

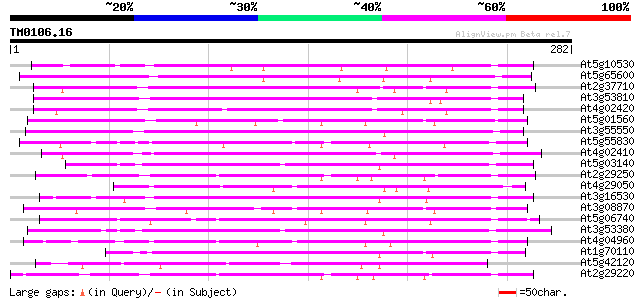
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.16
(282 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g10530 lectin-like protein kinase - like 131 4e-31
At5g65600 receptor protein kinase-like protein 129 2e-30
At2g37710 putative receptor-like protein kinase 113 1e-25
At3g53810 serine/threonine-specific kinase like protein 110 1e-24
At4g02420 102 2e-22
At5g01560 receptor like protein kinase 98 6e-21
At3g55550 probable serine/threonine-specific protein kinase 98 6e-21
At5g55830 serine/threonine-specific kinase like protein 96 2e-20
At4g02410 unknown protein 91 9e-19
At5g03140 receptor like protein kinase 89 3e-18
At2g29250 putative protein kinase 89 3e-18
At4g29050 serine/threonine-specific kinase like protein 85 5e-17
At3g16530 putative lectin 84 9e-17
At3g08870 putative serine/threonine protein kinase 84 9e-17
At5g06740 lectin-like protein kinase 83 1e-16
At3g53380 receptor lectin kinase -like protein 82 2e-16
At4g04960 unknown protein 80 2e-15
At1g70110 hypothetical protein 80 2e-15
At5g42120 receptor lectin kinase-like protein 78 5e-15
At2g29220 putative protein kinase 78 5e-15
>At5g10530 lectin-like protein kinase - like
Length = 651
Score = 131 bits (330), Expect = 4e-31
Identities = 95/264 (35%), Positives = 136/264 (50%), Gaps = 24/264 (9%)
Query: 12 SLLTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLPVY 71
S+L F+L++ V S + F ++F ++ +QGDA G ++LT ++
Sbjct: 4 SILLFSFVLVLPFVCS---VQFNISRFGSDVSEIAYQGDARA--NGAVELTNIDYTC--- 55
Query: 72 KSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVN--KIADGLAFFLAPVDTQ--P 127
G A Y V +W+ T + F T FSF ID+ NV G AFFLAP Q P
Sbjct: 56 -RAGWATYGKQVPLWNPGTSKPSDFSTRFSFRIDTRNVGYGNYGHGFAFFLAPARIQLPP 114
Query: 128 QKPGGLLGIFDN-DDPSQSNQVVAVEFDTHFNSNWDPKS--PHIGIDVGSIQSSNATSWG 184
GG LG+F+ ++ S + +V VEFDT N WDP H+GI+ S+ SSN TSW
Sbjct: 115 NSAGGFLGLFNGTNNQSSAFPLVYVEFDTFTNPEWDPLDVKSHVGINNNSLVSSNYTSWN 174
Query: 185 AAYG--EVANVFIHYQASTKELTVSLDHPSSKDTYTVSS---VVDLKNVLPEYVRVGFSA 239
A ++ V I Y ++ + L+VS + + D SS ++DL VLP V +GFSA
Sbjct: 175 ATSHNQDIGRVLIFYDSARRNLSVSWTYDLTSDPLENSSLSYIIDLSKVLPSEVTIGFSA 234
Query: 240 TTGLNPDHVETNDVLSWSFESDLK 263
T+G E N +LSW F S L+
Sbjct: 235 TSG---GVTEGNRLLSWEFSSSLE 255
>At5g65600 receptor protein kinase-like protein
Length = 675
Score = 129 bits (323), Expect = 2e-30
Identities = 95/270 (35%), Positives = 128/270 (47%), Gaps = 20/270 (7%)
Query: 6 SQKSLTSLLTLFFLLLVNKVNSTDYLSFTFNQFTPQQP-DLLFQGDALVAPTGKLQLTKV 64
S S S LF L + D L F F F P D+ + GDA G +
Sbjct: 11 SSSSSMSNSILFLSLFLFLPFVVDSLYFNFTSFRQGDPGDIFYHGDATPDEDGTVNFNNA 70
Query: 65 ENDLPVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVD 124
E V G Y V IW KTG + F TSFSF ID+ N++ G+ FFLAP+
Sbjct: 71 EQTSQV----GWITYSKKVPIWSHKTGKASDFSTSFSFKIDARNLSADGHGICFFLAPMG 126
Query: 125 TQ--PQKPGGLLGIFD-NDDPSQSNQVVAVEFDTHFNSNWDPK--SPHIGIDVGSIQSSN 179
Q GG L +F ++ S S +V VEFDT N WDP H+GI+ S+ SSN
Sbjct: 127 AQLPAYSVGGFLNLFTRKNNYSSSFPLVHVEFDTFNNPGWDPNDVGSHVGINNNSLVSSN 186
Query: 180 ATSWGAA--YGEVANVFIHYQASTKELTVSLDH-----PSSKDTYTVSSVVDLKNVLPEY 232
TSW A+ ++ + I Y + TK L+V+ + K++ ++S ++DL VLP
Sbjct: 187 YTSWNASSHSQDICHAKISYDSVTKNLSVTWAYELTATSDPKESSSLSYIIDLAKVLPSD 246
Query: 233 VRVGFSATTGLNPDHVETNDVLSWSFESDL 262
V GF A G N E + +LSW S L
Sbjct: 247 VMFGFIAAAGTN---TEEHRLLSWELSSSL 273
>At2g37710 putative receptor-like protein kinase
Length = 675
Score = 113 bits (282), Expect = 1e-25
Identities = 91/273 (33%), Positives = 137/273 (49%), Gaps = 31/273 (11%)
Query: 13 LLTLFFLLLVNKV--NSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLPV 70
LLT+FF N + +S+ L+F +N DL QG V P G L+LT
Sbjct: 5 LLTIFFFFFFNLIFQSSSQSLNFAYNNGFNPPTDLSIQGITTVTPNGLLKLTNT-----T 59
Query: 71 YKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDTQPQ-K 129
+ TG A Y P+ DS G V+SF TSF F I S G+AF +AP + P
Sbjct: 60 VQKTGHAFYTKPIRFKDSPNGTVSSFSTSFVFAIHSQIAILSGHGIAFVVAPNASLPYGN 119
Query: 130 PGGLLGIFD-NDDPSQSNQVVAVEFDTHFNSNW-DPKSPHIGIDVG---SIQSSNATSWG 184
P +G+F+ ++ +++N V AVE DT ++ + D H+GID+ S+QSS A W
Sbjct: 120 PSQYIGLFNLANNGNETNHVFAVELDTILSTEFNDTNDNHVGIDINSLKSVQSSPAGYWD 179
Query: 185 AAYGEVAN----------VFIHYQASTKELTVSLDHPSSKDTYT---VSSVVDLKNVLPE 231
G+ N V++ Y T ++ V++ P ++D T V++V DL +VL +
Sbjct: 180 EK-GQFKNLTLISRKPMQVWVDYDGRTNKIDVTM-APFNEDKPTRPLVTAVRDLSSVLLQ 237
Query: 232 YVRVGFSATTGLNPDHVETNDVLSWSFESDLKA 264
+ VGFS+ TG + + +L WSF + KA
Sbjct: 238 DMYVGFSSATG---SVLSEHYILGWSFGLNEKA 267
>At3g53810 serine/threonine-specific kinase like protein
Length = 677
Score = 110 bits (274), Expect = 1e-24
Identities = 87/265 (32%), Positives = 134/265 (49%), Gaps = 29/265 (10%)
Query: 13 LLTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLPVYK 72
L+ FFLL ++S+ L+FT+N F P D+ QG A V P G L+LT +
Sbjct: 7 LIFFFFLLCQIMISSSQNLNFTYNGFHPPLTDISLQGLATVTPNGLLKLTNTS-----VQ 61
Query: 73 STGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDTQP-QKPG 131
TG A + DS+ GNV+SF T+F F I S G+AF +AP P P
Sbjct: 62 KTGHAFCTERIRFKDSQNGNVSSFSTTFVFAIHSQIPTLSGHGIAFVVAPTLGLPFALPS 121
Query: 132 GLLGIFD-NDDPSQSNQVVAVEFDTHFNSNW-DPKSPHIGIDVGSIQSSNATSWGAAYGE 189
+G+F+ +++ + +N + AVEFDT +S + DP H+GID+ ++S+N ++ A Y +
Sbjct: 122 QYIGLFNISNNGNDTNHIFAVEFDTIQSSEFGDPNDNHVGIDLNGLRSANYST--AGYRD 179
Query: 190 VANVFIHYQ-ASTKELTVSLDH------------PSSKD---TYTVSSVVDLKNVLPEYV 233
+ F + S K + V +D+ P D VS V DL ++L E +
Sbjct: 180 DHDKFQNLSLISRKRIQVWIDYDNRSHRIDVTVAPFDSDKPRKPLVSYVRDLSSILLEDM 239
Query: 234 RVGFSATTGLNPDHVETNDVLSWSF 258
VGFS+ TG + + ++ WSF
Sbjct: 240 YVGFSSATG---SVLSEHFLVGWSF 261
>At4g02420
Length = 669
Score = 102 bits (255), Expect = 2e-22
Identities = 89/267 (33%), Positives = 128/267 (47%), Gaps = 33/267 (12%)
Query: 13 LLTLFFLLLV--NKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLPV 70
L T+FFL + +S+ + FT+N F P D+ G A + P G L+LT
Sbjct: 6 LFTIFFLSFFWQSLKSSSQIIDFTYNGFRPPPTDISILGIATITPNGLLKLTNT-----T 60
Query: 71 YKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDTQP-QK 129
+STG A Y P+ DS G V+SF T+F F I S IA G+AF +AP P
Sbjct: 61 MQSTGHAFYTKPIRFKDSPNGTVSSFSTTFVFAIHSQ--IPIAHGMAFVIAPNPRLPFGS 118
Query: 130 PGGLLGIFD-NDDPSQSNQVVAVEFDTHFNSNW-DPKSPHIGIDVGSIQSSNATSWGAAY 187
P LG+F+ ++ + N V AVE DT N + D + H+GID+ S+ S ++ A Y
Sbjct: 119 PLQYLGLFNVTNNGNVRNHVFAVELDTIMNIEFNDTNNNHVGIDINSLNSVKSSP--AGY 176
Query: 188 GEVANVFIH---YQASTKELTVSLDHPSSKDTYT-------------VSSVVDLKNVLPE 231
+ + F + + ++ V D P+ T VS V DL +VL +
Sbjct: 177 WDENDQFHNLTLISSKRMQVWVDFDGPTHLIDVTMAPFGEVKPRKPLVSIVRDLSSVLLQ 236
Query: 232 YVRVGFSATTGLNPDHVETNDVLSWSF 258
+ VGFS+ TG + V VL WSF
Sbjct: 237 DMFVGFSSATG---NIVSEIFVLGWSF 260
>At5g01560 receptor like protein kinase
Length = 685
Score = 97.8 bits (242), Expect = 6e-21
Identities = 83/277 (29%), Positives = 127/277 (44%), Gaps = 35/277 (12%)
Query: 10 LTSLLTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLP 69
+ SLL + FL+ + + F F+ F Q ++ QGD+ + G L+LT +D+
Sbjct: 1 MVSLLLVLFLVRAHVATTETTTEFIFHGFKGNQSEIHMQGDSTITSNGLLRLTDRNSDV- 59
Query: 70 VYKSTGRALYVAPVHIWDSKTGN--VASFITSFSFIIDSPNVNKIADGLAFFLAP----V 123
G A Y PV + DS + N V SF TSF FII S + + G F L+P
Sbjct: 60 ----VGTAFYHKPVRLLDSNSTNTTVRSFSTSFIFIIPSSSTSNGGFGFTFTLSPTPNRT 115
Query: 124 DTQPQKPGGLLGIFDNDDPSQSNQVVAVEFDT--HFNSNWDPKSPHIGIDVGSIQS---- 177
D P++ GLL + +D + SN V AVEFDT F + HIG++ S+ S
Sbjct: 116 DADPEQYMGLLN--ERNDGNSSNHVFAVEFDTVQGFKDGTNRIGNHIGLNFNSLSSDVQE 173
Query: 178 ---------SNATSWGAAYGEVANVFIHYQASTKELTVSLDHPS-----SKDTYTVSSVV 223
S + GE VF+ Y TK L +++ +P+ + V
Sbjct: 174 PVAYFNNNDSQKEEFQLVSGEPIQVFLDYHGPTKTLNLTV-YPTRLGYKPRIPLISREVP 232
Query: 224 DLKNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFES 260
L +++ + + VGF+A TG + + V+ WSF S
Sbjct: 233 KLSDIVVDEMFVGFTAATGRH-GQSSAHYVMGWSFAS 268
>At3g55550 probable serine/threonine-specific protein kinase
Length = 684
Score = 97.8 bits (242), Expect = 6e-21
Identities = 81/265 (30%), Positives = 122/265 (45%), Gaps = 23/265 (8%)
Query: 9 SLTSLLTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDL 68
S T + L L+ + + S+ F+F F P+L G A +APTG ++LT
Sbjct: 2 SQTFAVILLLLIFLTHLVSSLIQDFSFIGFKKASPNLTLNGVAEIAPTGAIRLT-----T 56
Query: 69 PVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPV-DTQP 127
+ G A Y P+ SF TSF+ + V GLAF + P D +
Sbjct: 57 ETQRVIGHAFYSLPIRFKPIGVNRALSFSTSFAIAMVPEFVTLGGHGLAFAITPTPDLRG 116
Query: 128 QKPGGLLGIFDNDDPSQSNQVVAVEFDTHFNSNW-DPKSPHIGIDVGSIQSSNATSWGAA 186
P LG+ ++ + S+ AVEFDT + + D H+GID+ S++SS +T G
Sbjct: 117 SLPSQYLGLLNSSRVNFSSHFFAVEFDTVRDLEFEDINDNHVGIDINSMESSISTPAGYF 176
Query: 187 Y------------GEVANVFIHYQASTKELTVSLDHPSSKDTYTVSSV-VDLKNVLPEYV 233
G V +I Y ++ K L V L S K ++ S VDL +VL + +
Sbjct: 177 LANSTKKELFLDGGRVIQAWIDYDSNKKRLDVKLSPFSEKPKLSLLSYDVDLSSVLGDEM 236
Query: 234 RVGFSATTGLNPDHVETNDVLSWSF 258
VGFSA+TGL ++ +L W+F
Sbjct: 237 YVGFSASTGL---LASSHYILGWNF 258
>At5g55830 serine/threonine-specific kinase like protein
Length = 681
Score = 95.9 bits (237), Expect = 2e-20
Identities = 87/275 (31%), Positives = 134/275 (48%), Gaps = 32/275 (11%)
Query: 6 SQKSLTSLLTLFFLLLVNK--VNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTK 63
S+K L T L ++K S+D ++FTF FT + +L F GD+ + G + LT+
Sbjct: 4 SRKLLVIFFTWITALSMSKPIFVSSDNMNFTFKSFTIR--NLTFLGDSHLR-NGVVGLTR 60
Query: 64 VENDLPVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDS--PNVNKIADGLAFFLA 121
E +P S+G +Y P+ +D + ASF T FSF + + P+ DGLAFFL+
Sbjct: 61 -ELGVPD-TSSGTVIYNNPIRFYDPDSNTTASFSTHFSFTVQNLNPDPTSAGDGLAFFLS 118
Query: 122 PVDTQPQKPGGLLGIFDNDDPSQSNQVVAVEFDT----HFNSNWDPKSPHIGIDVGSIQS 177
+ PGG LG+ ++ P N+ VA+EFDT HFN DP HIG+DV S+ S
Sbjct: 119 HDNDTLGSPGGYLGLVNSSQP-MKNRFVAIEFDTKLDPHFN---DPNGNHIGLDVDSLNS 174
Query: 178 SN------ATSWGAAYGEVANVFIHYQASTKELTVSLDHPSSKDTY------TVSSVVDL 225
+ ++ G+ +I Y+ + L V L + T +S +DL
Sbjct: 175 ISTSDPLLSSQIDLKSGKSITSWIDYKNDLRLLNVFLSYTDPVTTTKKPEKPLLSVNIDL 234
Query: 226 KNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFES 260
L + VGFS +T E + + +WSF++
Sbjct: 235 SPFLNGEMYVGFSGST---EGSTEIHLIENWSFKT 266
>At4g02410 unknown protein
Length = 674
Score = 90.5 bits (223), Expect = 9e-19
Identities = 84/272 (30%), Positives = 130/272 (46%), Gaps = 31/272 (11%)
Query: 17 FFLLLVNKV--NSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLPVYKST 74
FF++L++K +S+ L+FT+N F ++ QG A V G L+LT D V ST
Sbjct: 11 FFIILLSKPLNSSSQSLNFTYNSFHRPPTNISIQGIATVTSNGILKLT----DKTVI-ST 65
Query: 75 GRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDT-QPQKPGGL 133
G A Y P+ DS V+SF T+F I S G+AFF+AP
Sbjct: 66 GHAFYTEPIRFKDSPNDTVSSFSTTFVIGIYSGIPTISGHGMAFFIAPNPVLSSAMASQY 125
Query: 134 LGIFDN-DDPSQSNQVVAVEFDTHFNSNWDPKSP-HIGIDVGSIQSSNATSWGAAYGEVA 191
LG+F + ++ + +N ++AVEFDT N +D + H+GI++ S+ S ++ G Y +
Sbjct: 126 LGLFSSTNNGNDTNHILAVEFDTIMNPEFDDTNDNHVGININSLTSVKSSLVG--YWDEI 183
Query: 192 N--------------VFIHYQASTKELTVSL-DHPSSKDTYTVSSVV-DLKNVLPEYVRV 235
N V++ Y T ++ V++ K + SVV DL +V + + +
Sbjct: 184 NQFNNLTLISRKRMQVWVDYDDRTNQIDVTMAPFGEVKPRKALVSVVRDLSSVFLQDMYL 243
Query: 236 GFSATTGLNPDHVETNDVLSWSFESDLKAGGP 267
GFSA TG + + V WSF K P
Sbjct: 244 GFSAATGY---VLSEHFVFGWSFMVKGKTAPP 272
>At5g03140 receptor like protein kinase
Length = 711
Score = 89.0 bits (219), Expect = 3e-18
Identities = 76/243 (31%), Positives = 118/243 (48%), Gaps = 17/243 (6%)
Query: 29 DYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLPVYKST-GRALYVAPVHIWD 87
D+ + F+ T L GDA + G ++LT+ +L V ST G+ALY PV
Sbjct: 31 DFPATRFDLGTLTLSSLKLLGDAHLN-NGTIKLTR---ELSVPTSTAGKALYGKPVKFRH 86
Query: 88 SKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDTQPQKPGGLLGIFDNDDPSQSNQ 147
+T + ASF T FSF + + N + I GLAF ++P + GG LG+ ++ +
Sbjct: 87 PETKSPASFTTYFSFSVTNLNPSSIGGGLAFVISPDEDYLGSTGGFLGL--TEETGSGSG 144
Query: 148 VVAVEFDTHFNSNW-DPKSPHIGIDVGSIQSSNATSWG-----AAYGEVANVFIHYQAST 201
VAVEFDT + + D H+G+D+ ++ S+ G G N +I Y S
Sbjct: 145 FVAVEFDTLMDVQFKDVNGNHVGLDLNAVVSAAVADLGNVDIDLKSGNAVNSWITYDGSG 204
Query: 202 KELTVSLDHPSSKDTYTVSSV-VDLKNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFES 260
+ LTV + + + K + SV +DL + + + VGFS +T E + V WSF S
Sbjct: 205 RVLTVYVSYSNLKPKSPILSVPLDLDRYVSDSMFVGFSGST---QGSTEIHSVDWWSFSS 261
Query: 261 DLK 263
+
Sbjct: 262 SFE 264
>At2g29250 putative protein kinase
Length = 623
Score = 88.6 bits (218), Expect = 3e-18
Identities = 85/273 (31%), Positives = 130/273 (47%), Gaps = 37/273 (13%)
Query: 14 LTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLPVYKS 73
+++ FLL + V+S F + F + +LL G + + P+G L+LT +
Sbjct: 10 VSIIFLLYFSCVSSQQQTKFLNHGFL--EANLLKSGSSKIHPSGHLELTNTS-----MRQ 62
Query: 74 TGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAP-VDTQPQKPGG 132
G+A + P+ + + N+ SF TSF F I +P GLAF ++P +D P
Sbjct: 63 IGQAFHGFPIPFLNPNSSNLVSFPTSFVFAI-TPGPGAPGHGLAFVISPSLDFSGALPSN 121
Query: 133 LLGIFDNDDPSQS-NQVVAVEFDT----HFNSNWDPKSPHIGIDVG---SIQSSNA---- 180
LG+F+ + S N ++AVEFDT N D H+GID+ SI+S++A
Sbjct: 122 YLGLFNTSNNGNSLNCILAVEFDTVQAVELN---DIDDNHVGIDLNGVISIESTSAEYFD 178
Query: 181 ------TSWGAAYGEVANVFIHYQASTKELTVS---LDHPSSKDTYTVSSVVDLKNVLPE 231
S A G+ V+I Y A+ L V+ LD P K +S ++L ++ E
Sbjct: 179 DREAKNISLRLASGKPIRVWIEYNATETMLNVTLAPLDRPKPK-LPLLSRKLNLSGIISE 237
Query: 232 YVRVGFSATTGLNPDHVETNDVLSWSFESDLKA 264
VGFSA TG ++ VL WSF + KA
Sbjct: 238 ENYVGFSAATG---TVTSSHFVLGWSFSIEGKA 267
>At4g29050 serine/threonine-specific kinase like protein
Length = 669
Score = 84.7 bits (208), Expect = 5e-17
Identities = 75/229 (32%), Positives = 109/229 (46%), Gaps = 31/229 (13%)
Query: 53 VAPTGKLQLTKVENDLPVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKI 112
+A T L K+ N S G Y +PV +S G V+SF T+F F I S NVN +
Sbjct: 40 IAITNSKGLMKLTNSSEF--SYGHVFYNSPVRFKNSPNGTVSSFSTTFVFAIVS-NVNAL 96
Query: 113 -ADGLAFFLAPVDTQPQKPG----GLLGIFDNDDPSQSNQVVAVEFDTHFNSNWDP-KSP 166
GLAF ++P P GL + +N DP SN +VAVEFDT N +D +
Sbjct: 97 DGHGLAFVISPTKGLPYSSSSQYLGLFNLTNNGDP--SNHIVAVEFDTFQNQEFDDMDNN 154
Query: 167 HIGIDVGSIQSSNATSWGAAY---GEVANV----------FIHYQASTKELTVSLD--HP 211
H+GID+ S+ S A++ G G N+ +I Y +S ++L V++ H
Sbjct: 155 HVGIDINSLSSEKASTAGYYEDDDGTFKNIRLINQKPIQAWIEYDSSRRQLNVTIHPIHL 214
Query: 212 SSKDTYTVSSVVDLKNVLPEYVRVGFSATTG-LNPDHVETNDVLSWSFE 259
+S DL L + + VGF++ TG L H +L W+F+
Sbjct: 215 PKPKIPLLSLTKDLSPYLFDSMYVGFTSATGRLRSSHY----ILGWTFK 259
>At3g16530 putative lectin
Length = 276
Score = 84.0 bits (206), Expect = 9e-17
Identities = 78/279 (27%), Positives = 122/279 (42%), Gaps = 41/279 (14%)
Query: 16 LFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPT-------GKLQLTKVENDL 68
L FL+L N+ + F F+ F +LLF GDA + P+ G L +T+ EN
Sbjct: 6 LCFLVLF-LANAAFAVKFNFDSFDGS--NLLFLGDAELGPSSDGVSRSGALSMTRDENPF 62
Query: 69 PVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAP-VDTQP 127
S G+ LY+ + S T + SF TSF+F I G AF + P D
Sbjct: 63 ----SHGQGLYINQIPFKPSNTSSPFSFETSFTFSITPRTKPNSGQGFAFIITPEADNSG 118
Query: 128 QKPGGLLGIFD-NDDPSQSNQVVAVEFDTHFNSNW-DPKSPHIGIDVGSIQSSNATSWG- 184
GG LGI + +D N ++A+EFDT N + D H+G+++ S+ S A G
Sbjct: 119 ASDGGYLGILNKTNDGKPENHILAIEFDTFQNKEFLDISGNHVGVNINSMTSLVAEKAGY 178
Query: 185 ------------------AAYGEVANVFIHYQASTKELTVSL--DHPSSKDTYTVSSVVD 224
+ GE ++ ++ +TV+L ++ + +
Sbjct: 179 WVQTRVGKRKVWSFKDVNLSSGERFKAWVEFRNKDSTITVTLAPENVKKPKRALIEAPRV 238
Query: 225 LKNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFESDLK 263
L VL + + GF+ + G VE +D+ SWSFE+ K
Sbjct: 239 LNEVLLQNMYAGFAGSMG---RAVERHDIWSWSFENAAK 274
>At3g08870 putative serine/threonine protein kinase
Length = 693
Score = 84.0 bits (206), Expect = 9e-17
Identities = 81/289 (28%), Positives = 129/289 (44%), Gaps = 49/289 (16%)
Query: 8 KSLTSLLTLFFLLLVNKVNSTDYLS------FTFNQFTPQQPDLLFQGDALVAPTGKL-Q 60
+S+ S + FFL++++ + + L+ FTF F Q D+ +G + + L +
Sbjct: 5 RSINSFMFFFFLMILSNASKSSVLAEATTAKFTFIGFKENQTDIQTEGASTIQHDNDLLR 64
Query: 61 LTKVENDLPVYKSTGRALYVAPVHIWD---SKTGNVASFITSFSFIIDSPNVNKIADGLA 117
LT + ++ TG A Y P+ + + S V SF TSF F+I + G
Sbjct: 65 LTNRKQNV-----TGTAFYRKPIRLRELTNSSDIKVCSFSTSFVFVILPSSPGNGGFGFT 119
Query: 118 FFLAPVDTQPQKPG-------GLLGIFDNDDPSQSNQVVAVEFDT--HFNSNWDPKSPHI 168
F L+P P +PG GLL +N +P SN V AVEFDT F D + HI
Sbjct: 120 FTLSPT---PNRPGAESAQYLGLLNRTNNGNP--SNHVFAVEFDTVQGFKDGADRRGNHI 174
Query: 169 GIDVGSIQSS------------NATSWGAAYGEVANVFIHYQASTKELTVSLDHPS---- 212
G++ ++ S+ + GE V I Y S++ L V++ +P+
Sbjct: 175 GLNFNNLSSNVQEPLIYYDTEDRKEDFQLESGEPIRVLIDYDGSSETLNVTI-YPTRLEF 233
Query: 213 -SKDTYTVSSVVDLKNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFES 260
K V +L ++ + + VGF+A TG D + V+ WSF S
Sbjct: 234 KPKKPLISRRVSELSEIVKDEMYVGFTAATG--KDQSSAHYVMGWSFSS 280
>At5g06740 lectin-like protein kinase
Length = 652
Score = 83.2 bits (204), Expect = 1e-16
Identities = 78/263 (29%), Positives = 120/263 (44%), Gaps = 20/263 (7%)
Query: 16 LFFLLLVNKVNS-TDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLP---VY 71
L FL+L K+ + L F F F L + ++ + G +Q+T P +
Sbjct: 9 LLFLILTCKIETQVKCLKFDFPGFNVSNELELIRDNSYIV-FGAIQVTPDVTGGPGGTIA 67
Query: 72 KSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDTQPQKPG 131
GRALY P +W A+F T+F I S + +GLAF L P +T PQ
Sbjct: 68 NQAGRALYKKPFRLWSKHKS--ATFNTTFVINI-SNKTDPGGEGLAFVLTPEETAPQNSS 124
Query: 132 GLLGIFDNDDPSQSNQ--VVAVEFDTHFNSNWDPKSPHIGIDVGSIQS---SNATSWGAA 186
G+ N+ +++N+ +V+VEFDT + + D H+ ++V +I S + + G
Sbjct: 125 GMWLGMVNERTNRNNESRIVSVEFDTRKSHSDDLDGNHVALNVNNINSVVQESLSGRGIK 184
Query: 187 YGEVANVFIHYQASTKELTVSLDH---PSSKDTYTVSSVVDLKNVLPEYVRVGFSATTGL 243
++ H + K L+V + + S +DL LPE V VGF+A+T
Sbjct: 185 IDSGLDLTAHVRYDGKNLSVYVSRNLDVFEQRNLVFSRAIDLSAYLPETVYVGFTASTS- 243
Query: 244 NPDHVETNDVLSWSFESDLKAGG 266
+ E N V SWSFE LK G
Sbjct: 244 --NFTELNCVRSWSFEG-LKIDG 263
>At3g53380 receptor lectin kinase -like protein
Length = 715
Score = 82.4 bits (202), Expect = 2e-16
Identities = 78/271 (28%), Positives = 121/271 (43%), Gaps = 19/271 (7%)
Query: 10 LTSLLTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLP 69
++ L+ F +L+ N F F+ LL GDA ++ G + LT+ DL
Sbjct: 1 MSLFLSFFISILLCFFNGATTTQFDFSTLAISNLKLL--GDARLS-NGIVGLTR---DLS 54
Query: 70 VYKS-TGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDTQPQ 128
V S G+ LY P+ T SF + FSF I + N + I GLAF ++P
Sbjct: 55 VPNSGAGKVLYSNPIRFRQPGTHFPTSFSSFFSFSITNVNPSSIGGGLAFVISPDANSIG 114
Query: 129 KPGGLLGIFDNDDPSQSNQVVAVEFDTHFNSNW-DPKSPHIGIDVGSIQSSNATSWGAA- 186
GG LG+ ++ VAVEFDT + ++ D S H+G DV + SS + G
Sbjct: 115 IAGGSLGL--TGPNGSGSKFVAVEFDTLMDVDFKDINSNHVGFDVNGVVSSVSGDLGTVN 172
Query: 187 ----YGEVANVFIHYQASTKELTVSLDHPSSKDTYTVSSV-VDLKNVLPEYVRVGFSATT 241
G N +I Y T+ VS+ + + K + S +DL + +++ VGFS +T
Sbjct: 173 IDLKSGNTINSWIEYDGLTRVFNVSVSYSNLKPKVPILSFPLDLDRYVNDFMFVGFSGST 232
Query: 242 GLNPDHVETNDVLSWSFESDLKAGGPQAAGA 272
E + + WSF S + +G+
Sbjct: 233 ---QGSTEIHSIEWWSFSSSFGSSLGSGSGS 260
>At4g04960 unknown protein
Length = 686
Score = 79.7 bits (195), Expect = 2e-15
Identities = 78/272 (28%), Positives = 132/272 (47%), Gaps = 35/272 (12%)
Query: 8 KSLTSLLTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQLTKVEND 67
K+L LLTLF L+L N +++ D++ FN F ++ G A + + ++ + N
Sbjct: 2 KALLFLLTLF-LILPNPISAIDFI---FNGFNDSSSNVSLFGIATI----ESKILTLTNQ 53
Query: 68 LPVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKI-ADGLAFFLAPV--- 123
+TGRALY + D T +V F TSF F + +P N + G+ F AP
Sbjct: 54 TSF--ATGRALYNRTIRTKDPITSSVLPFSTSFIFTM-APYKNTLPGHGIVFLFAPSTGI 110
Query: 124 -DTQPQKPGGLLGIFDNDDPSQSNQVVAVEFDTHFNSNW-DPKSPHIGIDVGSIQS--SN 179
+ + GL + +N +P SN + VEFD N + D + H+GIDV S+ S SN
Sbjct: 111 NGSSSAQHLGLFNLTNNGNP--SNHIFGVEFDVFANQEFSDIDANHVGIDVNSLHSVYSN 168
Query: 180 ATSWGAAYGEV-----------ANVFIHYQASTKELTVSLDHPSSKDTYTVSSVVDLKNV 228
+ + + G V V+I Y+ +T+ + +S+ ++L +V
Sbjct: 169 TSGYWSDDGVVFKPLKLNDGRNYQVWIDYRDFVVNVTMQVAGKIRPKIPLLSTSLNLSDV 228
Query: 229 LPEYVRVGFSATTGLNPDHVETNDVLSWSFES 260
+ + + VGF+A TG V+++ +L+WSF +
Sbjct: 229 VEDEMFVGFTAATG---RLVQSHKILAWSFSN 257
>At1g70110 hypothetical protein
Length = 666
Score = 79.7 bits (195), Expect = 2e-15
Identities = 67/229 (29%), Positives = 108/229 (46%), Gaps = 27/229 (11%)
Query: 49 GDALVAPTGKLQLTKVENDLPVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPN 108
G A + G ++LT N P ++TG+ Y + +S G V+SF T+F F I+ N
Sbjct: 38 GSAYINNNGLIRLT---NSTP--QTTGQVFYNDQLRFKNSVNGTVSSFSTTFVFSIEFHN 92
Query: 109 VNKIADGLAFFLAPV-DTQPQKPGGLLGIFDNDDPSQ-SNQVVAVEFDTHFNSNWDPK-S 165
G+AF + P D P P LG+F+ + N +VAVE DT + ++ K +
Sbjct: 93 GIYGGYGIAFVICPTRDLSPTFPTTYLGLFNRSNMGDPKNHIVAVELDTKVDQQFEDKDA 152
Query: 166 PHIGIDVGSIQSSNATSWG------------AAYGEVANVFIHYQASTKELTVSLDHP-- 211
H+GID+ ++ S G G+ ++I Y + K++ V+L HP
Sbjct: 153 NHVGIDINTLVSDTVALAGYYMDNGTFRSLLLNSGQPMQIWIEYDSKQKQINVTL-HPLY 211
Query: 212 -SSKDTYTVSSVVDLKNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFE 259
+S DL L E + VGF++TTG D ++ +L W+F+
Sbjct: 212 VPKPKIPLLSLEKDLSPYLLELMYVGFTSTTG---DLTASHYILGWTFK 257
>At5g42120 receptor lectin kinase-like protein
Length = 691
Score = 78.2 bits (191), Expect = 5e-15
Identities = 79/248 (31%), Positives = 112/248 (44%), Gaps = 36/248 (14%)
Query: 14 LTLFFLLLVNKVNSTDYLSFTF----NQFTPQQPDLLFQGDALVAPTGKLQLTKVENDLP 69
L +F L+L +LS F ++F+P +L GDA + LT+ + P
Sbjct: 8 LVIFHLIL--------FLSLDFPTLSHRFSPPLQNLTLYGDAFFRDR-TISLTQQQPCFP 58
Query: 70 VYKST---------GRALYVAPVHIWDSKTGNVASFITSFSF-IIDSPNVNKIADGLAFF 119
+ GRALYV P+ + T ASF FSF II SP+ DG AF
Sbjct: 59 SVTTPPSKPSSSGIGRALYVYPIKFLEPSTNTTASFSCRFSFSIIASPSC-PFGDGFAFL 117
Query: 120 LAPVDTQPQKPGGLLGIFDNDDPSQSNQVVAVEFDTHFNS-NWDPKSPHIGIDVGSI--- 175
+ G LG+ P+ + +AVEFDT F+ + D H+GIDV SI
Sbjct: 118 ITSNADSFVFSNGFLGL-----PNPDDSFIAVEFDTRFDPVHGDINDNHVGIDVSSIFSV 172
Query: 176 QSSNATSWG--AAYGEVANVFIHYQASTKELTVSLDHPSSKDTYTV-SSVVDLKNVLPEY 232
S +A S G G+ +I Y K + V + + K T V S+ +DL + EY
Sbjct: 173 SSVDAISKGFDLKSGKKMMAWIEYSDVLKLIRVWVGYSRVKPTSPVLSTQIDLSGKVKEY 232
Query: 233 VRVGFSAT 240
+ VGFSA+
Sbjct: 233 MHVGFSAS 240
>At2g29220 putative protein kinase
Length = 627
Score = 78.2 bits (191), Expect = 5e-15
Identities = 84/285 (29%), Positives = 131/285 (45%), Gaps = 41/285 (14%)
Query: 1 MASFQSQKSLTSLLTLFFLLLVNKVNSTDYLSFTFNQFTPQQPDLLFQGDALVAPTGKLQ 60
M +F+S +LT + +F+ V+ T +L+ F +LL G + V P+G L+
Sbjct: 1 MITFKSI-ALTIIFLSYFVSCVSSQRETKFLNHGF-----LGANLLNFGSSKVYPSGLLE 54
Query: 61 LTKVENDLPVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFL 120
LT + G+A + P+ + + + N SF TSF F I + GLAF +
Sbjct: 55 LTNTS-----MRQIGQAFHGFPIPLSNPNSTNSVSFSTSFIFAI-TQGTGAPGHGLAFVI 108
Query: 121 AP-VDTQPQKPGGLLGIFDNDDPSQS-NQVVAVEFDT----HFNSNWDPKSPHIGIDVG- 173
+P +D P LG+F+ + S N+++A+EFDT N D H+GID+
Sbjct: 109 SPSMDFSGAFPSNYLGLFNTSNNGNSLNRILAIEFDTVQAVELN---DIDDNHVGIDLNG 165
Query: 174 --SIQSSNAT----------SWGAAYGEVANVFIHYQASTKELTVS---LDHPSSKDTYT 218
SI S+ A S A G+ V+I Y A+ L V+ LD P
Sbjct: 166 VISIASAPAAYFDDREAKNISLRLASGKPVRVWIEYNATETMLNVTLAPLDRP-KPSIPL 224
Query: 219 VSSVVDLKNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFESDLK 263
+S ++L + + VGFSA+TG ++ VL WSF + K
Sbjct: 225 LSRKMNLSGIFSQEHHVGFSASTG---TVASSHFVLGWSFNIEGK 266
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,525,175
Number of Sequences: 26719
Number of extensions: 287069
Number of successful extensions: 773
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 665
Number of HSP's gapped (non-prelim): 59
length of query: 282
length of database: 11,318,596
effective HSP length: 98
effective length of query: 184
effective length of database: 8,700,134
effective search space: 1600824656
effective search space used: 1600824656
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0106.16


