 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPTP 366
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 367 KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVD-----YK 421
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 422 SPPPPYVYSSPPPPYYSP 439
Score = 36.2 bits (82), Expect = 0.035
Identities = 33/138 (23%), Positives = 50/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S +D P + ++S P PYY+
Sbjct: 470 YKSPPPPYVYSSPPPPYYSPSPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 516
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 517 KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYNSPPPPYYSPSPKVD-----YK 571
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 572 SPPPPYVYSSPPPPYYSP 589
Score = 35.4 bits (80), Expect = 0.060
Identities = 32/138 (23%), Positives = 50/138 (36%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S + +D P + ++S P PYY+
Sbjct: 145 YKSPPPPYVYSSPPPPYYSPTPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 191
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 192 KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPTPKVD-----YK 246
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 247 SPPPPYVYSSPPPPYYSP 264
Score = 35.0 bits (79), Expect = 0.078
Identities = 32/138 (23%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S + +D P + ++S P PYY+
Sbjct: 345 YKSPPPPYVYSSPPPPYYSPTPKVDYKSPPP-----PYVYSSPP--------PPYYSPSP 391
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPP-PKADVLASAHS 130
+ + + PP + + P P + + P + P PK D +
Sbjct: 392 KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVD-----YK 446
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + SP P SP
Sbjct: 447 SPPPPYVYNSPPPPYYSP 464
>At1g24460 unknown protein
Length = 1791
Score = 37.0 bits (84), Expect = 0.021
Identities = 33/138 (23%), Positives = 63/138 (44%), Gaps = 17/138 (12%)
Query: 268 LSQQLDQIQA---DALVELQTLVVD---IVATLDKAIAVEQSIETKKAQVASNTNGLLPA 321
L+++ +QA +A +ELQ V D + + LD+ +A + ++E Q N + ++
Sbjct: 897 LTEENRNVQAAKENAELELQKAVADASSVASELDEVLATKSTLEAALMQAERNISDIISE 956
Query: 322 KEEVLKLTA----RRTLIAAELSSVDARLDELHREVARLEKQKAMLIDEGPAVNSRLNTL 377
KEE TA + ++ E S +L E H + LE+ A S +++L
Sbjct: 957 KEEAQGRTATAEMEQEMLQKEASIQKNKLTEAHSTINSLEETLAQ-------TESNMDSL 1009
Query: 378 AAECTGVMLSTSQLSKEV 395
+ + + T+ L E+
Sbjct: 1010 SKQIEDDKVLTTSLKNEL 1027
>At5g06630 putative protein
Length = 434
Score = 36.6 bits (83), Expect = 0.027
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 270 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 324
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP N+ + P V PPP H
Sbjct: 325 PPYVYSSPPPPYYS-PSP----NVYYKSPP--------PPYVYSSPPPPYYSPSPKVHYK 371
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 372 SPPPPYVYSSPPPPYYSP 389
Score = 36.2 bits (82), Expect = 0.035
Identities = 36/138 (26%), Positives = 48/138 (34%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P YY
Sbjct: 295 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPNVYYKSPP 349
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP H
Sbjct: 350 PPYVYSSPPPPYYS-PSP---------KVHYKSPP---PPYVYSSPPPPYYSPSPKVHYK 396
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 397 SPPPPYVYSSPPPPYYSP 414
Score = 33.9 bits (76), Expect = 0.17
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 19/138 (13%)
Query: 12 FSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKEC 71
+ S PPP SS P S S + P + ++S P + P + YY
Sbjct: 170 YKSPPPPYVYSSPPPPYYSPSPKVYYKSPPP-----PYVYSSPPPPYYSPSPKVYYKSPP 224
Query: 72 RDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAH-S 130
+ + PP + PSP +V P P V PPP +
Sbjct: 225 PPYVYSSPPPPYYS-PSP---------KVYYKSPP---PPYVYSSPPPPYYSPSPKVYYK 271
Query: 131 EPPPRQLQTSPQQPRLSP 148
PPP + +SP P SP
Sbjct: 272 SPPPPYVYSSPPPPYYSP 289
Score = 33.5 bits (75), Expect = 0.23
Identities = 37/143 (25%), Positives = 52/143 (35%), Gaps = 18/143 (12%)
Query: 7 PRATCFSSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPY 66
P + PPP + SSP L S S +S P + ++S P + P + Y
Sbjct: 114 PSPKVYYKSPPPPYVYSSPPPLYYSPSPKVYYKSPPP----PYVYSSPPPPYYSPSPKVY 169
Query: 67 YTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLA 126
Y + + PP + PSP +V P P V PPP
Sbjct: 170 YKSPPPPYVYSSPPPPYYS-PSP---------KVYYKSPP---PPYVYSSPPPPYYSPSP 216
Query: 127 SAH-SEPPPRQLQTSPQQPRLSP 148
+ PPP + +SP P SP
Sbjct: 217 KVYYKSPPPPYVYSSPPPPYYSP 239
Score = 30.4 bits (67), Expect = 1.9
Identities = 26/95 (27%), Positives = 33/95 (34%), Gaps = 21/95 (22%)
Query: 54 KPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQV 113
KP + P PYYT + N + PP + N P P P P
Sbjct: 50 KPYVYSSPP-PPYYTPSPKVNYKSPPPPYVYNSPPP----------------PYYSPSPK 92
Query: 114 IGVMPPPKADVLASAHSEPPPRQLQTSPQQPRLSP 148
+ PP V +S PPP SP+ SP
Sbjct: 93 VYYKSPPPPYV----YSSPPPPYYSPSPKVYYKSP 123
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 36.2 bits (82), Expect = 0.035
Identities = 35/134 (26%), Positives = 50/134 (37%), Gaps = 18/134 (13%)
Query: 13 SSKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECR 72
SS P P A SSS L S+ ++ P+ L +S P P +
Sbjct: 37 SSPPSPPADSSSTPPLSEPSTPPPDSQLPPLPSILPPLTDSPP--------PPSDSSPPV 88
Query: 73 DNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSEP 132
D+ + PP PSP + E P P++ PPP D+ + S P
Sbjct: 89 DSTPSPPPPTSNESPSPPEDS----------ETPPAPPNESNDNNPPPSQDLQSPPPSSP 138
Query: 133 PPRQLQTSPQQPRL 146
P T+P+ P L
Sbjct: 139 SPNVGPTNPESPPL 152
>At3g10170 hypothetical protein
Length = 634
Score = 35.8 bits (81), Expect = 0.046
Identities = 48/182 (26%), Positives = 80/182 (43%), Gaps = 26/182 (14%)
Query: 255 QLGQEVK--ELLAFLLSQQLDQIQADALVELQTLVVDIVATLDKAIAVEQSIETKKAQVA 312
+L QE + E +LS+ + ++D L ++ L DI + ++A E+ K +
Sbjct: 167 ELNQEAQKHETSLKMLSEHHESERSDLLSHIECLEKDIGSLSSSSLAKEKENLRKDFE-- 224
Query: 313 SNTNGLLPAKEEVLKLTAR-RTLIAAELSSVDARLDELHREVARLE-----------KQK 360
T L E LK + + +T + AE +S + L LH + A LE K++
Sbjct: 225 -KTKTKLKDTESKLKNSMQDKTKLEAEKASAERELKRLHSQKALLERDISKQESFAGKRR 283
Query: 361 AMLIDEGPAVNS------RLNTLAAECTGVMLSTSQLSKEVAAEQRMKQSLDCRIAAAGV 414
L+ E A S +L LA E + S L +E+AAE+ K+ CR G
Sbjct: 284 DSLLVERSANQSLQEEFKQLEVLAFEMETTIAS---LEEELAAERGEKEEALCRNDGLGS 340
Query: 415 QL 416
++
Sbjct: 341 EI 342
>At1g07360 unknown protein
Length = 481
Score = 35.4 bits (80), Expect = 0.060
Identities = 23/67 (34%), Positives = 33/67 (48%), Gaps = 15/67 (22%)
Query: 120 PKADVLASAHSEPPPRQL---------QTSPQQPRLSPQRIQQDARAALISPHAKGGSAS 170
P+A V++ H++PPP Q Q P P + PQR+ A+IS GGS++
Sbjct: 321 PRA-VISQQHNQPPPMQQYYMHPPPANQDKPYYPSMDPQRM-----GAVISTQEAGGSST 374
Query: 171 SKTAASS 177
ASS
Sbjct: 375 ENNGASS 381
>At3g19020 hypothetical protein
Length = 951
Score = 35.0 bits (79), Expect = 0.078
Identities = 54/228 (23%), Positives = 76/228 (32%), Gaps = 27/228 (11%)
Query: 14 SKPPPIALSSSPSGLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECRD 73
S PPP+ S P + S + S PV +S P Q P P ++
Sbjct: 693 SPPPPV--HSPPPPVHSPPPPVHSPPP-PV--------HSPPPPVQSPPPPPVFSPPPPA 741
Query: 74 NLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSEPP 133
+ + PP + + P P V P+ P + PPP HS PP
Sbjct: 742 PIYSPPPPPVHSPPPPVHSPP--PPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPP 799
Query: 134 PRQLQTSPQQPRLSPQRIQQDARAALISPHAKGGSASSKTAASSHT-------SSERLSA 186
P + SP P SP SP A + SS + T SE + A
Sbjct: 800 PSPIY-SPPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSEDIEA 858
Query: 187 LLVEDPLSIFQSFFDGTLDLES------PPRQAEIAETAQSGGVPDDS 228
+ +F+S + D E P + E+ QS P S
Sbjct: 859 PSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEPEVEAPKQSEATPSSS 906
Score = 28.5 bits (62), Expect = 7.3
Identities = 25/98 (25%), Positives = 33/98 (33%), Gaps = 12/98 (12%)
Query: 51 FNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNLPSPAREENILHDEVQGGEDPIIQP 110
+++ PI + P T+E + PP P P V P+ P
Sbjct: 609 YDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPP----------VHSPPPPVFSP 658
Query: 111 DQVIGVMPPPKADVLASAHSEPPPRQLQTSPQQPRLSP 148
+ PPP HS PPP SP P SP
Sbjct: 659 PPPMHSPPPPVYSPPPPVHSPPPPP--VHSPPPPVHSP 694
>At4g28300 predicted proline-rich protein
Length = 496
Score = 34.3 bits (77), Expect = 0.13
Identities = 35/146 (23%), Positives = 59/146 (39%), Gaps = 17/146 (11%)
Query: 79 PPQDMQNLPSPAREENILHDEVQGGEDPIIQPDQVIGVMPPPKADVL-ASAHSEPPPRQL 137
PP +QN P+P V + P MPPP A +SA ++ P+
Sbjct: 243 PPTQLQNTPAPV--------PVSTPPSQLQAPPAQSQFMPPPPAPSHPSSAQTQSFPQYQ 294
Query: 138 QTSPQQPRLSPQRIQQDARAALISPHAKGGSASSKTAASSHTSSERLSALLVEDPLSIFQ 197
Q P QP+ P Q SP G ++ S SS ++ + P Q
Sbjct: 295 QNWPPQPQARP---QSSGGYPTYSPAPPG----NQPPVESLPSSMQMQSPYSGPPQQSMQ 347
Query: 198 SF-FDGTLDLESPPRQAEIAETAQSG 222
++ + ++PP+Q +++ + Q+G
Sbjct: 348 AYGYGAAPPPQAPPQQTKMSYSPQTG 373
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,980,491
Number of Sequences: 26719
Number of extensions: 392638
Number of successful extensions: 2076
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 1673
Number of HSP's gapped (non-prelim): 283
length of query: 423
length of database: 11,318,596
effective HSP length: 102
effective length of query: 321
effective length of database: 8,593,258
effective search space: 2758435818
effective search space used: 2758435818
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0106.4


