

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0104.7
(76 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
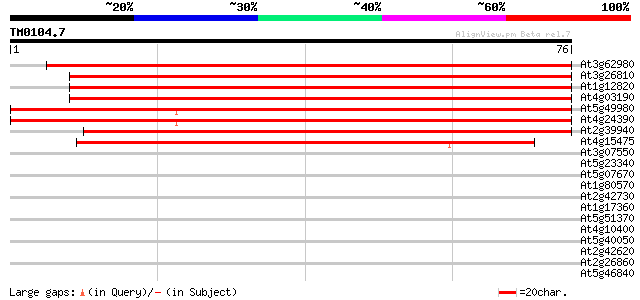
Score E
Sequences producing significant alignments: (bits) Value
At3g62980 transport inhibitor response 1 (TIR1) 95 7e-21
At3g26810 transport inhibitor response 1-like protein 92 4e-20
At1g12820 transport inhibitor response 1, putative 92 6e-20
At4g03190 putative homolog of transport inhibitor response 1 89 5e-19
At5g49980 transport inhibitor response 1 protein 79 3e-16
At4g24390 transport inhibitor response-like protein 77 1e-15
At2g39940 unknown protein 64 1e-11
At4g15475 unknown protein 43 2e-05
At3g07550 unknown protein 35 0.009
At5g23340 unknown protein 33 0.034
At5g07670 unknown protein 32 0.057
At1g80570 unknown protein 32 0.075
At2g42730 hypothetical protein 31 0.097
At1g17360 COP1-Interacting Protein 7, putative 30 0.22
At5g51370 putative protein 29 0.48
At4g10400 putative protein 29 0.48
At5g40050 putative protein 28 0.63
At2g42620 F-box containing protein ORE9 (ORE9) 28 0.63
At2g26860 unknown protein 28 0.63
At5g46840 unknown protein 28 0.83
>At3g62980 transport inhibitor response 1 (TIR1)
Length = 594
Score = 94.7 bits (234), Expect = 7e-21
Identities = 44/71 (61%), Positives = 51/71 (70%)
Query: 6 ASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRS 65
A FP+EVLE V ++ KDR+SVSLVCK WY ERW RR VFIGNCY+VSP + RR
Sbjct: 6 ALSFPEEVLEHVFSFIQLDKDRNSVSLVCKSWYEIERWCRRKVFIGNCYAVSPATVIRRF 65
Query: 66 SNIRSVTLKGK 76
+RSV LKGK
Sbjct: 66 PKVRSVELKGK 76
>At3g26810 transport inhibitor response 1-like protein
Length = 575
Score = 92.4 bits (228), Expect = 4e-20
Identities = 41/68 (60%), Positives = 53/68 (77%)
Query: 9 FPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNI 68
FPDEV+E V V S KDR+++SLVCK WY ER+SR+ VFIGNCY+++PE L RR +
Sbjct: 4 FPDEVIEHVFDFVTSHKDRNAISLVCKSWYKIERYSRQKVFIGNCYAINPERLLRRFPCL 63
Query: 69 RSVTLKGK 76
+S+TLKGK
Sbjct: 64 KSLTLKGK 71
>At1g12820 transport inhibitor response 1, putative
Length = 577
Score = 91.7 bits (226), Expect = 6e-20
Identities = 41/68 (60%), Positives = 53/68 (77%)
Query: 9 FPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNI 68
FPDEV+E V V S KDR+S+SLVCK W+ ER+SR+ VFIGNCY+++PE L RR +
Sbjct: 4 FPDEVIEHVFDFVASHKDRNSISLVCKSWHKIERFSRKEVFIGNCYAINPERLIRRFPCL 63
Query: 69 RSVTLKGK 76
+S+TLKGK
Sbjct: 64 KSLTLKGK 71
>At4g03190 putative homolog of transport inhibitor response 1
Length = 585
Score = 88.6 bits (218), Expect = 5e-19
Identities = 39/68 (57%), Positives = 52/68 (76%)
Query: 9 FPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNI 68
FP +VLE +L + S +DR+SVSLVCK W+ ER +R+ VF+GNCY+VSP +TRR +
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 69 RSVTLKGK 76
RS+TLKGK
Sbjct: 65 RSLTLKGK 72
>At5g49980 transport inhibitor response 1 protein
Length = 619
Score = 79.3 bits (194), Expect = 3e-16
Identities = 40/80 (50%), Positives = 52/80 (65%), Gaps = 4/80 (5%)
Query: 1 NILNQASPFPDEVLERVLGMV----KSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSV 56
N ++ + FPD VLE VL V SR DR++ SLVCK W+ E +R VFIGNCY++
Sbjct: 42 NCISNSQTFPDHVLENVLENVLQFLDSRCDRNAASLVCKSWWRVEALTRSEVFIGNCYAL 101
Query: 57 SPEILTRRSSNIRSVTLKGK 76
SP LT+R +RS+ LKGK
Sbjct: 102 SPARLTQRFKRVRSLVLKGK 121
>At4g24390 transport inhibitor response-like protein
Length = 623
Score = 77.4 bits (189), Expect = 1e-15
Identities = 41/80 (51%), Positives = 49/80 (61%), Gaps = 4/80 (5%)
Query: 1 NILNQASPFPDEVLERVLGMV----KSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSV 56
N + P PD VLE VL V SR DR++VSLVC+ WY E +R VFIGNCYS+
Sbjct: 42 NFKSSPPPCPDHVLENVLENVLQFLTSRCDRNAVSLVCRSWYRVEAQTRLEVFIGNCYSL 101
Query: 57 SPEILTRRSSNIRSVTLKGK 76
SP L R +RS+ LKGK
Sbjct: 102 SPARLIHRFKRVRSLVLKGK 121
>At2g39940 unknown protein
Length = 592
Score = 64.3 bits (155), Expect = 1e-11
Identities = 28/66 (42%), Positives = 44/66 (66%)
Query: 11 DEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNIRS 70
D+V+E+V+ + KDR S SLVC+ W+ + +R V + CY+ +P+ L+RR N+RS
Sbjct: 18 DDVIEQVMTYITDPKDRDSASLVCRRWFKIDSETREHVTMALCYTATPDRLSRRFPNLRS 77
Query: 71 VTLKGK 76
+ LKGK
Sbjct: 78 LKLKGK 83
>At4g15475 unknown protein
Length = 610
Score = 43.1 bits (100), Expect = 2e-05
Identities = 22/64 (34%), Positives = 39/64 (60%), Gaps = 2/64 (3%)
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSP--EILTRRSSN 67
P+E++ + ++S+ +R + SLVCK W + ER+SR ++ IG +S +L+RR
Sbjct: 12 PEELILEIFRRLESKPNRDACSLVCKRWLSLERFSRTTLRIGASFSPDDFISLLSRRFLY 71
Query: 68 IRSV 71
I S+
Sbjct: 72 ITSI 75
>At3g07550 unknown protein
Length = 395
Score = 34.7 bits (78), Expect = 0.009
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 1/71 (1%)
Query: 1 NILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSV-SPE 59
N+ PD+ L + + S D S L C W N + SRRS+ +SV +P
Sbjct: 10 NVETSIIHLPDDCLSFIFQRLDSVADHDSFGLTCHRWLNIQNISRRSLQFQCSFSVLNPS 69
Query: 60 ILTRRSSNIRS 70
L++ + ++ S
Sbjct: 70 SLSQTNPDVSS 80
>At5g23340 unknown protein
Length = 405
Score = 32.7 bits (73), Expect = 0.034
Identities = 18/61 (29%), Positives = 27/61 (43%), Gaps = 4/61 (6%)
Query: 11 DEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNIRS 70
D+ L VL + S KD+ LVCK W N + R+ + P +L R +S
Sbjct: 12 DDELRWVLSRLDSDKDKEVFGLVCKRWLNLQSTDRKKL----AARAGPHMLRRLASRFTQ 67
Query: 71 V 71
+
Sbjct: 68 I 68
>At5g07670 unknown protein
Length = 476
Score = 32.0 bits (71), Expect = 0.057
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 1/64 (1%)
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNIR 69
PD +L RV+ + + + R ++SLVCK W+ RS + + +S L R N+
Sbjct: 66 PDLILIRVIQKIPNSQ-RKNLSLVCKRWFRLHGRLVRSFKVSDWEFLSSGRLISRFPNLE 124
Query: 70 SVTL 73
+V L
Sbjct: 125 TVDL 128
>At1g80570 unknown protein
Length = 467
Score = 31.6 bits (70), Expect = 0.075
Identities = 14/42 (33%), Positives = 26/42 (61%)
Query: 10 PDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIG 51
PD ++ +L + + DR+S+SL CK +++ + R S+ IG
Sbjct: 5 PDHLVWDILSKLHTTDDRNSLSLSCKRFFSLDNEQRYSLRIG 46
>At2g42730 hypothetical protein
Length = 737
Score = 31.2 bits (69), Expect = 0.097
Identities = 26/91 (28%), Positives = 41/91 (44%), Gaps = 18/91 (19%)
Query: 1 NILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYN---------------AERWSR 45
N + S PDEVL R+L ++ S K+ S S++ K W N + +
Sbjct: 2 NAKDVISRLPDEVLGRILSLI-STKEAVSTSVLSKRWKNMFVLVSNLDIDDRQSVPKTKQ 60
Query: 46 RSVFIGNCYSVSPEIL--TRRSSNIRSVTLK 74
+ I Y + L T+R S+I+ +TLK
Sbjct: 61 NRIEIHRNYMAFVDKLLDTQRGSSIKKLTLK 91
>At1g17360 COP1-Interacting Protein 7, putative
Length = 1061
Score = 30.0 bits (66), Expect = 0.22
Identities = 19/49 (38%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Query: 16 RVLGMVKSRKDRSSVSLVCKEWYNAERW-SRRSVFIGNCYSVSPEILTR 63
RV+ S++D+SSV L ++ N E W +RR++ Y SPE+L R
Sbjct: 47 RVIESQASKRDQSSVRLEVEQSENGESWFTRRTLERFVQYVNSPEVLER 95
>At5g51370 putative protein
Length = 446
Score = 28.9 bits (63), Expect = 0.48
Identities = 17/46 (36%), Positives = 24/46 (51%)
Query: 28 SSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRSSNIRSVTL 73
+ VSLVCK W N + RS+ + + + E LT R N+ V L
Sbjct: 50 NDVSLVCKRWLNLQGQRLRSLKLLDFDFLLSERLTTRFPNLTHVDL 95
>At4g10400 putative protein
Length = 326
Score = 28.9 bits (63), Expect = 0.48
Identities = 18/55 (32%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Query: 3 LNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVS 57
+++ S PDEVL ++L V + K S S++ K W W + F YS S
Sbjct: 1 MDRISGLPDEVLVKILSFVPT-KVAVSTSILSKRWEFLWMWLTKLKFGSKRYSES 54
>At5g40050 putative protein
Length = 415
Score = 28.5 bits (62), Expect = 0.63
Identities = 11/35 (31%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query: 3 LNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEW 37
++ SP PDE+L +L + +++ +S S++ K W
Sbjct: 13 IDSISPLPDELLSHILSFLPTKR-AASTSILSKRW 46
>At2g42620 F-box containing protein ORE9 (ORE9)
Length = 693
Score = 28.5 bits (62), Expect = 0.63
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query: 7 SPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFI-GNCYSVS 57
S PD +L + +V + R+S+SLV ++ ER +R + I GN +S
Sbjct: 7 SDLPDVILSTISSLVSDSRARNSLSLVSHKFLALERSTRSHLTIRGNARDLS 58
>At2g26860 unknown protein
Length = 405
Score = 28.5 bits (62), Expect = 0.63
Identities = 18/67 (26%), Positives = 32/67 (46%), Gaps = 1/67 (1%)
Query: 3 LNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILT 62
+++ S PDE+L +L + + K+ S S++ K W W + F+ N Y I
Sbjct: 1 MDRISGLPDELLVEILHCLPT-KEVVSTSILSKRWEFLWLWVPKLTFVMNHYESDLPIQD 59
Query: 63 RRSSNIR 69
+ N+R
Sbjct: 60 FITKNLR 66
>At5g46840 unknown protein
Length = 501
Score = 28.1 bits (61), Expect = 0.83
Identities = 13/41 (31%), Positives = 25/41 (60%)
Query: 12 EVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGN 52
E+ E+ +G + + D + ++V KE ++ E R+VF+GN
Sbjct: 137 EMKEKKVGEKRKKADEVADTMVSKEGFDDESKLLRTVFVGN 177
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,637,321
Number of Sequences: 26719
Number of extensions: 50887
Number of successful extensions: 197
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 168
Number of HSP's gapped (non-prelim): 44
length of query: 76
length of database: 11,318,596
effective HSP length: 52
effective length of query: 24
effective length of database: 9,929,208
effective search space: 238300992
effective search space used: 238300992
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0104.7


