

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0105.6
(574 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
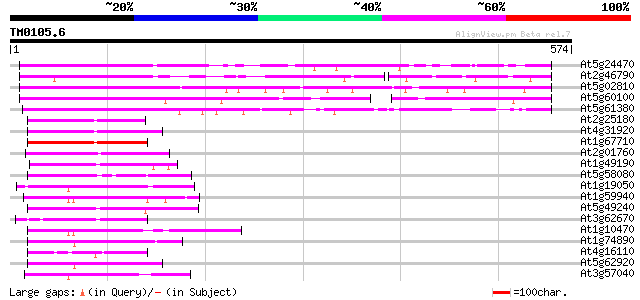
Score E
Sequences producing significant alignments: (bits) Value
At5g24470 pseudo-response regulator 5 (APRR5) 278 4e-75
At2g46790 hypothetical protein 201 9e-52
At5g02810 unknown protein 192 4e-49
At5g60100 pseudo-response regulator 3 (APRR3) 189 3e-48
At5g61380 pseudo-response regulator 1 128 8e-30
At2g25180 putative two-component response regulator protein 97 2e-20
At4g31920 predicted protein 97 3e-20
At1g67710 response regulator 11 (ARR11) 87 2e-17
At2g01760 putative two-component response regulator protein 85 1e-16
At1g49190 putative two-component response regulator gb|AAD12696.1 80 3e-15
At5g58080 ARR2 - like protein 79 5e-15
At1g19050 putative protein 79 9e-15
At1g59940 putative protein 76 4e-14
At5g49240 putative protein 75 8e-14
At3g62670 putative protein 74 2e-13
At1g10470 putative response regulator 3 74 2e-13
At1g74890 putative response regulator (two component phosphotran... 73 5e-13
At4g16110 hypothetical protein 70 3e-12
At5g62920 response regulator 6 (ARR6) 70 4e-12
At3g57040 responce reactor 4 68 1e-11
>At5g24470 pseudo-response regulator 5 (APRR5)
Length = 667
Score = 278 bits (712), Expect = 4e-75
Identities = 210/564 (37%), Positives = 287/564 (50%), Gaps = 96/564 (17%)
Query: 11 ERILPQTALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILT 70
ER LP+ ALR+LLVEADDSTRQIIAALLRKCSY+VAAV DGLKAWE LK K +DLILT
Sbjct: 151 ERFLPKIALRVLLVEADDSTRQIIAALLRKCSYRVAAVPDGLKAWEMLKGKPESVDLILT 210
Query: 71 EVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNEL 130
EVDLPSISG++LLTLI +HD CK+IPVIMMS+ DSV+ V+KCMLKGAAD+L+KP+RRNEL
Sbjct: 211 EVDLPSISGYALLTLIMEHDICKNIPVIMMSTQDSVNTVYKCMLKGAADYLVKPLRRNEL 270
Query: 131 RNLWQHVWRRQTITKPPQKLTLAHNNKLDAAIENKPASNDSTGSVASTQKNNETSEAQST 190
RNLWQHVWRRQT P + N + ++N+S G + +AQS+
Sbjct: 271 RNLWQHVWRRQTSLAPD---SFPWNESVGQQKAEGASANNSNGKRDDHVVSGNGGDAQSS 327
Query: 191 CTSPILGAESGSMENMQDATQLKSSSNLSDIDAVKQDKSTQLEMESPKHNSEAGELSKGN 250
CT P + ES +E +S DAV Q+E + N
Sbjct: 328 CTRPEMEGESADVE-------------VSARDAV------QMECAKSQFN---------- 358
Query: 251 PHISTEIRGCSDELLEPSSGAMDLI-AAFGNLPKRTIGNHSFHDGSTAKFEFDTQLELST 309
E R ++EL + A+D + A+F +R + S A++E +L+LS
Sbjct: 359 -----ETRLLANELQSKQAEAIDFMGASFRRTGRRN------REESVAQYESRIELDLSL 407
Query: 310 R----GDFPASEERLMLNHTNASAFSQY---------SSSKLFSENSPHESHELCENTNI 356
R + +S +R L+ ++ASAF++Y S+S + ++ + + +N +
Sbjct: 408 RRPNASENQSSGDRPSLHPSSASAFTRYVHRPLQTQCSASPVVTDQRKNVAASQDDNIVL 467
Query: 357 SHRYGGKNQNHNPENTNSMVIGQSGQVKTKFPNSQLGFFPA-----TGVSFDNVSTGDGD 411
++Y N N ++QL +P T +N+ D
Sbjct: 468 MNQYNTSEPPPNAPRRNDTSFYTGADSPGPPFSNQLNSWPGQSSYPTPTPINNIQFRD-- 525
Query: 412 VFPSMLYTQ-LTPKSVCQKESSPFPTSTSSQSNNESMKSEHHHWSDDATFTSDKCENDQS 470
P+ YT + P S+ SP P+S S H +S + K E Q
Sbjct: 526 --PNTAYTSAMAPASL-----SPSPSSVSP-----------HEYSSMFHPFNSKPEGLQD 567
Query: 471 NLDCAVNDSSATRQSSSTSFYHDSANHSSSGVNGSGGSGSDGNVTSTGVDDELRGTDSHR 530
DC++ D R SS + + NH + DG S G +
Sbjct: 568 R-DCSM-DVDERRYVSSATEHSAIGNHIDQLIEKKN---EDGYSLSVG--------KIQQ 614
Query: 531 TSQREAALTKFRLKRKDRCYDKKV 554
+ QREAALTKFR+KRKDRCY+KKV
Sbjct: 615 SLQREAALTKFRMKRKDRCYEKKV 638
>At2g46790 hypothetical protein
Length = 454
Score = 201 bits (511), Expect = 9e-52
Identities = 142/396 (35%), Positives = 187/396 (46%), Gaps = 84/396 (21%)
Query: 11 ERILPQTALRILLVEADDSTRQIIAALLRKCSYK----------VAAVRDGLKAWETLKN 60
E+ LP+T LR+LLVE+D STRQII ALLRKC YK AV DGL AWE LK
Sbjct: 29 EKYLPKTVLRVLLVESDYSTRQIITALLRKCCYKGRLVFPFISLFVAVSDGLAAWEVLKE 88
Query: 61 KAHDIDLILTEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADF 120
K+H+IDLILTE+DLPSISGF+LL L+ +H+ CK+IPVIMMSS DS+ MV KCML+GAAD+
Sbjct: 89 KSHNIDLILTELDLPSISGFALLALVMEHEACKNIPVIMMSSQDSIKMVLKCMLRGAADY 148
Query: 121 LIKPVRRNELRNLWQHVWRRQTITKPPQKLTLAHNNKLDAAIENKPASNDSTGSVASTQK 180
LIKP+R+NEL+NLWQHVWRR T+ P AH L A+ N
Sbjct: 149 LIKPMRKNELKNLWQHVWRRLTLRDDP----TAHAQSLPASQHN---------------- 188
Query: 181 NNETSEAQSTCTSPILGAESGSMENMQDATQLKSSSNLSDIDAVKQDKSTQLEMESPKHN 240
+ TC ++ G S A+ + +L ME+ K
Sbjct: 189 ---LEDTDETCEDSRYHSDQG-----------------SGAQAINYNGHNKL-MENGKSV 227
Query: 241 SEAGELSKGNPHISTEIRGCSDELLEPSSGAMDLIAAFGNLPKRTIGNHSFHDGSTAKFE 300
E DE E MDLI P + S + +
Sbjct: 228 DE------------------RDEFKETFDVTMDLIGGIDKRPDSIYKDKSRDECVGPELG 269
Query: 301 FDTQLELSTRGDFPASEERLMLNHTNASAFSQYSSSKLFSE-------------NSPHES 347
+ S + + L+ ++ASAFS++ SK + +P ES
Sbjct: 270 LSLKRSCSVSFENQDESKHQKLSLSDASAFSRFEESKSAEKAVVALEESTSGEPKTPTES 329
Query: 348 HELCENTNISHRYGGKNQNHNPENTNSMVIGQSGQV 383
HE + ++ G + N EN S + QV
Sbjct: 330 HE--KLRKVTSDQGSATTSSNQENIGSSSVSFRNQV 363
Score = 53.5 bits (127), Expect = 3e-07
Identities = 54/192 (28%), Positives = 80/192 (41%), Gaps = 35/192 (18%)
Query: 388 PNSQLGFFPATGVSFDN--------VSTGDGDVFPSMLYTQLTPKSVCQKESSPFPTSTS 439
P L + VSF+N +S D F ++ K+V E S TS
Sbjct: 266 PELGLSLKRSCSVSFENQDESKHQKLSLSDASAFSRFEESKSAEKAVVALEES---TSGE 322
Query: 440 SQSNNESMKSEHHHWSDDATFTSDKCENDQSNLDCA-------VNDSSATRQSSSTSFYH 492
++ ES + SD + T+ ++Q N+ + V S+ T Q +
Sbjct: 323 PKTPTESHEKLRKVTSDQGSATTS---SNQENIGSSSVSFRNQVLQSTVTNQKQDSPIPV 379
Query: 493 DSANHSSSGVNGSGGSGSDGNVTSTGVDDELRGTDSHRT----------SQREAALTKFR 542
+S ++ GS S T+ G+ + T+ + SQREAAL KFR
Sbjct: 380 ESNREKAASKEVEAGSQS----TNEGIAGQSSSTEKPKEEESAKQRWSRSQREAALMKFR 435
Query: 543 LKRKDRCYDKKV 554
LKRKDRC+DKKV
Sbjct: 436 LKRKDRCFDKKV 447
>At5g02810 unknown protein
Length = 727
Score = 192 bits (488), Expect = 4e-49
Identities = 175/630 (27%), Positives = 275/630 (42%), Gaps = 96/630 (15%)
Query: 11 ERILPQTALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILT 70
ER L +R+LLVE DD TR I+ ALLR CSY+V +G++AW+ L++ + ID++LT
Sbjct: 70 ERFLHVRTIRVLLVENDDCTRYIVTALLRNCSYEVVEASNGIQAWKVLEDLNNHIDIVLT 129
Query: 71 EVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNEL 130
EV +P +SG LL I +H ++IPVIMMSS+DS+ +VFKC+ KGA DFL+KP+R+NEL
Sbjct: 130 EVIMPYLSGIGLLCKILNHKSRRNIPVIMMSSHDSMGLVFKCLSKGAVDFLVKPIRKNEL 189
Query: 131 RNLWQHVWRRQTITKPPQKLTLAHNNKLDAAIENKPASNDSTGSVASTQKNNETSEAQST 190
+ LWQHVWRR + + H + ++ S+ +GS + +N S
Sbjct: 190 KILWQHVWRRCQSSSGSGSESGTHQTQKSVKSKSIKKSDQDSGS-SDENENGSIGLNASD 248
Query: 191 CTSPILGAESGSMENMQDATQLKSSSNLSD---------IDAVKQDKSTQL-----EMES 236
+S GA+S + D + +L D + + + S QL E E+
Sbjct: 249 GSSDGSGAQSSWTKKAVDVDDSPRAVSLWDRVDSTCAQVVHSNPEFPSNQLVAPPAEKET 308
Query: 237 PKHNSEAGELSKGNPHISTEIRGC------SDELLEPSSGAMDLIAAF---GNLPKRTIG 287
+H+ + +++ G + R C DE L ++G M +F + K +G
Sbjct: 309 QEHDDKFEDVTMGRDLEISIRRNCDLALEPKDEPLSKTTGIMRQDNSFEKSSSKWKMKVG 368
Query: 288 NHSFHDGSTAKFEFDTQLELSTRGDFPASEERLMLN------HTNASAFSQYSSSKLFSE 341
S + Q+ F A L N +T+ +S SE
Sbjct: 369 KGPLDLSSES--PSSKQMHEDGGSSFKAMSSHLQDNREPEAPNTHLKTLDTNEASVKISE 426
Query: 342 NSPHESHE-------------LCENTNISHR-YGGKNQNHNP-ENTNSMVIGQSGQVKTK 386
H H + ++ N+ R G +NP N N + G G +
Sbjct: 427 ELMHVEHSSKRHRGTKDDGTLVRDDRNVLRRSEGSAFSRYNPASNANKISGGNLGSTSLQ 486
Query: 387 FPNSQLGFFPATGVSFD---------------NVSTGDGDVFPSMLYTQLTPKSVCQKES 431
NSQ T ++D +V + + D+ + T + K
Sbjct: 487 DNNSQ-DLIKKTEAAYDCHSNMNESLPHNHRSHVGSNNFDM------SSTTENNAFTKPG 539
Query: 432 SPFPTSTSSQSNNES----MKSEHH--HWSDDATFTSDK----------------CENDQ 469
+P +S S S S + +HH H S + +++ E +
Sbjct: 540 APKVSSAGSSSVKHSSFQPLPCDHHNNHASYNLVHVAERKKLPPQCGSSNVYNETIEGNN 599
Query: 470 SNLDCAVNDSSATRQSSSTSFYHDSANHSSSGVN--GSGGSGSDGNVTSTGVDDELRG-- 525
+ ++ +VN S + S Y S ++ G+N G+G +GN +G
Sbjct: 600 NTVNYSVNGSVSGSGHGSNGPYGSSNGMNAGGMNMGSDNGAGKNGNGDGSGSGSGSGSGN 659
Query: 526 -TDSHRTSQREAALTKFRLKRKDRCYDKKV 554
D ++ SQREAALTKFR KRK+RC+ KKV
Sbjct: 660 LADENKISQREAALTKFRQKRKERCFRKKV 689
>At5g60100 pseudo-response regulator 3 (APRR3)
Length = 495
Score = 189 bits (481), Expect = 3e-48
Identities = 133/385 (34%), Positives = 197/385 (50%), Gaps = 38/385 (9%)
Query: 11 ERILPQTALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILT 70
ER LP +L++LLVE DDSTR I+ ALL+ CSY+V AV D L+AW L+++ IDL+LT
Sbjct: 56 ERYLPVRSLKVLLVENDDSTRHIVTALLKNCSYEVTAVPDVLEAWRILEDEKSCIDLVLT 115
Query: 71 EVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNEL 130
EVD+P SG LL+ I H K+IPVIMMSS+DS+ +VFKC+ GA DFL+KP+R+NEL
Sbjct: 116 EVDMPVHSGTGLLSKIMSHKTLKNIPVIMMSSHDSMVLVFKCLSNGAVDFLVKPIRKNEL 175
Query: 131 RNLWQHVWRRQTITKPPQKLTLAHNNKL-----------DAAI--ENKPASNDSTGSVAS 177
+NLWQHVWRR + + H+ K DA+I E++ S S G
Sbjct: 176 KNLWQHVWRRCHSSSGSGSESGIHDKKSVKPESTQGSENDASISDEHRNESGSSGGLSNQ 235
Query: 178 TQKNNETSEAQSTCTSPILGAESGSMEN-MQDATQLKSS--------SNLSDIDAVKQDK 228
++ S QS+ T +S S N DA K + + L + + K+
Sbjct: 236 DGGSDNGSGTQSSWTKRASDTKSTSPSNQFPDAPNKKGTYENGCAHVNRLKEAEDQKEQI 295
Query: 229 ST--QLEMESPKHNSEAGELSKGNPHISTEIRGCSDELLEPSSGAMDLIAAFGNLPKRTI 286
T Q M K E G+L K + + +D+ L SSG + + P
Sbjct: 296 GTGSQTGMSMSKKAEEPGDLEKNAKYSVQALERNNDDTLNRSSGNSQVES---KAPSSNR 352
Query: 287 GNHSFHDGSTAKFEFDTQLELSTRGDFPASEERLMLNHTNASAFSQYSSSKLFSENSPHE 346
+ + + K D ++ +R +L H+N SAFS+Y++ ++ +P E
Sbjct: 353 EDLQSLEQTLKKTREDRDYKVG---------DRSVLRHSNLSAFSKYNNGATSAKKAPEE 403
Query: 347 SHELC--ENTNISHRYGGKNQNHNP 369
+ E C ++ I+ G + + NP
Sbjct: 404 NVESCSPHDSPIAKLLGSSSSSDNP 428
Score = 55.1 bits (131), Expect = 1e-07
Identities = 50/175 (28%), Positives = 78/175 (44%), Gaps = 17/175 (9%)
Query: 391 QLGFFPATGVSFDNVSTGDGDVFPSMLYTQLTPKSVCQKESSPFPTSTSSQSNNESMKSE 450
Q+G TG+S + GD+ + Y SV E + T S N++
Sbjct: 294 QIGTGSQTGMSMSKKAEEPGDLEKNAKY------SVQALERNNDDTLNRSSGNSQVESKA 347
Query: 451 HHHWSDDATFTSDKCENDQSNLDCAVNDSSATRQSSSTSF--YHDSANHSSSGV--NGSG 506
+D + + + D V D S R S+ ++F Y++ A + N
Sbjct: 348 PSSNREDLQSLEQTLKKTREDRDYKVGDRSVLRHSNLSAFSKYNNGATSAKKAPEENVES 407
Query: 507 GSGSDGNV-----TSTGVDDELR--GTDSHRTSQREAALTKFRLKRKDRCYDKKV 554
S D + +S+ D+ L+ + S R +QREAAL KFRLKRK+RC++KKV
Sbjct: 408 CSPHDSPIAKLLGSSSSSDNPLKQQSSGSDRWAQREAALMKFRLKRKERCFEKKV 462
>At5g61380 pseudo-response regulator 1
Length = 618
Score = 128 bits (322), Expect = 8e-30
Identities = 146/602 (24%), Positives = 246/602 (40%), Gaps = 123/602 (20%)
Query: 14 LPQTALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVD 73
+ ++ +RILL + D ++ + LL +CSY+V AV+ + + L + DID+IL E+D
Sbjct: 14 IDRSRVRILLCDNDSTSLGEVFTLLSECSYQVTAVKSARQVIDALNAEGPDIDIILAEID 73
Query: 74 LPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNL 133
LP G +L IT + IPVIMMS D V +V KC+ GAAD+L+KP+R NEL NL
Sbjct: 74 LPMAKGMKMLRYITRDKDLRRIPVIMMSRQDEVPVVVKCLKLGAADYLVKPLRTNELLNL 133
Query: 134 WQHVWRRQTITKPPQKLTLAHNNKLDAAIENKPASNDST--------GSVAST--QKNNE 183
W H+WRR+ + +K L+++ L + ++ P +N + S+ ST Q+ N
Sbjct: 134 WTHMWRRRRMLGLAEKNMLSYDFDLVGSDQSDPNTNSTNLFSDDTDDRSLRSTNPQRGNL 193
Query: 184 TSEAQ--STCTSPI------LGAESGSMENMQDAT--------------QLKSSSNLSDI 221
+ + S T+P+ LGA+ + ++ +K +SN +
Sbjct: 194 SHQENEWSVATAPVHARDGGLGADGTATSSLAVTAIEPPLDHLAGSHHEPMKRNSNPAQF 253
Query: 222 DAVKQDKSTQLEMESP----------KHNSEAGELSKGNPHISTEIRGCSDEL-LEPSSG 270
+ + ++ S + N + L GN + RG +++ + S G
Sbjct: 254 SSAPKKSRLKIGESSAFFTYVKSTVLRTNGQDPPLVDGNGSLHLH-RGLAEKFQVVASEG 312
Query: 271 AMDLIAAFGNLPKRTI--------------GNHSFHDGSTAKFEFDTQLELSTRGDFPAS 316
+ A PK T+ G+H H G+ KF+ AS
Sbjct: 313 INNTKQARRATPKSTVLRTNGQDPPLVNGNGSHHLHRGAAEKFQV------------VAS 360
Query: 317 EERLMLNHTNASAFS----QYSSSKLFSENSPHESHELCENTNISHRYGGKNQNHNPENT 372
E +N+T + S QY S +N H L + + +N+ N
Sbjct: 361 EG---INNTKQAHRSRGTEQYHSQGETLQNGASYPHSLERSRTLPTSMESHGRNYQEGNM 417
Query: 373 NSMVIGQSGQVKTKFPNSQLGFFPATGVSFDNVSTGDGDVFPSMLYTQLTPKSVCQKESS 432
N I Q ++K +SQ+ G F + + M + ++ +
Sbjct: 418 N---IPQVAMNRSK-DSSQVD-----GSGFSAPNAYPYYMHGVMNQVMMQSAAMMPQYGH 468
Query: 433 PFPTSTSSQSNNESMKSEHHHWSDDATFTSDKCENDQSNLDCAVNDSSATRQSSSTSFYH 492
P + N + +HH ++ ++ S + Q+ S H
Sbjct: 469 QIPHCQPNHPNGMTGYPYYHH-----------------PMNTSLQHSQMSLQNGQMSMVH 511
Query: 493 DSANHSSSGVNGSGGSGSDGNVTSTGVDDELRGTDSHRTSQREAALTKFRLKRKDRCYDK 552
S + + GN S +E+R ++ +RE AL KFR KR RC+DK
Sbjct: 512 HSWSPA-------------GNPPS----NEVR---VNKLDRREEALLKFRRKRNQRCFDK 551
Query: 553 KV 554
K+
Sbjct: 552 KI 553
>At2g25180 putative two-component response regulator protein
Length = 573
Score = 97.1 bits (240), Expect = 2e-20
Identities = 48/121 (39%), Positives = 73/121 (59%), Gaps = 3/121 (2%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPSIS 78
+R+L V+ D + +I+ +LLR C Y V KA E L+ + DL++++VD+P +
Sbjct: 17 MRVLAVDDDQTCLKILESLLRHCQYHVTTTNQAQKALELLRENKNKFDLVISDVDMPDMD 76
Query: 79 GFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNLWQHVW 138
GF LL L+ +PVIM+S++ V K + GA D+L+KPVR EL+N+WQHV
Sbjct: 77 GFKLLELV---GLEMDLPVIMLSAHSDPKYVMKGVTHGACDYLLKPVRIEELKNIWQHVV 133
Query: 139 R 139
R
Sbjct: 134 R 134
>At4g31920 predicted protein
Length = 552
Score = 96.7 bits (239), Expect = 3e-20
Identities = 48/138 (34%), Positives = 78/138 (55%), Gaps = 3/138 (2%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPSIS 78
+R+L V+ D + +I+ LL++C Y V A E L+ + DL++++VD+P +
Sbjct: 17 MRVLAVDDDQTCLRILQTLLQRCQYHVTTTNQAQTALELLRENKNKFDLVISDVDMPDMD 76
Query: 79 GFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNLWQHVW 138
GF LL L+ +PVIM+S++ V K + GA D+L+KPVR EL+N+WQHV
Sbjct: 77 GFKLLELV---GLEMDLPVIMLSAHSDPKYVMKGVKHGACDYLLKPVRIEELKNIWQHVV 133
Query: 139 RRQTITKPPQKLTLAHNN 156
R+ + K ++ N
Sbjct: 134 RKSKLKKNKSNVSNGSGN 151
>At1g67710 response regulator 11 (ARR11)
Length = 521
Score = 87.4 bits (215), Expect = 2e-17
Identities = 44/123 (35%), Positives = 75/123 (60%), Gaps = 3/123 (2%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPSIS 78
LR+L+V+ D + +I+ +L+KCSY+V +A L+ + D+++++V++P +
Sbjct: 11 LRVLVVDDDPTWLKILEKMLKKCSYEVTTCGLAREALRLLRERKDGYDIVISDVNMPDMD 70
Query: 79 GFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNLWQHVW 138
GF LL + +PVIMMS + S V K + GA D+L+KP+R EL+ +WQHV
Sbjct: 71 GFKLLEHV---GLELDLPVIMMSVDGETSRVMKGVQHGACDYLLKPIRMKELKIIWQHVL 127
Query: 139 RRQ 141
R++
Sbjct: 128 RKK 130
>At2g01760 putative two-component response regulator protein
Length = 382
Score = 85.1 bits (209), Expect = 1e-16
Identities = 50/148 (33%), Positives = 84/148 (55%), Gaps = 3/148 (2%)
Query: 17 TALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPS 76
+ LRIL+V+ D S I+ +L + Y+V A L+ + DL+L++V +P
Sbjct: 9 SGLRILVVDDDTSCLFILEKMLLRLMYQVTICSQADVALTILRERKDSFDLVLSDVHMPG 68
Query: 77 ISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNLWQH 136
++G++LL + + +PVIMMS + + V + GA D+LIKP+R EL+N+WQH
Sbjct: 69 MNGYNLLQQVGLLEM--DLPVIMMSVDGRTTTVMTGINHGACDYLIKPIRPEELKNIWQH 126
Query: 137 VWRRQTITKPPQKLTLA-HNNKLDAAIE 163
V RR+ + K + + A +NK ++E
Sbjct: 127 VVRRKCVMKKELRSSQALEDNKNSGSLE 154
>At1g49190 putative two-component response regulator gb|AAD12696.1
Length = 608
Score = 80.1 bits (196), Expect = 3e-15
Identities = 44/161 (27%), Positives = 88/161 (54%), Gaps = 13/161 (8%)
Query: 21 ILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPSISGF 80
+L+V+ + +T + ++++ +Y+V+ D KA L + H+I++++ + +P I G
Sbjct: 36 VLVVDTNFTTLLNMKQIMKQYAYQVSIETDAEKALAFLTSCKHEINIVIWDFHMPGIDGL 95
Query: 81 SLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNLWQHVWRR 140
L IT +PV++MS ++ V K GA D+++KPV+ + N+WQH+ R+
Sbjct: 96 QALKSITSK---LDLPVVIMSDDNQTESVMKATFYGACDYVVKPVKEEVMANIWQHIVRK 152
Query: 141 QTITK----PPQKLTLAHNNKLDAA------IENKPASNDS 171
+ I K PP + A +++LD +E++P N++
Sbjct: 153 RLIFKPDVAPPVQSDPARSDRLDQVKADFKIVEDEPIINET 193
>At5g58080 ARR2 - like protein
Length = 632
Score = 79.3 bits (194), Expect = 5e-15
Identities = 47/168 (27%), Positives = 88/168 (51%), Gaps = 8/168 (4%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPSIS 78
+R+L V+ + + + + LL +C Y V + KA E L+ ++ DL++++V++P
Sbjct: 18 MRVLAVDDNPTCLRKLEELLLRCKYHVTKTMESRKALEMLRENSNMFDLVISDVEMPDTD 77
Query: 79 GFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNLWQHVW 138
GF LL + + D +PVI S DS V K ++ GA D+L+KPV EL+N+W HV
Sbjct: 78 GFKLLEIGLEMD----LPVITHSDYDS---VMKGIIHGACDYLVKPVGLKELQNIWHHVV 130
Query: 139 RRQTITKPPQKLTLAHNNKLDAAIENKPASNDSTGSVASTQKNNETSE 186
++ I + L + ++ + +A + + +G + + + E
Sbjct: 131 KK-NIKSYAKLLPPSESDSVPSASRKRKDKVNDSGDEDDSDREEDDGE 177
>At1g19050 putative protein
Length = 206
Score = 78.6 bits (192), Expect = 9e-15
Identities = 54/193 (27%), Positives = 99/193 (50%), Gaps = 17/193 (8%)
Query: 8 GDVERILPQTALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETL--------- 58
GD+ P+ L +L V+ R++I LLR S KV V G +A + L
Sbjct: 15 GDLTVTTPE--LHVLAVDDSIVDRKVIERLLRISSCKVTTVESGTRALQYLGLDGGKGAS 72
Query: 59 KNKAHDIDLILTEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAA 118
K ++LI+T+ +P +SG+ LL I + + +PV++MSS + + + +C+ +GA
Sbjct: 73 NLKDLKVNLIVTDYSMPGLSGYDLLKKIKESSAFREVPVVIMSSENILPRIQECLKEGAE 132
Query: 119 DFLIKPVRRNELRNLWQHVWRRQTITKPPQKLTLAHNN--KLDAAIENKPASNDSTGSVA 176
+FL+KPV+ +++ + Q + R + + L+H+N KL + +S+D T
Sbjct: 133 EFLLKPVKLADVKRIKQLIMRNEA----EECKILSHSNKRKLQEDSDTSSSSHDDTSIKD 188
Query: 177 STQKNNETSEAQS 189
S+ SE+++
Sbjct: 189 SSCSKRMKSESEN 201
>At1g59940 putative protein
Length = 231
Score = 76.3 bits (186), Expect = 4e-14
Identities = 59/203 (29%), Positives = 106/203 (52%), Gaps = 24/203 (11%)
Query: 15 PQTALRILLVEADDST--RQIIAALLRKCSYKVAAVRDGLKAWETL---KNKAH------ 63
P + ++ ++ DDS R +I LLR S KV AV G +A E L +KA
Sbjct: 27 PLDSDQVHVLAVDDSLVDRIVIERLLRITSCKVTAVDSGWRALEFLGLDDDKAAVEFDRL 86
Query: 64 DIDLILTEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIK 123
+DLI+T+ +P ++G+ LL I + K +PV++MSS + ++ + +C+ +GA DFL+K
Sbjct: 87 KVDLIITDYCMPGMTGYELLKKIKESTSFKEVPVVIMSSENVMTRIDRCLEEGAEDFLLK 146
Query: 124 PVRRNELRNLWQHVWR---------RQTITKPPQKLTLAHNNKL---DAAIENKPASNDS 171
PV+ +++ L ++ R ++ +T PP L+ + + D+ +E+ + D
Sbjct: 147 PVKLADVKRLRSYLTRDVKVAAEGNKRKLTTPPPPPPLSATSSMESSDSTVESPLSMVDD 206
Query: 172 TGSVASTQKNNETSEAQSTCTSP 194
S+ + + + TS S SP
Sbjct: 207 EDSLTMSPE-SATSLVDSPMRSP 228
>At5g49240 putative protein
Length = 292
Score = 75.5 bits (184), Expect = 8e-14
Identities = 54/181 (29%), Positives = 89/181 (48%), Gaps = 9/181 (4%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPSIS 78
LR+L+ + D S I+ L+K Y+V + KA TL+N + DL + +V+
Sbjct: 42 LRVLVFDEDPSYLLILERHLQKFQYQVTICNEVNKAMHTLRNHRNRFDLAMIQVNNAEGD 101
Query: 79 GFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNELRNLWQHV- 137
F L+ I +P+I++S +DSV V K M+ GAAD+LIKP+R +LR +++H+
Sbjct: 102 IFRFLSEIGSE---MDLPIIIISEDDSVKSVKKWMINGAADYLIKPIRPEDLRIVFKHLV 158
Query: 138 ----WRRQTITKPPQKLTLAHNNKL-DAAIENKPASNDSTGSVASTQKNNETSEAQSTCT 192
RR +T +K ++ + D+ I N S S+ A + + C
Sbjct: 159 KKMRERRSVVTGEAEKAAGEKSSSVGDSTIRNPNKSKRSSCLEAEVNEEDRHDHNDRACA 218
Query: 193 S 193
S
Sbjct: 219 S 219
>At3g62670 putative protein
Length = 426
Score = 73.9 bits (180), Expect = 2e-13
Identities = 44/135 (32%), Positives = 73/135 (53%), Gaps = 6/135 (4%)
Query: 7 NGDVERILPQTALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDID 66
N D E L ++ R+LLV AD ++ + L+ + SY+V G +A L H+ID
Sbjct: 30 NDDDEEFLTKSN-RVLLVGADSNSS--LKNLMTQYSYQVTKYESGEEAMAFLMKNKHEID 86
Query: 67 LILTEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVR 126
L++ + +P I+G L +I +PV++MS V + + GA DFL+KPV
Sbjct: 87 LVIWDFHMPDINGLDALNIIGKQ---MDLPVVIMSHEYKKETVMESIKYGACDFLVKPVS 143
Query: 127 RNELRNLWQHVWRRQ 141
+ + LW+HV+R++
Sbjct: 144 KEVIAVLWRHVYRKR 158
>At1g10470 putative response regulator 3
Length = 259
Score = 73.9 bits (180), Expect = 2e-13
Identities = 62/228 (27%), Positives = 102/228 (44%), Gaps = 23/228 (10%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETL---KNKAH------DIDLIL 69
+ +L V+ R +I LLR S KV AV G +A E L KA +DLI+
Sbjct: 34 VHVLAVDDSLVDRIVIERLLRITSCKVTAVDSGWRALEFLGLDNEKASAEFDRLKVDLII 93
Query: 70 TEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNE 129
T+ +P ++G+ LL I + + +PV++MSS + ++ + +C+ +GA DFL+KPV+ +
Sbjct: 94 TDYCMPGMTGYELLKKIKESSNFREVPVVIMSSENVLTRIDRCLEEGAQDFLLKPVKLAD 153
Query: 130 LRNLWQHVWRRQTITKPPQKLTLAHNNKLDAAIENKPASNDSTGSVASTQKNNETSEAQS 189
++ L H+ + + L++ NK P + S S T +S
Sbjct: 154 VKRLRSHL---------TKDVKLSNGNK-----RKLPEDSSSVNSSLPPPSPPLTISPES 199
Query: 190 TCTSPILGAESGSMENMQDATQLKSSSNLSDIDAVKQDKSTQLEMESP 237
+ + S S + +S S ID D T ESP
Sbjct: 200 SPPLTVSTESSDSSPPLSPVEIFSTSPLSSPIDDEDDDVLTSSSEESP 247
>At1g74890 putative response regulator (two component
phosphotransfer signaling)
Length = 206
Score = 72.8 bits (177), Expect = 5e-13
Identities = 46/168 (27%), Positives = 89/168 (52%), Gaps = 10/168 (5%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHD---------IDLIL 69
L +L V+ R++I LL+ + KV V G +A + L + ++LI+
Sbjct: 18 LHVLAVDDSFVDRKVIERLLKISACKVTTVESGTRALQYLGLDGDNGSSGLKDLKVNLIV 77
Query: 70 TEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNE 129
T+ +P ++G+ LL I + + IPV++MSS + + +CM++GA +FL+KPV+ +
Sbjct: 78 TDYSMPGLTGYELLKKIKESSALREIPVVIMSSENIQPRIEQCMIEGAEEFLLKPVKLAD 137
Query: 130 LRNLWQHVWRRQTITKPPQKLTLAHNNKLDAAIENKPASNDSTGSVAS 177
++ L + + R + K L+ L I++ P+S+ ++ S +S
Sbjct: 138 VKRLKELIMRGGEAEEGKTK-KLSPKRILQNDIDSSPSSSSTSSSSSS 184
>At4g16110 hypothetical protein
Length = 644
Score = 70.1 bits (170), Expect = 3e-12
Identities = 45/131 (34%), Positives = 74/131 (56%), Gaps = 14/131 (10%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHDIDLILTEVDLPSIS 78
LR+L+V+ D + I+ +L C Y+ R + KN D+++++V +P +
Sbjct: 28 LRVLVVDDDPTCLMILERMLMTCLYREQ--RAHCLCFGRTKN---GFDIVISDVHMPDMD 82
Query: 79 GFSLLTLI--------TDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNEL 130
GF LL + + + K + VI+MS++DS S+V K + GA D+LIKPVR L
Sbjct: 83 GFKLLEHVGLEMDLPVINLNVLKPL-VIVMSADDSKSVVLKGVTHGAVDYLIKPVRIEAL 141
Query: 131 RNLWQHVWRRQ 141
+N+WQHV R++
Sbjct: 142 KNIWQHVVRKK 152
>At5g62920 response regulator 6 (ARR6)
Length = 186
Score = 69.7 bits (169), Expect = 4e-12
Identities = 43/149 (28%), Positives = 78/149 (51%), Gaps = 11/149 (7%)
Query: 19 LRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETLKNKAHD---------IDLIL 69
L +L V+ R+ I LLR S KV V +A + L + ++LI+
Sbjct: 25 LHVLAVDDSHVDRKFIERLLRVSSCKVTVVDSATRALQYLGLDVEEKSVGFEDLKVNLIM 84
Query: 70 TEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKGAADFLIKPVRRNE 129
T+ +P ++G+ LL I + + +PV++MSS + + + +C+ +GA DFL+KPV+ ++
Sbjct: 85 TDYSMPGMTGYELLKKIKESSAFREVPVVIMSSENILPRIDRCLEEGAEDFLLKPVKLSD 144
Query: 130 LRNLWQHVWRRQ--TITKPPQKLTLAHNN 156
++ L + + + + TK QK L N
Sbjct: 145 VKRLRDSLMKVEDLSFTKSIQKRELETEN 173
>At3g57040 responce reactor 4
Length = 234
Score = 68.2 bits (165), Expect = 1e-11
Identities = 48/189 (25%), Positives = 86/189 (45%), Gaps = 30/189 (15%)
Query: 16 QTALRILLVEADDSTRQIIAALLRKCSYKVAAVRDGLKAWETL----------------- 58
++ +L V+ R++I LL+K S +V V G KA E L
Sbjct: 6 ESQFHVLAVDDSLFDRKLIERLLQKSSCQVTTVDSGSKALEFLGLRQSTDSNDPNAFSKA 65
Query: 59 --KNKAHDIDLILTEVDLPSISGFSLLTLITDHDFCKHIPVIMMSSNDSVSMVFKCMLKG 116
++ +++LI+T+ +P ++G+ LL + + + IPV++MSS + + + +C+ +G
Sbjct: 66 PVNHQVVEVNLIITDYCMPGMTGYDLLKKVKESSAFRDIPVVIMSSENVPARISRCLEEG 125
Query: 117 AADFLIKPVRRNELRNLWQHVWRRQTITKPPQKLTLAHNNKLDAAIENKPASNDSTGSVA 176
A +F +KPVR +L L KP T N KL+ N +VA
Sbjct: 126 AEEFFLKPVRLADLNKL-----------KPHMMKTKLKNQKLEEIETTSKVENGVPTAVA 174
Query: 177 STQKNNETS 185
+ + T+
Sbjct: 175 DPEIKDSTN 183
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.126 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,222,652
Number of Sequences: 26719
Number of extensions: 575116
Number of successful extensions: 2305
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 2159
Number of HSP's gapped (non-prelim): 171
length of query: 574
length of database: 11,318,596
effective HSP length: 105
effective length of query: 469
effective length of database: 8,513,101
effective search space: 3992644369
effective search space used: 3992644369
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0105.6


