

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
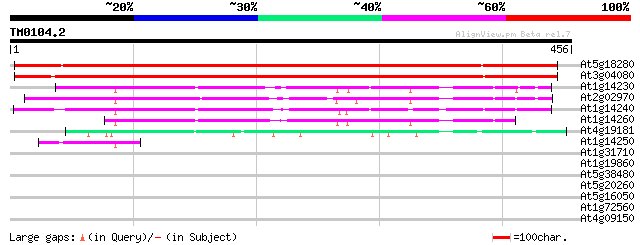
Query= TM0104.2
(456 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g18280 apyrase (gb|AAF00612.1) 479 e-135
At3g04080 apyrase (Atapy1) 478 e-135
At1g14230 putative nucleoside triphosphatase 157 1e-38
At2g02970 nucleoside triphosphatase like protein 155 3e-38
At1g14240 putative nucleoside triphosphatase 152 3e-37
At1g14260 hypothetical protein, 5' partial 134 1e-31
At4g19181 putative protein 81 1e-15
At1g14250 putative nucleoside triphosphatase, 3' partial 50 3e-06
At1g31710 putative protein 31 1.6
At1g19860 unknown protein 30 3.6
At5g38480 14-3-3 protein homolog RCI1 (pir||S47969) 29 4.7
At5g20260 putative protein 29 4.7
At5g16050 14-3-3-LIKE PROTEIN GF14 UPSILON 29 4.7
At1g72560 PAUSED 29 4.7
At4g09150 unknown protein 28 8.0
>At5g18280 apyrase (gb|AAF00612.1)
Length = 472
Score = 479 bits (1233), Expect = e-135
Identities = 243/443 (54%), Positives = 324/443 (72%), Gaps = 4/443 (0%)
Query: 5 ISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNF 64
I L+ V +LMP S S + ++ + P ++ P +YAVIFDAGS+GSRVHVY F
Sbjct: 31 IVLIGLVLLLMPGRSISDSVVEEYSVHNRKGGPNSRGP-KNYAVIFDAGSSGSRVHVYCF 89
Query: 65 DQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKL 124
DQNLDL+P+ NELE + +KPGLS+Y +P +AA SL+ LL +AE VP +P T V++
Sbjct: 90 DQNLDLIPLGNELELFLQLKPGLSAYPTDPRQAANSLVSLLDKAEASVPRELRPKTHVRV 149
Query: 125 GATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGK 184
GATAGLR L +A+ENILQAVR++L +RS L +++AV++LDGTQEGSY WVTINYLL
Sbjct: 150 GATAGLRTLGHDASENILQAVRELLRDRSMLKTEANAVTVLDGTQEGSYQWVTINYLLRN 209
Query: 185 LGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHS 244
LGK ++ TVGVVDLGGGSVQM YA+S A +APK EGED Y++++ L+G+KY LYVHS
Sbjct: 210 LGKPYSDTVGVVDLGGGSVQMAYAISEEDAASAPKPLEGEDSYVREMYLKGRKYFLYVHS 269
Query: 245 YLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALK 304
YL YG A RAEI KV+ S NPCI+AG+DG Y Y G E+K A SG++L++CR+I +
Sbjct: 270 YLHYGLLAARAEILKVSEDSENPCIVAGYDGMYKYGGKEFKAPASQSGASLDECRRITIN 329
Query: 305 ALKVN-APCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVNKPNAKIRPVD 362
ALKVN C + CTFGG+WNGG G GQKN+F+ S F+ + + G + +P A +RP+D
Sbjct: 330 ALKVNDTLCTHMKCTFGGVWNGGRGGGQKNMFVASFFFDRAAEAGFVDPKQPVATVRPMD 389
Query: 363 LKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANE 422
+ AAK AC LE+ KS +P L E++++ Y+C+DLVY YTLL+DGFGL+P Q +T+ +
Sbjct: 390 FEKAAKKACSMKLEEGKSTFP-LVEEENLPYLCMDLVYQYTLLIDGFGLEPSQTITLVKK 448
Query: 423 IEYQDALVEAAWPLGTAIEAISS 445
++Y D VEAAWPLG+AIEA+SS
Sbjct: 449 VKYGDQAVEAAWPLGSAIEAVSS 471
>At3g04080 apyrase (Atapy1)
Length = 471
Score = 478 bits (1230), Expect = e-135
Identities = 242/443 (54%), Positives = 320/443 (71%), Gaps = 5/443 (1%)
Query: 5 ISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNF 64
I L+ V +LMP S+S + + N + N +YAVIFDAGS+GSRVHVY F
Sbjct: 31 IVLIALVLLLMPGTSTSVSVIEYTMKNHEG--GSNSRGPKNYAVIFDAGSSGSRVHVYCF 88
Query: 65 DQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKL 124
DQNLDL+P+ENELE + +KPGLS+Y +P ++A SL+ LL +AE VP +P TPV++
Sbjct: 89 DQNLDLVPLENELELFLQLKPGLSAYPNDPRQSANSLVTLLDKAEASVPRELRPKTPVRV 148
Query: 125 GATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGK 184
GATAGLR L A+ENILQAVR++L RS L +++AV++LDGTQEGSY WVTINYLL
Sbjct: 149 GATAGLRALGHQASENILQAVRELLKGRSRLKTEANAVTVLDGTQEGSYQWVTINYLLRT 208
Query: 185 LGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHS 244
LGK ++ TVGVVDLGGGSVQM YA+ A APK EGED Y++++ L+G+KY LYVHS
Sbjct: 209 LGKPYSDTVGVVDLGGGSVQMAYAIPEEDAATAPKPVEGEDSYVREMYLKGRKYFLYVHS 268
Query: 245 YLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALK 304
YL YG A RAEI KV+ S NPCI G+ G Y Y G +K +A SG++L++CR++A+
Sbjct: 269 YLHYGLLAARAEILKVSEDSNNPCIATGYAGTYKYGGKAFKAAASPSGASLDECRRVAIN 328
Query: 305 ALKV-NAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVNKPNAKIRPVD 362
ALKV N+ C + CTFGG+WNGGGG GQK +F+ S F+ + + G + N+P A++RP+D
Sbjct: 329 ALKVNNSLCTHMKCTFGGVWNGGGGGGQKKMFVASFFFDRAAEAGFVDPNQPVAEVRPLD 388
Query: 363 LKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANE 422
+ AA AC +E+ KSK+P + E+D++ Y+CLDLVY YTLLVDGFGL P Q +T+ +
Sbjct: 389 FEKAANKACNMRMEEGKSKFPRV-EEDNLPYLCLDLVYQYTLLVDGFGLKPSQTITLVKK 447
Query: 423 IEYQDALVEAAWPLGTAIEAISS 445
++Y D VEAAWPLG+AIEA+SS
Sbjct: 448 VKYGDYAVEAAWPLGSAIEAVSS 470
>At1g14230 putative nucleoside triphosphatase
Length = 503
Score = 157 bits (396), Expect = 1e-38
Identities = 125/430 (29%), Positives = 201/430 (46%), Gaps = 54/430 (12%)
Query: 38 KNQEPVTSYAVIFDAGSTGSRVHVYNFDQNLDLLPVENELEFYDSVK--PGLSSYAANPE 95
+N+ Y+VI D GS+G+RVHV+ + + E Y S+K PGLS+YA NPE
Sbjct: 73 RNRRVSLHYSVIIDGGSSGTRVHVFGYRIESGKPVFDFGEENYASLKLSPGLSAYADNPE 132
Query: 96 EAAESLIPLLKEAENVVPVSQQPNTPVKLGATAGLRLLEGNAAENILQAVRDMLSNRSAL 155
+ES+ L++ A+ V + + ++L ATAG+RLLE E IL R +L + S
Sbjct: 133 GVSESVTELVEFAKKRVHKGKLKKSDIRLMATAGMRLLELPVQEQILDVTRRVLRS-SGF 191
Query: 156 NVQSDAVSILDGTQEGSYLWVTINYLLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAK 215
+ + + S++ G+ EG Y WV N+ LG LG KT G+V+LGG S Q+T+
Sbjct: 192 DFRDEWASVISGSDEGVYAWVVANHALGSLGGEPLKTTGIVELGGASAQVTFV------- 244
Query: 216 NAPKVPEGEDPYIKKLVLQGKKYDLYVHSYLRYGREAFRAEIFKVAGGSA---------- 265
+ VP + + L Y+LY HS+L +G++A + ++ + SA
Sbjct: 245 STELVP---SEFSRTLAYGNVSYNLYSHSFLDFGQDAAQEKLSESLYNSAANSTGEGIVP 301
Query: 266 NPCILAGFD---------GAYTYSGAEYKVSAPASGSNLNQCRKIALKAL-KVNAPCPYQ 315
+PCI G+ + ++ + A+G N ++CR A L + C Y+
Sbjct: 302 DPCIPKGYILETNLQKDLPGFLADKGKFTATLQAAG-NFSECRSAAFAMLQEEKGKCTYK 360
Query: 316 NCTFGGIWN---GGGGSGQKNLFLTSSFYYLSEDVGIFVNKPNAKIRPVDLKTAAKLACK 372
C+ G I+ G +N F TS F+ L E + ++ A K C
Sbjct: 361 RCSIGSIFTPNLQGSFLATENFFHTSKFFGLGEKEWL-----------SEMILAGKRFCG 409
Query: 373 TNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFG--LDPFQEVTVANEIEYQDALV 430
K KYP +++ + Y C Y+ ++L D G LD + + A++ +D +
Sbjct: 410 EEWSKLKVKYPTFKDENLLRY-CFSSAYIISMLHDSLGVALDD-ERIKYASKAGEED--I 465
Query: 431 EAAWPLGTAI 440
W LG I
Sbjct: 466 PLDWALGAFI 475
>At2g02970 nucleoside triphosphatase like protein
Length = 555
Score = 155 bits (393), Expect = 3e-38
Identities = 136/461 (29%), Positives = 210/461 (45%), Gaps = 62/461 (13%)
Query: 13 MLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNFDQNLDLLP 72
+L+ S S LG L + N Y+V+ D GSTG+R+HV+ +
Sbjct: 54 LLLTVGSISVVLGVLFLCYSILFSGGNLRGSLRYSVVIDGGSTGTRIHVFGYRIESGKPV 113
Query: 73 VENELEFYDSVK--PGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLGATAGL 130
E Y S+K PGLS++A +P+ A+ SL L++ A+ VP T V+L ATAG+
Sbjct: 114 FEFRGANYASLKLHPGLSAFADDPDGASVSLTELVEFAKGRVPKGMWIETEVRLMATAGM 173
Query: 131 RLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGKLGKRFT 190
RLLE E IL R +L + L + + S++ G+ EG Y WV N+ LG LG
Sbjct: 174 RLLELPVQEKILGVARRVLKSSGFL-FRDEWASVISGSDEGVYAWVVANFALGSLGGDPL 232
Query: 191 KTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHSYLRYGR 250
KT G+V+LGG S Q+T+ S P PE + + + Y+LY HS+L +G+
Sbjct: 233 KTTGIVELGGASAQVTFVSSE------PMPPE----FSRTISFGNVTYNLYSHSFLHFGQ 282
Query: 251 EAFRAEIFKVAGGS------------------ANPCILAGFD-GAYTYS------GAEYK 285
A +++ GS +PC G++ A T E +
Sbjct: 283 NAAHDKLW----GSLLSRDHNSAVEPTREKIFTDPCAPKGYNLDANTQKHLSGLLAEESR 338
Query: 286 VS-APASGSNLNQCRKIALKALK-VNAPCPYQNCTFGGIWN---GGGGSGQKNLFLTSSF 340
+S + +G N +QCR AL L+ N C YQ+C+ G + G +N F TS F
Sbjct: 339 LSDSFQAGGNYSQCRSAALTILQDGNEKCSYQHCSIGSTFTPKLRGRFLATENFFYTSKF 398
Query: 341 YYLSEDVGIFVNKPNAKIRPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVY 400
+ L E + ++ +A + C + + K P L+E+D + Y C Y
Sbjct: 399 FGLGEKAWL-----------SNMISAGERFCGEDWSKLRVKDPSLHEEDLLRY-CFSSAY 446
Query: 401 VYTLLVDGFGLDPFQEVTVANEIEYQDALVEAAWPLGTAIE 441
+ +LL D G+ P + + + D + W LG I+
Sbjct: 447 IVSLLHDTLGI-PLDDERIGYANQAGD--IPLDWALGAFIQ 484
>At1g14240 putative nucleoside triphosphatase
Length = 483
Score = 152 bits (385), Expect = 3e-37
Identities = 123/458 (26%), Positives = 212/458 (45%), Gaps = 51/458 (11%)
Query: 4 LISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYN 63
++S+ + +L+ +S+ + + L++R+ L Y+V+ DAGS+G+RVHV+
Sbjct: 36 VVSVTITLGLLLYVFNSNSVISSGSLLSRRCKL--------RYSVLIDAGSSGTRVHVFG 87
Query: 64 FDQNLDLLPVENELEFYDSVK--PGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTP 121
+ + + Y ++K PGLSSYA NPE A+ S+ L++ A+ +P +
Sbjct: 88 YWFESGKPVFDFGEKHYANLKLTPGLSSYADNPEGASVSVTKLVEFAKQRIPKRMFRRSD 147
Query: 122 VKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYL 181
++L ATAG+RLLE E IL+ R +L + S + + +++ G+ EG Y W+T NY
Sbjct: 148 IRLMATAGMRLLEVPVQEQILEVTRRVLRS-SGFMFRDEWANVISGSDEGIYSWITANYA 206
Query: 182 LGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLY 241
LG LG +T G+V+LGG S Q+T+ S + PE Y + + Y +Y
Sbjct: 207 LGSLGTDPLETTGIVELGGASAQVTFVSSEHVP------PE----YSRTIAYGNISYTIY 256
Query: 242 VHSYLRYGREAFRAEIFKVAGGSAN---------PCILAG---------FDGAYTYSGAE 283
HS+L YG++A ++ + SAN PC G + + ++
Sbjct: 257 SHSFLDYGKDAALKKLLEKLQNSANSTVDGVVEDPCTPKGYIYDTNSKNYSSGFLADESK 316
Query: 284 YKVSAPASGSNLNQCRKIALKALKVNAP-CPYQNCTFGGIWNGGGGSGQKNLFLTSSFYY 342
K S A+G N ++CR LK C Y++C+ G + Q + T+SFYY
Sbjct: 317 LKGSLQAAG-NFSKCRSATFALLKEGKENCLYEHCSIGSTFT---PDLQGSFLATASFYY 372
Query: 343 LSEDVGIFVNKPNAKIRPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVY 402
++ + K +L A K C +YP +++ + C Y
Sbjct: 373 TAKFFEL-----EEKGWLSELIPAGKRYCGEEWSKLILEYPTT-DEEYLRGYCFSAAYTI 426
Query: 403 TLLVDGFGLDPFQEVTVANEIEYQDALVEAAWPLGTAI 440
++L D G+ + ++ + + + W LG I
Sbjct: 427 SMLHDSLGI-ALDDESITYASKAGEKHIPLDWALGAFI 463
>At1g14260 hypothetical protein, 5' partial
Length = 669
Score = 134 bits (337), Expect = 1e-31
Identities = 104/361 (28%), Positives = 163/361 (44%), Gaps = 51/361 (14%)
Query: 78 EFYDSVK--PGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLGATAGLRLLEG 135
E Y S+K PGLSSYA NPE A+ S+ L++ A+ +P + + ++L ATAG+RLL+
Sbjct: 20 EHYASLKLSPGLSSYADNPEGASVSVTKLVEFAKGRIPKGKLKKSDIRLMATAGMRLLDV 79
Query: 136 NAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGKLGKRFTKTVGV 195
E IL R +L + S Q + +++ GT EG Y WV N+ LG LG KT G+
Sbjct: 80 PVQEQILDVTRRVLRS-SGFKFQDEWATVISGTDEGIYAWVVANHALGSLGGDPLKTTGI 138
Query: 196 VDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGK-KYDLYVHSYLRYGREAFR 254
V+LGG S Q+T+ S + VP P + + G Y +Y HS+L +G++A
Sbjct: 139 VELGGASAQVTFVPSEH-------VP----PEFSRTISYGNVSYTIYSHSFLDFGQDAAE 187
Query: 255 AEIFKVAGGSA----------NPCILAGF---------DGAYTYSGAEYKVSAPA-SGSN 294
++ + S +PC G+ + +++K S + +
Sbjct: 188 DKLLESLQNSVAASTGDGIVEDPCTPKGYIYDTHSQKDSSGFLSEESKFKASLQVQAAGD 247
Query: 295 LNQCRKIALKALKVNAP-CPYQNCTFGGIWN---GGGGSGQKNLFLTSSFYYLSEDVGIF 350
+CR L L+ C Y++C+ G + G +N F TS F+ L E +
Sbjct: 248 FTKCRSATLAMLQEGKENCAYKHCSIGSTFTPNIQGSFLATENFFHTSKFFGLGEKEWL- 306
Query: 351 VNKPNAKIRPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFG 410
++ A K C K KYP +K Y C Y+ ++L D G
Sbjct: 307 ----------SEMILAGKRFCGEEWSKLKEKYPTTKDKYLHRY-CFSSAYIISMLHDSLG 355
Query: 411 L 411
+
Sbjct: 356 V 356
>At4g19181 putative protein
Length = 740
Score = 81.3 bits (199), Expect = 1e-15
Identities = 104/453 (22%), Positives = 177/453 (38%), Gaps = 65/453 (14%)
Query: 46 YAVIFDAGSTGSRVHVY----NFDQNLDLLPVENEL----------EFYD--SVKPGLSS 89
Y V+FD GSTG+R +VY N+ ++ L V L YD +PG
Sbjct: 145 YYVVFDCGSTGTRAYVYQASINYKKDSSLPIVMKSLTEGISRKSRGRAYDRMETEPGFDK 204
Query: 90 YAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLGATAGLRLLEGNAAENILQAVRDML 149
N ++ PL++ AE +P + T + + ATAG+R L + IL V +L
Sbjct: 205 LVNNRTGLKTAIKPLIQWAEKQIPKNAHRTTSLFVYATAGVRRLRPADSSWILGNVWSIL 264
Query: 150 SNRSALNVQSDAVSILDGTQEGSYLWVTINY---LLGKLGKRFTKTVGVVDLGGGSVQMT 206
+ +S + + V I+ GT+E + W +NY +LG L K+ T G +DLGG S+Q+T
Sbjct: 265 A-KSPFTCRREWVKIISGTEEAYFGWTALNYQTSMLGALPKK--ATFGALDLGGSSLQVT 321
Query: 207 YAVSRNT--AKNAPKVPEGEDPYIKKLVLQG----KKYDLYVHSYLRYGREAFRAEIFKV 260
+ T N + ++ L G +D V L+ ++++ +
Sbjct: 322 FENEERTHNETNLNLRIGSVNHHLSAYSLAGYGLNDAFDRSVVHLLKKLPNVNKSDLIEG 381
Query: 261 AGGSANPCILAGFDGAYTYSGAEYKVSAPASGS-----------NLNQCRKIALKAL--- 306
+PC+ +G++G Y S V G N +C +A A+
Sbjct: 382 KLEMKHPCLNSGYNGQYICSQCASSVQGGKKGKSGVSIKLVGAPNWGECSALAKNAVNSS 441
Query: 307 -----KVNAPCPYQNCTFGGIWNGGGGS--GQKNLFLTSSFYYLSEDVGIFVNKPNAKIR 359
K C Q C + G F+ F+ LS + +
Sbjct: 442 EWSNAKHGVDCDLQPCALPDGYPRPHGQFYAVSGFFVVYRFFNLSAEASL---------- 491
Query: 360 PVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTV 419
D+ + C + A++ + + +E C Y+ +LL +G + Q +
Sbjct: 492 -DDVLEKGREFCDKAWQVART---SVSPQPFIEQYCFRAPYIVSLLREGLYITDKQIIIG 547
Query: 420 ANEIEYQDALVEAAWPLGTAIEAISSLPKFERL 452
+ I + L A G A+ + L +E L
Sbjct: 548 SGSITW--TLGVALLESGKALSSTLGLKSYETL 578
>At1g14250 putative nucleoside triphosphatase, 3' partial
Length = 128
Score = 49.7 bits (117), Expect = 3e-06
Identities = 33/85 (38%), Positives = 49/85 (56%), Gaps = 5/85 (5%)
Query: 24 LGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNFDQNLDLLPVENELEFYDSV 83
LG+N +M L ++ Y+VI DAGS+G+R+HV+ + + E Y S+
Sbjct: 46 LGSNSVMFSASFLRRSS---LHYSVIIDAGSSGTRIHVFGYWFESGKPVFDFGEEHYASL 102
Query: 84 K--PGLSSYAANPEEAAESLIPLLK 106
K PGLSSYA NPE A+ S+ L++
Sbjct: 103 KLSPGLSSYADNPEGASVSVTKLVE 127
>At1g31710 putative protein
Length = 422
Score = 30.8 bits (68), Expect = 1.6
Identities = 28/77 (36%), Positives = 38/77 (48%), Gaps = 9/77 (11%)
Query: 73 VENELEFYDSVK----PGLSSYAANPEEA---AESLIPLLKEAENVVPVSQQPNTPVKLG 125
V NEL S K P + + P+ A AE+ + L +AE +V V+ PN K G
Sbjct: 231 VRNELVTTRSPKSVNTPRKTYWTTKPKTAKTEAEARVKLGLKAEELVVVN--PNRKTKHG 288
Query: 126 ATAGLRLLEGNAAENIL 142
G RLL G+AA +L
Sbjct: 289 NEVGYRLLHGSAAGPLL 305
>At1g19860 unknown protein
Length = 413
Score = 29.6 bits (65), Expect = 3.6
Identities = 24/107 (22%), Positives = 45/107 (41%), Gaps = 14/107 (13%)
Query: 74 ENELEFYDSVKPGLSSYAANPEEAAE-----------SLIPLLKEAENVVPVSQQPNTPV 122
E EL ++ PG SS NP A+ +IP+L ++ + + + P
Sbjct: 107 ERELRVLEAFYPGASSIPPNPSVPADVEDSHHDDQQTIVIPILPVEDDDIAMDSASDFPT 166
Query: 123 KLGATAGLR---LLEGNAAENILQAVRDMLSNRSALNVQSDAVSILD 166
+ G G E + + L A D+++ SA++ + S++D
Sbjct: 167 QSGVDVGTEPSITDENTSTSSTLPAGPDIMAALSAISNSKEQGSMID 213
>At5g38480 14-3-3 protein homolog RCI1 (pir||S47969)
Length = 255
Score = 29.3 bits (64), Expect = 4.7
Identities = 28/121 (23%), Positives = 47/121 (38%), Gaps = 16/121 (13%)
Query: 72 PVENELEFYDSVKPGLSSY------AANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLG 125
P E+++ FY +K Y A +EAAES + K A ++ P P++LG
Sbjct: 116 PAESKV-FYLKMKGDYHRYLAEFKAGAERKEAAESTLVAYKSASDIATAELAPTHPIRLG 174
Query: 126 ATAGLRLL-------EGNAAENILQAVRDMLSNRSALNVQS--DAVSILDGTQEGSYLWV 176
+ A QA D ++ L +S D+ I+ ++ LW
Sbjct: 175 LALNFSVFYYEILNSPDRACSLAKQAFDDAIAELDTLGEESYKDSTLIMQLLRDNLTLWT 234
Query: 177 T 177
+
Sbjct: 235 S 235
>At5g20260 putative protein
Length = 334
Score = 29.3 bits (64), Expect = 4.7
Identities = 17/44 (38%), Positives = 26/44 (58%), Gaps = 3/44 (6%)
Query: 76 ELEFYDSVKPGLSSYAA-NPEEAAESLIPLLKEAENVVPVSQQP 118
E +F D ++ G+S +AA NPEEA L+P+ N+V +P
Sbjct: 30 EGQFMDEIETGMSPFAANNPEEAHAFLLPV--SVANIVHYLYRP 71
>At5g16050 14-3-3-LIKE PROTEIN GF14 UPSILON
Length = 268
Score = 29.3 bits (64), Expect = 4.7
Identities = 29/116 (25%), Positives = 46/116 (39%), Gaps = 10/116 (8%)
Query: 92 ANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLGATAGLRLL-------EGNAAENILQA 144
A +EAAES + K A+++ P P++LG + A QA
Sbjct: 144 AERKEAAESTLVAYKSAQDIALADLAPTHPIRLGLALNFSVFYYEILNSSDRACSLAKQA 203
Query: 145 VRDMLSNRSALNVQS--DAVSILDGTQEGSYLWVT-INYLLGKLGKRFTKTVGVVD 197
+ +S L +S D+ I+ ++ LW + +N G K K V VD
Sbjct: 204 FDEAISELDTLGEESYKDSTLIMQLLRDNLTLWTSDLNDEAGDDIKEAPKEVQKVD 259
>At1g72560 PAUSED
Length = 988
Score = 29.3 bits (64), Expect = 4.7
Identities = 15/45 (33%), Positives = 27/45 (59%), Gaps = 3/45 (6%)
Query: 139 ENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLG 183
+ ILQ ++D LS + +N S L GT++GS+++ I ++G
Sbjct: 570 DKILQNLQDTLSQLTTMNFASRE---LTGTEDGSHIFEAIGIIIG 611
>At4g09150 unknown protein
Length = 1097
Score = 28.5 bits (62), Expect = 8.0
Identities = 33/130 (25%), Positives = 57/130 (43%), Gaps = 10/130 (7%)
Query: 67 NLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLGA 126
N+D+ + N LEF + LS+ AN EE + L+ E +VP N+ +
Sbjct: 705 NVDMGYLGNILEFSLGILLKLSA-PANEEEIRVTHHKLMTELGEIVPTDGHSNSSYAVLM 763
Query: 127 TAGLR-------LLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGT--QEGSYLWVT 177
GLR +L+ +++ L+ + +L + L + S G+ Q S L +T
Sbjct: 764 VKGLRFVLQQIQILKKEISKSRLKLLEPLLKGPAGLEYLKKSFSSRHGSPDQASSSLPLT 823
Query: 178 INYLLGKLGK 187
+LL G+
Sbjct: 824 KRWLLSVRGE 833
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,926,703
Number of Sequences: 26719
Number of extensions: 437999
Number of successful extensions: 1246
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1214
Number of HSP's gapped (non-prelim): 16
length of query: 456
length of database: 11,318,596
effective HSP length: 103
effective length of query: 353
effective length of database: 8,566,539
effective search space: 3023988267
effective search space used: 3023988267
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0104.2


