

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.13
(445 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
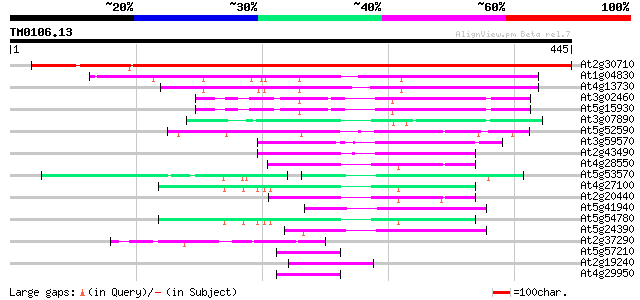
Score E
Sequences producing significant alignments: (bits) Value
At2g30710 unknown protein 667 0.0
At1g04830 unknown protein 125 4e-29
At4g13730 unknown protein 100 3e-21
At3g02460 putative plant adhesion molecule 83 3e-16
At5g15930 plant adhesion molecule 1 (PAM1) 82 6e-16
At3g07890 putative GTPase activator protein 79 4e-15
At5g52590 putative protein 77 3e-14
At3g59570 putative protein 74 2e-13
At2g43490 hypothetical protein 74 2e-13
At4g28550 putative protein 64 2e-10
At5g53570 GTPase activator protein of Rab-like small GTPases-lik... 58 9e-09
At4g27100 unknown protein 58 1e-08
At2g20440 unknown protein 58 1e-08
At5g41940 GTPase activator protein of Rab-like small GTPases-lik... 54 2e-07
At5g54780 putative protein 53 4e-07
At5g24390 GTPase activator-like protein of Rab-like small GTPases 52 9e-07
At2g37290 hypothetical protein 45 1e-04
At5g57210 microtubule-associated protein-like 43 3e-04
At2g19240 hypothetical protein 43 3e-04
At4g29950 unknown protein 42 5e-04
>At2g30710 unknown protein
Length = 440
Score = 667 bits (1721), Expect = 0.0
Identities = 326/431 (75%), Positives = 374/431 (86%), Gaps = 6/431 (1%)
Query: 18 DSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTYTRSNSYNDAGTSDHASE 77
DSRF+QTL++VQG LKGRS+PGK+LL++R DP SPTY RS S NDAG ++
Sbjct: 13 DSRFNQTLKNVQGFLKGRSIPGKVLLTRRSDPPPYP--ISPTYQRSLSENDAGRNELFES 70
Query: 78 TVEVEVHSSSGIPGEN---KLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGT 134
VEVE H+SS KL+ S S E ++V+ +G R++DSARVMKF KVLS T
Sbjct: 71 PVEVEDHNSSKKHDNTYAGKLR-SNSSAERSVKEVQNLKIGVRSSDSARVMKFNKVLSET 129
Query: 135 VVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIP 194
VIL+KLRELAW+GVP YMRP VWRLLLGYAPPNSDRRE VLRRKRLEYL+ V Q+YD+P
Sbjct: 130 TVILEKLRELAWNGVPHYMRPDVWRLLLGYAPPNSDRREAVLRRKRLEYLESVGQFYDLP 189
Query: 195 DTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGIND 254
D+ERS+DEINMLRQIAVDCPRTVPDVSFFQQ+QVQKSLERILY WAIRHPASGYVQGIND
Sbjct: 190 DSERSDDEINMLRQIAVDCPRTVPDVSFFQQEQVQKSLERILYTWAIRHPASGYVQGIND 249
Query: 255 LVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGI 314
LVTPF V+FLSEYL+G +D+WSM DLS++++S+VEADCYWCL+KLLDGMQDHYTFAQPGI
Sbjct: 250 LVTPFLVIFLSEYLDGGVDSWSMDDLSAEKVSDVEADCYWCLTKLLDGMQDHYTFAQPGI 309
Query: 315 QRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAE 374
QRLVFKLKELVRRID+PVS HME GLEFLQFAFRW+NCLLIREIPFN++ RLWDTYLAE
Sbjct: 310 QRLVFKLKELVRRIDEPVSRHMEEHGLEFLQFAFRWYNCLLIREIPFNLINRLWDTYLAE 369
Query: 375 GDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHS 434
GDALPDFLVYI+ASFLLTWSD+L+KLDFQ++VMFLQHLPT +W+ QELEMVLSRA+MWHS
Sbjct: 370 GDALPDFLVYIYASFLLTWSDELKKLDFQEMVMFLQHLPTHNWSDQELEMVLSRAYMWHS 429
Query: 435 MFNNSPNHLAT 445
MFNNSPNHLA+
Sbjct: 430 MFNNSPNHLAS 440
>At1g04830 unknown protein
Length = 448
Score = 125 bits (315), Expect = 4e-29
Identities = 118/427 (27%), Positives = 180/427 (41%), Gaps = 85/427 (19%)
Query: 64 NSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSHVENPSEDVRKS---------- 112
+SY+D G H+ T ++ E S S P N +S+ PS R
Sbjct: 22 SSYDD-GLLRHSPVTKMKEEAESISVAPRLNSAPPPSSNTSVPSPSHRPRNGNSISGGSG 80
Query: 113 ----SMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDY--MRPTVWRLLLGYAP 166
S+G A D +R + LS V+ + +LR LA +PD +R TVW+LLLGY P
Sbjct: 81 EYGHSVGPSAEDFSRQAHVSAELSKKVINMKELRSLALQSLPDSPGIRSTVWKLLLGYLP 140
Query: 167 PNSDRREGVLRRKRLEYLDCVSQY-------------------YDIPDTER--------- 198
P L++KR +Y + YD+ R
Sbjct: 141 PERSLWSTELKQKRSQYKHYKDELLTSPSEITWKMVRSKGFDNYDLKSESRCMLARSRIT 200
Query: 199 SED----------------EINMLRQIAVDCPRTVPDVSFFQQQQV-----QKSLERILY 237
ED + + QI D RT PD+ FF + Q+S++ IL
Sbjct: 201 DEDHPLSLGKASIWNTYFQDTETIEQIDRDVKRTHPDIPFFSGESSFARSNQESMKNILL 260
Query: 238 AWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLS 297
+A + YVQG+N+++ P F VF +D D S+ EAD ++C
Sbjct: 261 VFAKLNQGIRYVQGMNEILAPIFYVF-------------RNDPDEDSSSHAEADAFFCFV 307
Query: 298 KLLDGMQDHYTF----AQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQF-AFRWFN 352
+LL G +D Y + GI+ + +L +LVR+ D+ + H+E QF AFRW
Sbjct: 308 ELLSGFRDFYCQQLDNSVVGIRSAITRLSQLVRKHDEELWRHLEITTKVNPQFYAFRWIT 367
Query: 353 CLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHL 412
LL +E F +WD L++ + + L+ I + L+ +L DF + LQH
Sbjct: 368 LLLTQEFSFFDSLHIWDALLSDPEGPLESLLGICCAMLVLVRRRLIAGDFTSNMKLLQHY 427
Query: 413 PTQDWTH 419
PT + +H
Sbjct: 428 PTTNISH 434
>At4g13730 unknown protein
Length = 449
Score = 99.8 bits (247), Expect = 3e-21
Identities = 90/356 (25%), Positives = 151/356 (42%), Gaps = 69/356 (19%)
Query: 120 DSARVMKFTKVLSGTVVILDKLRELAWSGVPDY--MRPTVWRLLLGYAPPNSDRREGVLR 177
D +R + LS V+ L +LR++A G+PD +R VW+LLL Y P+ L
Sbjct: 96 DVSRKAQVVAELSKKVIDLKELRKIASQGLPDDAGIRSIVWKLLLDYLSPDRSLWSSELA 155
Query: 178 RKRLEYLDCVSQYYDIPDT------------------------ERSE------------- 200
+KR +Y + P RSE
Sbjct: 156 KKRSQYKQFKEELLMNPSEVTRKMDKSKGGDSNDPKIESPGALSRSEITHEDHPLSLGTT 215
Query: 201 -------DEINMLRQIAVDCPRTVPDVSFFQQQQV-----QKSLERILYAWAIRHPASGY 248
+ +L QI D RT PD+ FF Q +L+ IL +A +P Y
Sbjct: 216 SLWNNFFKDTEVLEQIERDVMRTHPDMHFFSGDSAVAKSNQDALKNILTIFAKLNPGIRY 275
Query: 249 VQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYT 308
VQG+N+++ P F +F ++ +G+ + E+D ++C +L+ G +D++
Sbjct: 276 VQGMNEILAPIFYIFKNDPDKGNA-------------AYAESDAFFCFVELMSGFRDNFC 322
Query: 309 F----AQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQF-AFRWFNCLLIREIPFNM 363
+ GI+ + +L L++ D+ + H+E QF AFRW LL +E F
Sbjct: 323 QQLDNSVVGIRYTITRLSLLLKHHDEELWRHLEVTTKINPQFYAFRWITLLLTQEFNFVE 382
Query: 364 VTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTH 419
+WDT L++ + + L+ I + L+ +L DF + LQ+ P + +H
Sbjct: 383 SLHIWDTLLSDPEGPQETLLRICCAMLILVRRRLLAGDFTSNLKLLQNYPPTNISH 438
>At3g02460 putative plant adhesion molecule
Length = 353
Score = 83.2 bits (204), Expect = 3e-16
Identities = 69/273 (25%), Positives = 122/273 (44%), Gaps = 53/273 (19%)
Query: 148 GVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERSEDEINMLR 207
G+PD +R VW+L+ G R+ +L + Y + E S E++++R
Sbjct: 88 GIPDCLRGLVWQLISG-------SRDLLLMNPGV--------YEQLVIYETSASELDIIR 132
Query: 208 QIAVDCPRTVPDVSFFQQQQV--QKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLS 265
I+ RT P FFQ++ Q+SL +L A+++ GYVQG+ + ++++S
Sbjct: 133 DIS----RTFPSHVFFQKRHGPGQRSLYNVLKAYSVYDRDVGYVQGMG-FIAGLLLLYMS 187
Query: 266 EYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDG-----MQDHYTFAQPGIQRLVFK 320
E D +W L LL G M+ Y P +Q+ +F+
Sbjct: 188 E-----------------------EDAFWLLVALLKGAVHAPMEGLYHAGLPLVQQYLFQ 224
Query: 321 LKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPD 380
L+ LV+ + + H + + +A +WF + PF + R+WD +L+EG +
Sbjct: 225 LESLVKELIPKLGEHFTQEMINPSMYASQWFITVFSYSFPFPLALRIWDVFLSEGVKI-- 282
Query: 381 FLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLP 413
+ + + L D+L KL F+ L+ L+ P
Sbjct: 283 -VFKVGLALLKYCQDELVKLPFEKLIHALKTFP 314
>At5g15930 plant adhesion molecule 1 (PAM1)
Length = 356
Score = 82.0 bits (201), Expect = 6e-16
Identities = 68/273 (24%), Positives = 122/273 (43%), Gaps = 53/273 (19%)
Query: 148 GVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERSEDEINMLR 207
G+PD +R VW+L+ G R+ +L + Y + E S E++++R
Sbjct: 85 GIPDCLRGLVWQLISG-------SRDLLLMNPGV--------YVQLVIYETSASELDIIR 129
Query: 208 QIAVDCPRTVPDVSFFQQQQV--QKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLS 265
I+ RT P FFQ++ Q+SL +L A+++ GYVQG+ + ++++S
Sbjct: 130 DIS----RTFPSHVFFQKRHGPGQRSLYNVLKAYSVYDRDVGYVQGMG-FIAGLLLLYMS 184
Query: 266 EYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDG-----MQDHYTFAQPGIQRLVFK 320
E D +W L LL G ++ Y P +Q+ + +
Sbjct: 185 E-----------------------EDAFWLLVALLKGAVHSPIEGLYQAGLPLVQQYLLQ 221
Query: 321 LKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPD 380
+LVR + + H + + +A +WF + +PF+ R+WD +LAEG +
Sbjct: 222 FDQLVRELMPKLGEHFTQEMINPSMYASQWFITVFSYSLPFHSALRIWDVFLAEGVKI-- 279
Query: 381 FLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLP 413
+ + + L D L KL F++L+ L++ P
Sbjct: 280 -VFKVGLALLKHCHDDLLKLPFEELMHALRNFP 311
>At3g07890 putative GTPase activator protein
Length = 400
Score = 79.3 bits (194), Expect = 4e-15
Identities = 74/287 (25%), Positives = 115/287 (39%), Gaps = 48/287 (16%)
Query: 141 LRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERSE 200
L+ L G+P +RP VW L G A S E S Y D+ T+ E
Sbjct: 109 LKRLIRKGIPPVLRPKVWFSLSGAAKKKSTVPE--------------SYYSDL--TKAVE 152
Query: 201 DEIN-MLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPF 259
+ RQI D PRT P + + +L R+L ++ R GY QG+N +
Sbjct: 153 GMVTPATRQIDHDLPRTFPGHPWLDTPEGHAALRRVLVGYSFRDSDVGYCQGLNYVAALL 212
Query: 260 FVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGM--QDHYTFAQPG--IQ 315
+V E D +W L+ LL+ + +D YT G ++
Sbjct: 213 LLVM-----------------------KTEEDAFWMLAVLLENVLVRDCYTTNLSGCHVE 249
Query: 316 RLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEG 375
+ VFK +L+ + ++TH+E+ G + A WF CL + +P R+WD EG
Sbjct: 250 QRVFK--DLLAQKCSRIATHLEDMGFDVSLVATEWFLCLFSKSLPSETTLRVWDVLFYEG 307
Query: 376 DALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQEL 422
+ A F + ++ L D++ LQ Q + EL
Sbjct: 308 AKV--LFHAALAIFKMKENELLMTHQVGDVINILQKTSHQLFDPDEL 352
>At5g52590 putative protein
Length = 338
Score = 76.6 bits (187), Expect = 3e-14
Identities = 77/303 (25%), Positives = 128/303 (41%), Gaps = 45/303 (14%)
Query: 126 KFTKVLS--GTVVILDKLRE-LAWSGVPDYMRPTVWRLLLGYAPPNSD--RREGVLRRKR 180
+FT +L G VV LRE + + G+ +R VW LLGY +S RE + KR
Sbjct: 14 EFTALLDSEGRVVESKALRERVFYGGIEHQLRREVWPFLLGYYAYDSTYAEREYLRSVKR 73
Query: 181 LEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQK----SLERIL 236
+EY Q+ I + R+ +D D +F + S+ IL
Sbjct: 74 MEYATLKQQWQSISPEQAKRFTKYRERKGLIDKDVVRTDRAFEYYEGDDNLHVNSMRDIL 133
Query: 237 YAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCL 296
++ + GY QG++D ++P V M D E++ +WC
Sbjct: 134 LTYSFYNFDLGYCQGMSDYLSPILFV--------------MED---------ESESFWCF 170
Query: 297 SKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHM-ENQGLEFLQFAFRWFNCLL 355
L++ + ++ Q G+ +F L +LV +D P+ + EN L + F FRW
Sbjct: 171 VALMERLGPNFNRDQNGMHTQLFALSKLVELLDSPLHNYFKENDCLNYF-FCFRWILIQF 229
Query: 356 IREIPFNMVTRLWDT----YLAEGDALPDFLVYIFASFLLTWSDKL--QKLDFQDLVMFL 409
RE + +LW+ YL+E F +Y+ + L K+ +++DF L+ F+
Sbjct: 230 KREFEYEKTMQLWEVMWTHYLSE-----HFHLYVCVAVLKRCRSKIMGEQMDFDTLLKFI 284
Query: 410 QHL 412
L
Sbjct: 285 NEL 287
>At3g59570 putative protein
Length = 549
Score = 73.9 bits (180), Expect = 2e-13
Identities = 56/196 (28%), Positives = 90/196 (45%), Gaps = 27/196 (13%)
Query: 197 ERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLV 256
+R + + L +I VD RT + F++ + IL +A PA+GY QG++DLV
Sbjct: 380 DRVSEWLWTLHRIVVDVVRTDSHLEFYEDPGNLGRMSDILAVYAWVDPATGYCQGMSDLV 439
Query: 257 TPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQP-GIQ 315
+PF VFL E DN AD +WC L+ + ++ P G+
Sbjct: 440 SPF--VFLFE------DN---------------ADAFWCFEMLIRRTRANFQMEGPTGVM 476
Query: 316 RLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEG 375
+ L +++ D + +H+ G E L FAFR L RE+ FN R+W+
Sbjct: 477 DQLQSLWRILQLTDKEMFSHLSRIGAESLHFAFRMLLVLFRRELSFNKALRMWE---VGN 533
Query: 376 DALPDFLVYIFASFLL 391
+ F++ + SF+L
Sbjct: 534 SSAVCFVLPVSCSFIL 549
>At2g43490 hypothetical protein
Length = 756
Score = 73.9 bits (180), Expect = 2e-13
Identities = 50/174 (28%), Positives = 80/174 (45%), Gaps = 24/174 (13%)
Query: 197 ERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLV 256
+R + + L +I VD RT + F++ + IL +A PA+GY QG++DLV
Sbjct: 350 DRVSEWLWTLHRIVVDVVRTDSHLEFYEDPGNLGRMSDILAVYAWVDPATGYCQGMSDLV 409
Query: 257 TPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQP-GIQ 315
+PF V+F DN AD +WC L+ + ++ P G+
Sbjct: 410 SPFVVLF--------EDN---------------ADAFWCFEMLIRRTRANFQMEGPTGVM 446
Query: 316 RLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWD 369
+ L +++ D + +H+ G E L FAFR L RE+ FN R+W+
Sbjct: 447 DQLQSLWHILQITDKDIFSHLSRIGAESLHFAFRMLLVLFRRELSFNEALRMWE 500
>At4g28550 putative protein
Length = 424
Score = 63.5 bits (153), Expect = 2e-10
Identities = 44/168 (26%), Positives = 77/168 (45%), Gaps = 27/168 (16%)
Query: 205 MLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFL 264
+L QI +D RT + F++ + Q L IL + +P GYVQG+ND+ +P ++
Sbjct: 166 VLSQIGLDVVRTDRYLCFYESESNQARLWDILSIYTWLNPDIGYVQGMNDICSPMIILL- 224
Query: 265 SEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLK 322
EAD +WC + + +++++ T G+Q + L
Sbjct: 225 ----------------------EDEADAFWCFERAMRRLRENFRTTATSMGVQTQLGMLS 262
Query: 323 ELVRRIDDPVSTHMEN-QGLEFLQFAFRWFNCLLIREIPFNMVTRLWD 369
++++ +D + H+E+ G E+L FA R L RE F LW+
Sbjct: 263 QVIKTVDPRLHQHLEDLDGGEYL-FAIRMLMVLFRREFSFLDALYLWE 309
>At5g53570 GTPase activator protein of Rab-like small GTPases-like
protein
Length = 550
Score = 58.2 bits (139), Expect = 9e-09
Identities = 42/195 (21%), Positives = 77/195 (38%), Gaps = 42/195 (21%)
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
L IL A+A+ P GY QG++DL++P V ++ +
Sbjct: 340 LVAILEAYAMYDPEIGYCQGMSDLLSPILAVISEDH-----------------------E 376
Query: 292 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWF 351
+WC + + ++ + GIQR + + ++++ D + H+EN E F +R
Sbjct: 377 AFWCFVGFMKKARHNFRLDEAGIQRQLSIVSKIIKNKDSQLYKHLENLQAEDCSFVYRMV 436
Query: 352 NCLLIREIPFNMVTRLWDTYLAEGDAL-------------------PDFLVYIFASFLLT 392
+ RE+ F LW+ A+ A+ D L+Y A+ +L
Sbjct: 437 LVMFRRELSFEQTLCLWEVMWADQAAIRAGVGKSPWSRIRQQAPPTDDLLLYAIAALVLR 496
Query: 393 WSDKLQKLDFQDLVM 407
+QK D ++
Sbjct: 497 RKLIIQKYSSMDEIV 511
Score = 43.9 bits (102), Expect = 2e-04
Identities = 54/213 (25%), Positives = 85/213 (39%), Gaps = 22/213 (10%)
Query: 26 RSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTYTRSNSYNDAGTSDHASETVEV-EVH 84
RS G S L S + +S+ SSP + SNSY+++ +S +S + + V
Sbjct: 6 RSYTSTSSGNSSSSSSLPSSSSSSLPSSSSSSPPSSNSNSYSNSNSSSSSSSWIHLRSVL 65
Query: 85 SSSGIPGENKLKISTSHVENP-SEDVRKSSMGARATDSARVMKFTKVLSGTVVILDKLRE 143
+ + + + S ++P S RK ++ S + K+ G V L K+R
Sbjct: 66 FVANLSSPSSVTSSDRRRKSPWSRRKRKWALTPHQWRSLFTPE-GKLRDGGVGFLKKVRS 124
Query: 144 LAWSGVPDYMRPTVWRLLLGYAPPN--SDRREGVLRRKRLEY----------LDC----V 187
GV +R VW LLG N S+ RE V +KR EY L C
Sbjct: 125 ---RGVDPSIRAEVWLFLLGVYDLNSTSEEREAVKTQKRKEYEKLQRRCQMLLKCGNGST 181
Query: 188 SQYYDIPDTERSEDEINMLRQIAVDCPRTVPDV 220
++P E + + + + P T DV
Sbjct: 182 DNLEELPSDEANSQCVRFVDDYKITGPMTSQDV 214
>At4g27100 unknown protein
Length = 436
Score = 57.8 bits (138), Expect = 1e-08
Identities = 68/305 (22%), Positives = 113/305 (36%), Gaps = 77/305 (25%)
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNS--DRREGVL 176
T S R + V G++ I LR + G+ +R VW LLG P S + RE +
Sbjct: 34 TLSVRKWQAVFVQEGSLHIGKTLRRIRRGGIHPSIRGEVWEFLLGCYDPMSTFEEREQIR 93
Query: 177 RRKRLEYL----DCVSQYYDIPD---------TERSE--------DEINM---------- 205
+R+RL+Y +C + I TE + EIN+
Sbjct: 94 QRRRLQYASWKEECKQMFPVIGSGRFTTAPVITENGQPNYDPLVLQEINLGTNSNGSVFF 153
Query: 206 -------------------LRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPAS 246
L QI +D RT + F+++++ L IL +A
Sbjct: 154 KELTSRGPLDKKIIQWLLTLHQIGLDVNRTDRALVFYEKKENLSKLWDILSVYAWIDNDV 213
Query: 247 GYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDH 306
GY QG++DL +P ++ EAD +WC +L+ ++ +
Sbjct: 214 GYCQGMSDLCSPMIILL-----------------------EDEADAFWCFERLMRRLRGN 250
Query: 307 Y--TFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMV 364
+ T G++ + L + + +D + H++ G FA R RE F
Sbjct: 251 FRSTGRSVGVEAQLTHLSSITQVVDPKLHQHLDKLGGGDYLFAIRMLMVQFRREFSFCDS 310
Query: 365 TRLWD 369
LW+
Sbjct: 311 LYLWE 315
>At2g20440 unknown protein
Length = 371
Score = 57.8 bits (138), Expect = 1e-08
Identities = 41/169 (24%), Positives = 75/169 (44%), Gaps = 29/169 (17%)
Query: 206 LRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLS 265
L QI +D RT + F++ + Q L +L + + GYVQG+ND+ +P ++F
Sbjct: 51 LHQIGLDVARTDRYLCFYENDRNQSKLWDVLAIYTWLNLDIGYVQGMNDICSPMIILF-- 108
Query: 266 EYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLKE 323
+ E D +WC + + +++++ T G+Q + L +
Sbjct: 109 ---------------------DDEGDAFWCFERAMRRLRENFRATATSMGVQTQLGVLSQ 147
Query: 324 LVRRIDDPVSTHMENQGL---EFLQFAFRWFNCLLIREIPFNMVTRLWD 369
+++ +D + H+ + L E+L FA R L RE F LW+
Sbjct: 148 VIKTVDPRLHQHLGKKDLDGGEYL-FAIRMLMVLFRREFSFLDALYLWE 195
>At5g41940 GTPase activator protein of Rab-like small GTPases-like
protein
Length = 506
Score = 53.5 bits (127), Expect = 2e-07
Identities = 34/144 (23%), Positives = 61/144 (41%), Gaps = 23/144 (15%)
Query: 235 ILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYW 294
IL A+A+ P GY QG++DL++P V + L +W
Sbjct: 307 ILEAYAVYDPEIGYCQGMSDLLSPLIAVMEDDVL-----------------------AFW 343
Query: 295 CLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCL 354
C + + ++ + GI+R + + ++++ D + H+EN E F +R L
Sbjct: 344 CFVGFMSKARHNFRLDEVGIRRQLSMVSKIIKFKDIHLYRHLENLEAEDCFFVYRMVVVL 403
Query: 355 LIREIPFNMVTRLWDTYLAEGDAL 378
RE+ F LW+ A+ A+
Sbjct: 404 FRRELTFEQTLCLWEVMWADQAAI 427
Score = 28.9 bits (63), Expect = 6.0
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 5/57 (8%)
Query: 129 KVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGY--APPNSDRREGVLRRKRLEY 183
++ G V L K+R GV +RP VW LLG N + R+ + + K EY
Sbjct: 60 RLSDGGVKFLKKVRS---GGVHPSIRPEVWPFLLGVYDLKSNKEERDSIRQLKLTEY 113
>At5g54780 putative protein
Length = 435
Score = 52.8 bits (125), Expect = 4e-07
Identities = 67/303 (22%), Positives = 109/303 (35%), Gaps = 75/303 (24%)
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNS--DRREGVL 176
T S R + G + I L + G+ +R VW LLG P S + RE +
Sbjct: 34 TLSVRKWQAAFTTEGFLDIGKTLSRIQRGGIHPSIRGEVWEFLLGCYDPKSTFEEREQIR 93
Query: 177 RRKRLEYL----DCVSQYYDIPD---------TERSE--------DEINM---------- 205
+R+RL+Y +C + I T + E E N+
Sbjct: 94 QRRRLQYASWKEECKQMFPVIGSGGFITAPVITNKGEPIYDPIVLQETNLGANGSDFFKD 153
Query: 206 -----------------LRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGY 248
L QI +D RT + F+++++ L IL +A GY
Sbjct: 154 LASRGPLDQKVIQWLLTLHQIGLDVNRTDRTLVFYEKKENLSKLWDILALYAWIDNDVGY 213
Query: 249 VQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHY- 307
QG++DL +P ++ EAD +WC +L+ ++ ++
Sbjct: 214 CQGMSDLCSPMIMLL-----------------------EDEADAFWCFERLMRRLRGNFR 250
Query: 308 -TFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTR 366
T G++ + L + + ID + H+E G FA R RE F
Sbjct: 251 DTGRSVGVEAQLTHLASITQIIDPKLHHHLEKLGGGDYLFAIRMIMVQFRREFSFCDSLY 310
Query: 367 LWD 369
LW+
Sbjct: 311 LWE 313
>At5g24390 GTPase activator-like protein of Rab-like small GTPases
Length = 528
Score = 51.6 bits (122), Expect = 9e-07
Identities = 33/162 (20%), Positives = 67/162 (40%), Gaps = 25/162 (15%)
Query: 219 DVSFFQQQQVQKS--LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWS 276
D S+ + ++ + L +L A+A+ P GY QG++DL++P V +Y
Sbjct: 304 DYSYLEPSKIFHAARLVAVLEAYALHDPEIGYCQGMSDLLSPILSVIPDDY--------- 354
Query: 277 MSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHM 336
+ +WC + + ++ + GI R + + ++++ D + H+
Sbjct: 355 --------------EAFWCFVGFMKKARQNFRVDEVGITRQLNIVSKIIKSKDSQLYKHL 400
Query: 337 ENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDAL 378
E E F +R + RE+ LW+ A+ A+
Sbjct: 401 EKVKAEDCFFVYRMVLVMFRRELTLEQTLHLWEVIWADQAAI 442
>At2g37290 hypothetical protein
Length = 544
Score = 44.7 bits (104), Expect = 1e-04
Identities = 49/176 (27%), Positives = 74/176 (41%), Gaps = 28/176 (15%)
Query: 81 VEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVI--- 137
V+ H+SS +K S S +E E+ R S +T + +K G+V
Sbjct: 124 VDDHASS-------IKESLSSIEESGENDRDSET---STSRSHSIKEENEAQGSVSPEPF 173
Query: 138 ---LDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIP 194
++L L GVP +R VW+ +G RR Y D ++Q +
Sbjct: 174 FPWYEELEVLVRLGVPKDLRGEVWQAFVGVKA----------RRVERYYQDLLAQITN-S 222
Query: 195 DTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQ 250
D S+ + +QI D PRT P + + SL RIL A+A +P+ GY Q
Sbjct: 223 DENSSDVQRKWKKQIEKDIPRTFPGHPALNENG-RDSLRRILLAYACHNPSVGYCQ 277
>At5g57210 microtubule-associated protein-like
Length = 737
Score = 43.1 bits (100), Expect = 3e-04
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Query: 212 DCPRTVPDV-SFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVV 262
D R P+ S+FQ Q L RIL W ++HP GY QG+++L+ P V
Sbjct: 120 DLSRLYPEHGSYFQSSGCQGMLRRILLLWCLKHPEIGYRQGMHELLAPLLYV 171
Score = 34.7 bits (78), Expect = 0.11
Identities = 16/49 (32%), Positives = 25/49 (50%)
Query: 321 LKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWD 369
L L+ +D + +H+ G+E FA RW L RE P + + +WD
Sbjct: 321 LYHLLSLVDASLHSHLVELGVEPQYFALRWLRVLFGREFPLSNLLIVWD 369
>At2g19240 hypothetical protein
Length = 840
Score = 43.1 bits (100), Expect = 3e-04
Identities = 21/67 (31%), Positives = 34/67 (50%)
Query: 222 FFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLS 281
+FQ + Q L RIL W ++HP GY QG+++L+ P V + + S S D
Sbjct: 121 YFQTPRYQGMLRRILLLWCLKHPEYGYRQGMHELLAPLLYVLHVDIMRLSEVRKSYEDYF 180
Query: 282 SDEISNV 288
+D ++
Sbjct: 181 TDRFDSL 187
Score = 29.3 bits (64), Expect = 4.6
Identities = 16/54 (29%), Positives = 24/54 (43%), Gaps = 1/54 (1%)
Query: 328 IDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWD-TYLAEGDALPD 380
+D + +H+ G+E F RW L RE + +WD LA+ A D
Sbjct: 309 VDSSLHSHLVELGVEPQYFGLRWLRVLFGREFLLQDLLLVWDEIILADNSARTD 362
>At4g29950 unknown protein
Length = 828
Score = 42.4 bits (98), Expect = 5e-04
Identities = 21/52 (40%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Query: 212 DCPRTVPDV-SFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVV 262
D R P+ S+FQ Q L RIL W ++HP GY QG+++L+ P V
Sbjct: 106 DLSRLYPEHWSYFQAPGCQGMLRRILLLWCLKHPEYGYRQGMHELLAPLLYV 157
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,366,656
Number of Sequences: 26719
Number of extensions: 439233
Number of successful extensions: 1214
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1143
Number of HSP's gapped (non-prelim): 45
length of query: 445
length of database: 11,318,596
effective HSP length: 102
effective length of query: 343
effective length of database: 8,593,258
effective search space: 2947487494
effective search space used: 2947487494
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0106.13


